Abstract
Mycobacterium tuberculosis is dependent on cysteine biosynthesis, and reduced sulfur compounds such as mycothiol synthesized from cysteine serve in first-line defense mechanisms against oxidative stress imposed by macrophages. Two biosynthetic routes to l-cysteine, each with its own specific cysteine synthase (CysK1 and CysM), have been described in M. tuberculosis, but the function of a third putative sulfhydrylase in this pathogen, CysK2, has remained elusive. We present biochemical and biophysical evidence that CysK2 is an S-sulfocysteine synthase, utilizing O-phosphoserine (OPS) and thiosulfate as substrates. The enzyme uses a mechanism via a central aminoacrylate intermediate that is similar to that of other members of this pyridoxal phosphate-dependent enzyme family. The apparent second-order rate of the first half-reaction with OPS was determined as kmax/Ks = (3.97 × 103) ± 619 M−1 s−1, which compares well to the OPS-specific mycobacterial cysteine synthase CysM with a kmax/Ks of (1.34 × 103) ± 48.2. Notably, CysK2 does not utilize thiocarboxylated CysO as a sulfur donor but accepts thiosulfate and sulfide as donor substrates. The specificity constant kcat/Km for thiosulfate is 40-fold higher than for sulfide, suggesting an annotation as S-sulfocysteine synthase. Mycobacterial CysK2 thus provides a third metabolic route to cysteine, either directly using sulfide as donor or indirectly via S-sulfocysteine. Hypothetically, S-sulfocysteine could also act as a signaling molecule triggering additional responses in redox defense in the pathogen upon exposure to reactive oxygen species during dormancy.
INTRODUCTION
Mycobacterium tuberculosis infects macrophages and establishes a niche within the host, evading the bactericidal action of immune cells (1). The intracellular bacteria are exposed to reactive oxygen and reactive nitrogen species (ROS and RNS) generated by macrophages (2), and redox defense mechanisms are therefore crucial for the survival of this pathogen. In actinobacteria, mycothiol and other low-molecular-weight thiols, i.e., ergothioneine and small cysteine-containing redox proteins, serve as powerful and indispensable radical scavengers (3). Mutations leading to mycothiol depletion cause increased sensitivity to oxidative stress (2, 4).
Mycothiol contains a cysteine-derived sulfhydryl group as active moiety (5), and the redox homeostasis in M. tuberculosis is therefore linked to the availability of l-cysteine. Genes related to sulfur acquisition and cysteine biosynthesis are significantly upregulated under conditions simulating the persistent phase (6–8). Thus, sulfur metabolism and cysteine biosynthesis provide promising targets for novel antibacterial compounds aiming at dormant M. tuberculosis (9, 10).
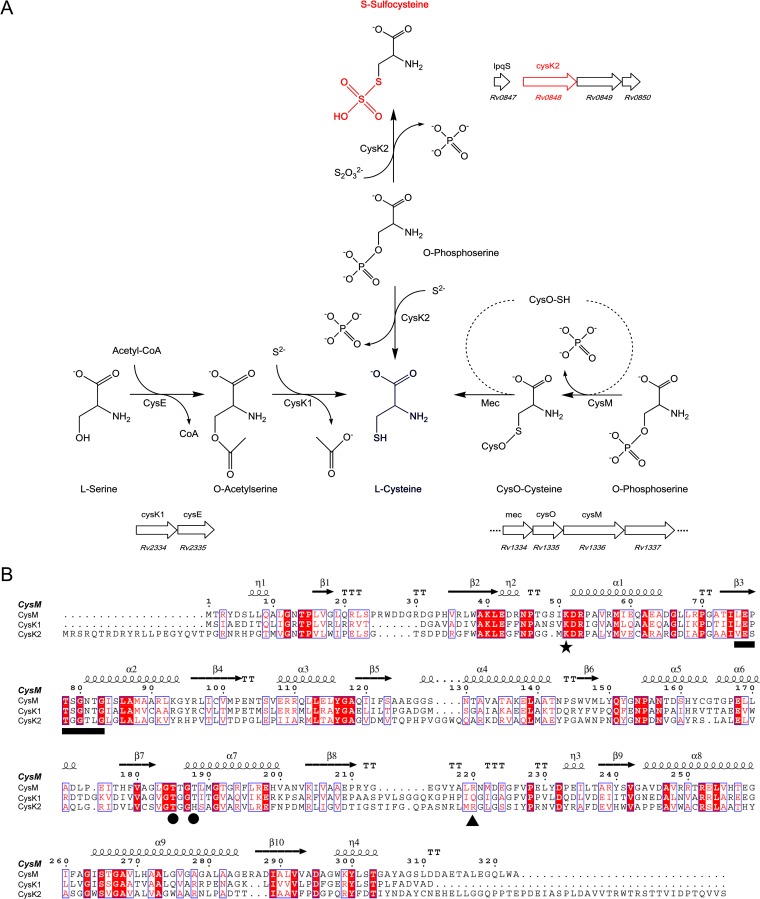
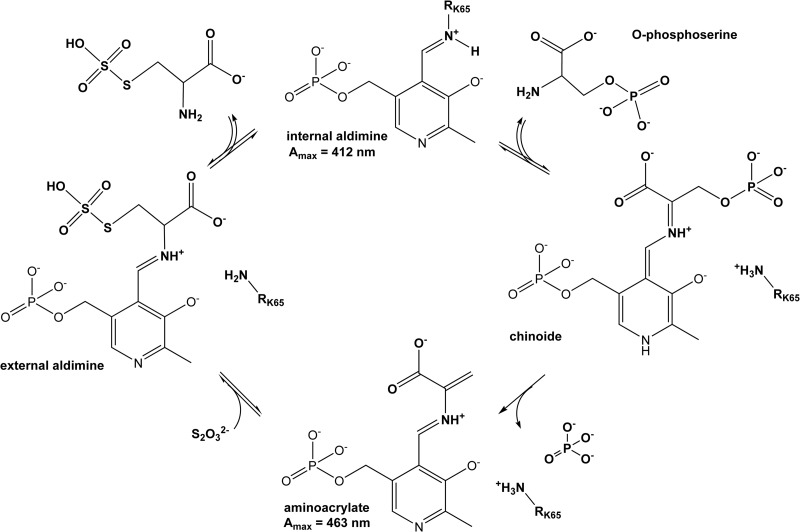
To date, two biosynthetic pathways leading to l-cysteine have been characterized in M. tuberculosis (Fig. 1A). The classical pathway is based on the serine acetyltransferase CysE and the cysteine synthase CysK1, whereby acetylation of l-serine catalyzed by CysE results in O-acetyl-l-serine (OAS) (11), which is converted to l-cysteine by CysK1 using hydrogen sulfide as the sulfur donor (12). The alternative pathway based on CysM uses O-phospho-l-serine (OPS) as the acceptor substrate (13, 14) and thiocarboxylated CysO as a sulfur donor (15, 16) to produce cysteine. Protection of the unstable enzyme-aminoacrylate intermediate by CysM from small oxidants such as H2O2 secures cysteine biosynthesis under oxidative stress and likely supports the long-term intracellular survival within the host macrophages (17). The two pyridoxal phosphate (PLP)-dependent cysteine synthases share a reaction cycle (Fig. 2) that is divided into two half-reactions with characteristic spectral changes (18, 19).
FIG 1.
Comparison of cysteine synthases and CysK2 in M. tuberculosis. (A) l-Cysteine production in M. tuberculosis through three pathways. The classical pathway (left), present in most eubacteria and plants, uses CysK1Mtb for the conversion of O-acetyl-l-serine (OAS) and hydrogen sulfide to l-cysteine. The alternative pathway (right) in M. tuberculosis is based on CysMMtb for l-cysteine formation from O-phospho-l-serine (OPS) and thiocarboxylated CysO. The third pathway (middle), based on CysK2, uses OPS and hydrogen sulfide or thiosulfate for cysteine or S-sulfocysteine production, respectively. The organization of the operons including the three cysteine synthases (CysK1, CysM, and CysK2) in the M. tuberculosis H37Rv genome is also shown (http://tbdb.org). (B) Structure-based sequence alignment of CysK1Mtb, CysMMtb, and CysK2 using CysMMtb as the reference structure, with the secondary structure elements displayed above the alignments. The position of the invariant lysine residue that covalently binds the cofactor *), residues binding the cofactor phosphate group (●), the motif involved in H-bonding interactions with the substrate carboxylate (black horizontal bar), and the position of the arginine residue responsible for OPS specificity in CysMMtb (▲) are indicated. The image in panel B was created with ESPript (35).
FIG 2.
Catalytic cycle of CysK2 and related cysteine synthases. The α-amino group of the acceptor substrate OPS displaces the ε-amino group of the active-site lysine residue (Lys65 in CysK2), resulting in the formation of an external aldimine. Elimination of the OPS β-phosphate and deprotonation of the Cα atom lead to the formation of the reactive aminoacrylate intermediate, the result of the first half-reaction. In the second half-reaction, a nucleophilic attack by the sulfur donor and reprotonation of Cα occur, with subsequent release of the reaction product.
The genome of M. tuberculosis contains a third gene, cysK2 (rv0848), which encodes a protein annotated as a putative cysteine synthase that displays 26% sequence identity to CysK1 and CysM, respectively. The cysK2 gene is significantly overexpressed in various dormancy models, in particular under conditions simulating oxidative stress and hypoxia (8). Expression of cysK2 was proposed to be regulated by the dormancy survival regulator DosR (rv3133c), the primary mediator of hypoxia-related response mechanisms in M. tuberculosis (8, 20). So far, the function and biochemical properties of CysK2 have, however, been elusive.
Here we report the biochemical characterization of CysK2 from Mycobacterium tuberculosis. We show that CysK2 utilizes OPS and thiosulfate as acceptor and preferred sulfur donor substrates in a pyridoxal phosphate-dependent reaction resulting in the formation of S-sulfocysteine. The biochemical data thus suggest an annotation of CysK2 as an S-sulfocysteine synthase.
MATERIALS AND METHODS
l-Cysteine S-sulfate (S-sulfocysteine) was obtained from Sigma. All other chemicals and reagents were of the purest grade commercially available.
Gene cloning and site-directed mutagenesis.
The DNA fragment coding for CysK2 (rv0848) (kindly provided by Mahavir Singh) was cloned into the expression vector pET28a (Novagen) using upstream NdeI and downstream HindIII restriction sites. This construct (pET-His6CysK2) results in a recombinant protein carrying a thrombin-cleavable six-histidine tag at the N terminus. The site-directed mutagenesis to introduce the R243A mutation was carried out using the Quick-Change (Stratagene) approach starting from pET-His6CysK2 as a template. The expression construct CysK2NT, including the range of residues T20 to S372, was amplified by PCR using pET-His6CysK2 as a template and cloned in the pET28a (Novagen) expression vector using upstream NdeI and downstream HindIII restriction sites. PCR amplification primers are listed in Table S1 in the supplemental material. All expression constructs were verified by DNA sequencing.
Recombinant protein production and purification.
Escherichia coli BL21(DE3) carrying the pET-His6CysK2 plasmid was cultivated in 6 liters of LB medium at 37°C supplemented with kanamycin (30 μg μl−1). At an optical density at 600 nm (OD600) value of 0.5 to 0.7, the culture was cooled to 21°C, pyridoxal was added to a final concentration of 10 μM, and gene expression was induced by the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside. After 24 h of cultivation at 21°C, the cells were harvested and resuspended in a buffer consisting of 10 mM Tris HCl (pH 8.0), 300 mM NaCl, and 10 mM imidazole. The cells were disrupted by freeze-thaw cycles and lysozyme and DNase I treatment followed by sonication. The clarified lysates were loaded onto a HisPur Ni2+-nitrilotriacetic acid (NTA) column (Thermo Scientific) with a column volume of 1.2 ml. After washing with 10 mM imidazole, protein was eluted with an imidazole step gradient (25 to 1,000 mM). Fractions containing His6-CysK2 were pooled and loaded onto a HighPrep 26/10 desalting column (GE Healthcare) and eluted in a buffer of 25 mM Tris (pH 8.0)–150 mM NaCl. The N-terminal His6 tag was removed by thrombin cleavage. Uncleaved His-CysK2 was retained by passage through an Ni-NTA column in the presence of 10 mM imidazole and 300 mM NaCl. The CysK2 containing flowthrough was pooled and loaded onto a Superdex-200 column (GE Healthcare) preequilibrated with 25 mM Tris, pH 8.0, and 150 mM NaCl. CysK2-containing fractions were collected and concentrated to 20 mg ml−1 (determined by the Bradford assay [Thermo Scientific]) using a Vivaspin centrifugation device with a 10-kDa molecular mass cutoff. Aliquots of purified CysK2 were flash frozen in liquid nitrogen and stored at −80°C until further use.
The CysK2R243A mutant and the truncated variant CysK2NT were produced and purified using the same protocol as described above for the wild-type enzyme. Homogeneity of purified proteins was analyzed by SDS-PAGE and native gel chromatography. Mass spectrometry revealed that the recombinant CysK2 construct, due to unspecific thrombin cleavage at position Arg2, lacked five amino acids (Gly-Ser-His-Met-Arg) at the N terminus, leaving Ser3 as the first residue of the final recombinant protein product.
Production and purification of M. tuberculosis CysK1 (CysK1Mtb), CysO, and CysM (CysMMtb) followed protocols described previously (12, 13).
UV-vis spectrophotometry.
For UV-visible (UV-vis) spectrophotometry, absorption spectra of CysK2 were recorded at 12 μM enzyme concentration in 100 mM Na-HEPES, pH 7.0, using a Jasco-V65 spectrophotometer. Spectra of reaction intermediates were recorded at time points 3, 6, 9, 14, 20, 30, 40, and 55 min after the addition of the substrates OPS and OAS to a 2 mM final concentration. The reverse reaction starting from l-cysteine or S-sulfocysteine was carried out under similar conditions.
Kinetic parameters of the first half-reaction.
The first half-reactions, catalyzed by CysK2, CysK2R243A, and CysK2NT, respectively, using OPS as the substrate, were investigated by stopped-flow spectrophotometry, monitoring the appearance of the aminoacrylate intermediate at a wavelength of 475 nm. Reactions were carried out in 100 mM Na-HEPES (pH 7.0) at 21°C in 100-μl reaction volumes, with OPS concentrations of 0.05, 0.1, 0.2, 0.4, 0.5, 1.0, 2.0, 4.0, and 5.0 mM and an enzyme concentration of 30 μM.
The derivation of the kinetic parameters of the first half-reaction was carried out as described previously (21, 22). First-order rate constants (kobs) were derived from an exponential fit to the recorded absorbance data. The kobs values were plotted against substrate concentrations to derive the second-order rate constants (kmax/Ks) from the linear fit.
Decomposition of the aminoacrylate intermediate and reverse reaction with S-sulfocysteine.
The spontaneous decomposition of the aminoacrylate intermediate was followed in 100-μl reaction volumes with 100 μM enzyme and 100 μM OPS in 100 mM Na-HEPES, pH 7.0, at 21°C. The decomposition of this intermediate in the presence of sulfur donors was monitored using the same conditions, but with the addition of 2 mM Na2S or 0.3 mM Na2S2O3, respectively.
The back reaction, i.e., formation of the aminoacrylate intermediate from S-sulfocysteine, was followed in 100-μl reaction volumes with 50 μM enzyme and 0.2 mM S-sulfocysteine in 100 mM Na-HEPES, pH 7.0, at 21°C.
Kinetic analysis of phosphate release from OPS.
The malachite green colorimetric assay was used to measure inorganic phosphate release, following the protocol described previously (23). Malachite green working reagent (WR) was prepared freshly by mixing 10 parts of malachite green stock (0.44 g malachite green hydrochloride dissolved in 60 ml H2SO4 and 300 ml H2O), 2.5 parts of ammonium molybdate stock (7.5% [wt/vol]), and 0.2 parts of 11% (vol/vol) Tween 20. From a 100 μM (KH2PO4) phosphate stock solution, a seven-point phosphate standard curve using 2-fold dilution steps in assay buffer (100 mM Na-HEPES, pH 7.0), was determined with phosphate concentrations ranging from 0.39 to 50.0 μM. Typically, enzyme reactions were carried out in triplicates at 22°C in a 1.0-ml total volume, containing 300 nM enzyme in 100 mM Na-HEPES, pH 7.0, and OPS concentrations ranging from 0.05 to 8.0 mM. The reactions were started with the addition of the substrate OPS. For determination of kinetic parameters, 100-μl aliquots were transferred to a microtiter plate at different time points (1 to 16 min). The reactions were stopped with the addition of 40 μl WR and developed for 15 min at 22°C. The absorbance values at 620 nm were recorded using the Synergy HT Multi-Mode microplate reader (BioTek).
Kinetic analysis of the overall reaction.
Recombinant CysK2 was assayed spectrophotometrically at 560 nm for cysteine synthase activity by monitoring the formation of cysteine using the acid-ninhydrin method at 22°C (12, 24). The kinetic parameters for the reaction of CysK2 with Na2S as the sulfur donor were determined using OPS concentrations ranging from 9.9 μM to 5.08 mM in the presence of 1 mM Na2S and an enzyme concentration of 1 μM. Reactions were carried out in a total volume of 1.0 ml in 100 mM Na-HEPES buffer at pH 7.0. For the determination of the Michaelis-Menten parameters for the reaction with the sulfur donor, the Na2S concentrations were varied in the interval from 50 μM to 6 mM in the presence of 8 mM OPS and a final enzyme concentration of 0.5 μM. For the determination of the OPS dependence of the reaction of the CysK2R243A mutant, the assays were carried out in a total volume of 1.0 ml in 100 mM Na-HEPES buffer at pH 7.0. OPS was used in a concentration range from 39.7 μM to 2.54 mM in the presence of 1 mM Na2S and an enzyme final concentration of 5 μM.
For reactions with thiosulfate as the sulfur donor, the acid-ninhydrin method could also be employed. Under the highly acidic conditions of this derivatization method, the product S-sulfocysteine is quantitatively hydrolyzed into cysteine, which can be detected using this assay (25). Analysis of the commercially available S-sulfocysteine using Ellman's reagent, DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)], showed that the compound did not contain free thiol groups, i.e., no cysteine impurities, which could have interfered with the acid-ninhydrin method.
The reactions of CysK2 with Na2S2O3 were carried out in a total volume of 0.9 ml in 100 mM Na-HEPES buffer at pH 7.0. Increasing amounts of OPS ranging from 57.8 μM to 5.0 mM were used with 300 μM Na2S2O3 at an enzyme concentration of 1 μM. Determination of the kinetic parameters for Na2S2O3 was carried out using the same enzyme concentration and increasing amounts of Na2S2O3 ranging from 13 μM to 5.0 mM in the presence of 5 mM OPS. Michaelis-Menten parameters for the CysK2R243A mutant were determined in a total volume of 0.9 ml in 100 mM Na-HEPES buffer at pH 7.0 and an enzyme concentration of 2 μM. The assays used OPS concentrations in the range of 92.5 μM to 8.0 mM (Na2S2O3 fixed at 300 μM) or Na2S2O3 ranging from 57.8 μM to 5.0 mM OPS (OPS fixed at 5 mM).
Aliquots of 100 μl were taken at various time points, and the reaction was stopped by addition of 20 μl trichloroacetic acid. One hundred twenty microliters of ninhydrin reagent (250 mg ninhydrin in acetic acid-HCl [6:4]) was added, and the samples were heated for 8 min at 96°C and thereafter cooled on ice for 5 min and diluted with an equal amount of 95% ethanol. Absorbance values at 560 nm were measured using a Synergy HT Multi-Mode microplate reader, and the amount of l-cysteine and/or S-sulfocysteine was determined using an l-cysteine or S-sulfocysteine standard curve. Km and Vmax were derived using Origin 8.5 and GraphPad Prism.
CD spectroscopy.
Circular dichroism (CD) measurements were performed with a JASCO J-810 spectropolarimeter at 22°C in 0.2-cm path length quartz cuvettes between 190 to 260 nm with 0.2-nm steps. Protein samples were diluted in water to a final concentration of 0.2 mg ml−1 protein and 20 mM NaCl.
TLC.
The enzymatic reactions of CysK2 were monitored by thin-layer chromatography (TLC) using a silica-covered aluminum plate (Merck) with butan-2-ol–acetic acid–water (4:2:1) as the mobile phase. The reaction was started by mixing 25 μM CysK2 in 100 mM Na-HEPES (pH 7.0) with 6 mM Na2S2O3 and 6 mM OPS in a final volume of 100 μl at 20°C. After 120 min, the reaction was stopped by removing the enzyme with a 10-min filtration step at 4°C using a centrifugal device with a 3-kDa cutoff (Pall, Port Washington, NY). Twenty microliters of the filtrate was applied to the silica plate, which was subsequently transferred to the TLC chamber. After 75 min, the plate was dried, soaked in 1% (wt/vol) ninhydrin in propan-2-ol, and heated to 175°C.
Sample preparation for ESI-MS.
Reactions that were further analyzed by electrospray ionization mass spectrometry (ESI-MS) or ESI tandem MS (ESI-MS/MS) were started by mixing 100 μM CysK2 in double-distilled water (ddH2O) with 10 mM Na2S2O3 and 10 mM OPS in a final volume of 200 μl at 22°C. After 30 min, the reaction was stopped by removing the enzyme by filtration (10 min at 4°C) using a 3-kDa cutoff. The filtrate was analyzed directly by ESI-MS.
The reaction mixture for the extraction from the TLC plate was prepared as described above for direct ESI-MS. Fifty microliters of the filtrate was applied to the silica plate in multiple spots and treated as described above. In order to identify the location of the reaction product on the plate, a 50-μl S-sulfocysteine control spot and one spot of the reaction were cut from the rest of the plate, soaked in 1% (wt/vol) ninhydrin in propan-2-ol, and heated to 175°C. Untreated lanes were then used to cut out the area corresponding to the reaction product, followed by extraction with 100 μl (vol/vol) methanol-ddH2O (1:1). The extracted product was analyzed by ESI-MS.
Small-molecule ESI-MS and ESI-MS/MS.
The reaction product was identified by ESI-MS using a Micromass quadrupole/orthogonal acceleration-time of flight (Q-TOF) II (Waters Corp., Manchester, United Kingdom) mass spectrometer. Direct injections of 10-μl samples in 5% (vol/vol) acetonitrile containing 0.1% (vol/vol) formic acid were performed through a capillary LC system into the mass spectrometer, operated in positive-ion mode with a capillary voltage of 3.3 kV, a cone voltage of 35 V, and a desolvation temperature of 140°C. For MS/MS experiments, precursor ions were fragmented by collision-induced dissociation (CID) using collision energies optimized between 10 and 25 V.
Protein mass spectrometry.
The reactions between thiocarboxylated CysO and CysK2 or CysMMtb were carried out in 20 mM Na-HEPES (pH 7.0) using 10 μg CysO-SH (106 μM), 0.5 mM OPS, and 1 μM enzyme in a total volume of 10 μl. The reaction mixture and a sample of unreacted CysO-SH were diluted in 2.5% (vol/vol) acetonitrile containing 0.05% (vol/vol) formic acid, loaded on a CapLC system, and analyzed by a Q-TOF mass spectrometer (Waters Corp., Manchester, United Kingdom) as described previously (26).
RESULTS
Characterization of M. tuberculosis CysK2.
CysK2 displays about 26% sequence identity to CysMMtb and CysK1Mtb, including conservation of functionally important sequence motifs of PLP-dependent enzymes (Fig. 1B). The invariant lysine residue that binds the PLP cofactor corresponds to Lys65 in CysK2; furthermore, the sequence fingerprint (201GTGGT205) involved in H bonding to the phosphate group of PLP is retained. The motif implicated in coordinating the carboxylate group of the substrate by H bonding via the two threonine side chains and main-chain amines shows minor alteration in CysK2 (90ESTGGTLG97) in comparison to CysK1Mtb and CysMMtb (Fig. 1B). The sequence alignment also shows that CysK2 contains significant N- and C-terminal extensions of 20 and 41 residues, respectively, compared to the other mycobacterial cysteine synthases CysMMtb and CysK1Mtb.
Recombinant wild-type CysK2 was produced and purified as N-terminally hexahistidine-tagged protein by affinity chromatography, subsequent tag removal by thrombin, and size exclusion chromatography. The elution profile from analytical size exclusion chromatography suggests that CysK2 forms a dimer in solution (see Fig. S1 in the supplemental material). The CD spectrum of CysK2 is characteristic of a folded protein with a mixed α/β secondary structure and is very similar to that of the related cysteine synthases CysK1Mtb and CysMMtb (see Fig. S2 in the supplemental material).
OPS is an acceptor substrate for CysK2 in the first half-reaction.
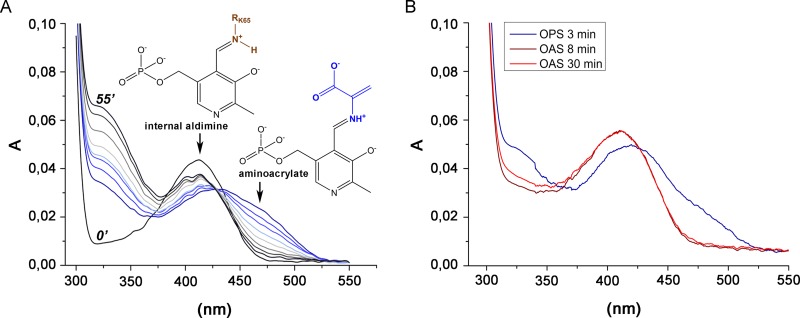
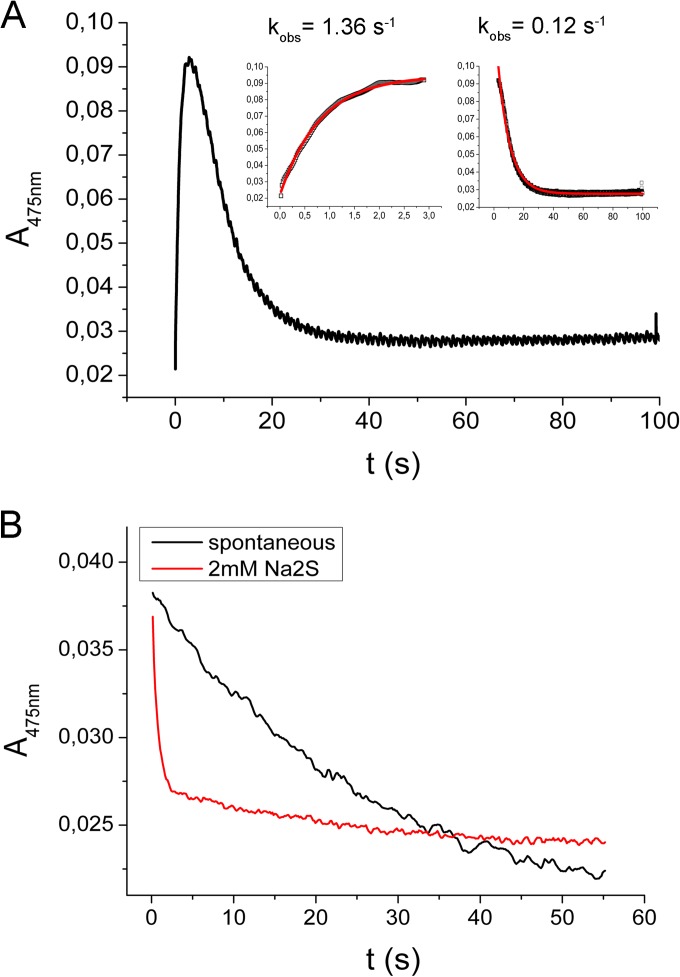
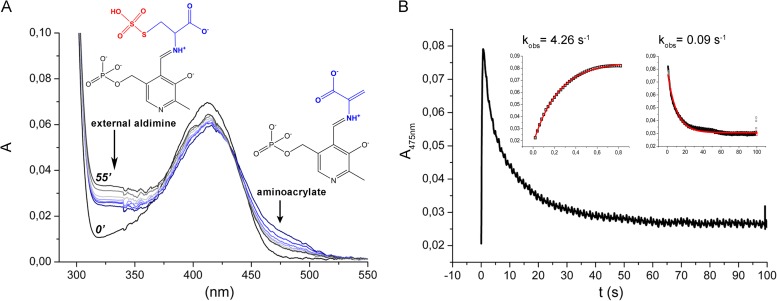
The UV-vis absorption spectrum of the recombinant CysK2 shows the characteristic absorbance maximum at 412 nm indicative of the internal aldimine bond formed between Lys65 and the PLP cofactor (Fig. 2 and 3A). In a search for potential acceptor substrates of CysK2, spectral changes upon addition of various amino acids such as O-acetyl-l-serine (OAS), O-phospho-l-serine (OPS), Asp, Val, Gln, Glu, Ser, Asn, Cys, Ser, Leu, homocysteine, ketoacids such as pyruvate and α-ketoglutarate, amino acid precursors like 3-phosphoglycerate and succinate, and derivatives like N-acetylcysteine and diaminopimelic acid were tested. The absorption maximum appearing at 330 nm and the characteristic shift from 412 nm to approximately 465 to 470 nm were observed upon OPS addition (Fig. 3A). These spectral changes are consistent with the formation of the enzyme-aminoacrylate intermediate (Fig. 2) commonly observed in this class of PLP-dependent enzymes (12, 13, 27). None of the other tested compounds, in particular not OAS, induced significant absorbance shifts indicative of a specific reaction with CysK2 (Fig. 3B). The formation of the aminoacrylate was monitored by stopped-flow spectrophotometry at 475 nm by mixing 100 μM CysK2 and 100 μM OPS, resulting in a pseudo-first-order rate constant (kobs) of 1.36 s−1 (Fig. 4A). A more detailed kinetic analysis gave a second-order rate constant (kmax/Ks) of (3.97 × 103) ± 619 M−1 s−1 (Fig. 5A).
FIG 3.
UV-visible spectra of holo-CysK2 reactivity and enzyme-substrate complexes. (A) The UV-visible spectrum of CysK2 shows a maximum at 412 nm (black line), typical of the internal aldimine. Addition of 2 mM OPS results in fast enzyme-aminoacrylate intermediate formation indicated by an absorption increase at 330 nm and the shift of the PLP peak from 412 nm to 460 to 470 nm. Spontaneous decay of the aminoacrylate intermediate is demonstrated by the set of spectra recorded after 3, 6, 9, 14, 20, 30, 40, and 55 min following addition of OPS (blue-gray). Spectra at time points 0 and 55 min are labeled. (B) Comparison of the UV-visible spectra of CysK2 with OPS or OAS as the substrate. OPS addition results in spectral changes consistent with the formation of the enzyme-aminoacrylate intermediate (blue trace). The addition of 2 mM OAS does not indicate enzyme-aminoacrylate intermediate formation after 3 min or 30 min of incubation (red and orange traces).
FIG 4.
Formation and spontaneous decay of the aminoacrylate intermediate at the CysK2 active site monitored by stopped-flow spectrophotometry. (A) Aminoacrylate formation and decay were measured with 100 μM enzyme and 100 μM OPS in a stopped-flow spectrophotometer at 475 nm. The fast formation of the enzyme-aminoacrylate intermediate (inset on left) is followed by a spontaneous decomposition (inset on right) with a half-life of 8.49 s. (B) In the presence of 2 mM Na2S, the consumption of the reaction intermediate is approximately 14 times faster than the spontaneous decomposition.
FIG 5.
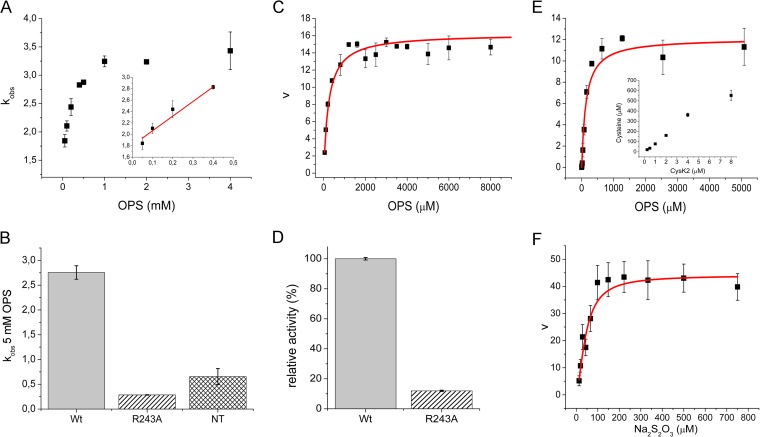
Kinetic characterization of CysK2, CysK2R243A mutant, and CysK2NT. (A) First half-reaction parameters investigated by stopped-flow spectrophotometry. Pseudo-first-order rate constants (kobs) at different OPS concentrations. The linear fit used to derive the second-order rate constant (kmax/Ks) is shown in the inset. (B) Comparison of kobs at 5 mM OPS between wild-type CysK2 (wt), CysK2R243A (R243A), and CysK2NT (NT) constructs. (C) Michaelis-Menten kinetics of CysK2 based on phosphate release using the malachite green calorimetric assay. (D) Relative activity of wild-type CysK2 compared to CysK2R243A based on inorganic phosphate release. (E) Michaelis-Menten kinetics of CysK2 in the presence of sulfide as a sulfur donor substrate. The inset shows the dependence of l-cysteine production on enzyme concentration. (F) Michaelis-Menten kinetics of CysK2 in the presence of thiosulfate as an S donor substrate.
Enzymatic conversion of OPS to the aminoacrylate is accompanied by the release of a phosphate ion (Fig. 2). Kinetic analysis using the malachite green assay showed that CysK2 dephosphorylates OPS with the kinetic constants Km(OPS) of 233 μM and kcat of 16.2 min−1 (Fig. 5C) and a specificity constant (kcat/Km) of 0.070 × 103 M−1 min−1 at 22°C. The pH optimum for this reaction is around pH 9.0, most likely because the spontaneous hydrolysis of the aminoacrylate intermediate is faster at this pH, resulting in an increased turnover rate in the absence of the sulfur donor substrate (27) (see Fig. S3B in the supplemental material).
Thiosulfate is the preferred sulfur donor in the second half-reaction.
The amino acrylate intermediate is not stable and decomposes with a pseudo-first-order rate constant kobs of 0.12 s−1 (Fig. 4A). However, it is stable enough to be used for the screening of potential sulfur donors in the enzyme reaction. Several potential sulfur donors were tested: Na2S, Na2S2O3, Na2S2O5, NaSCN, Na2SO4, and Na2SO3. With the exception of Na2S and Na2S2O3, none of these compounds resulted in an increase in the rate of disappearance of the aminoacrylate or the formation of l-cysteine. However, the addition of 2 mM Na2S in the substrate syringe together with 100 μM OPS led to a faster disappearance of the aminoacrylate signal, with a half-life of 0.59 s, which is about 14 times faster than spontaneous amino acrylate hydrolysis (Fig. 4B). The decomposition of the enzyme-aminoacrylate intermediate in the presence of 0.3 mM Na2S2O3 was too rapid to detect by stopped-flow spectrophotometry.
The sulfur donor S2− led to l-cysteine formation, which was assayed spectrophotometrically at 560 nm using the acid-ninhydrin method (12, 24) (Fig. 5E). The determination of the kinetic parameters for the enzyme reaction in the presence of S2− as a sulfur donor resulted in a kcat of 12.1 min−1 and a Km(Na2S) of 374 μM and a Km(OPS) of 135 μM (Fig. 5E and Table 1).
TABLE 1.
Kinetic parameters of M. tuberculosis CysK2
| Cysteine synthase |
O-Phosphoserine sulfhydrylase |
O-Phosphoserine S-sulfocysteine synthase |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Km (μM) |
kcat (min−1) |
kcat/Km (103 M−1 min−1) |
Km (μM) |
kcat (min−1) |
kcat/Km (103 M−1 min−1) |
|||||
| OPS | Na2S | OPS | Na2S | OPS | Na2S2O3 | OPS | Na2S2O3 | |||
| CysK2 | 135 ± 39.4 | 374 ± 233.6 | 12.1 ± 1.44 | 0.089 | 0.032 | 1,085.5 ± 82.3 | 43.9 ± 7.2 | 54.8 ± 1.8 | 0.050 | 1.248 |
| CysK2R243A | 1,085.4 ± 306.8 | NDa | 0.4 ± 0.075 | 0.0061 | 485.1 ± 96.0 | 213.7 ± 13.1 | 27.4 ± 1.6 | 0.056 | 0.128 | |
ND, not determined.
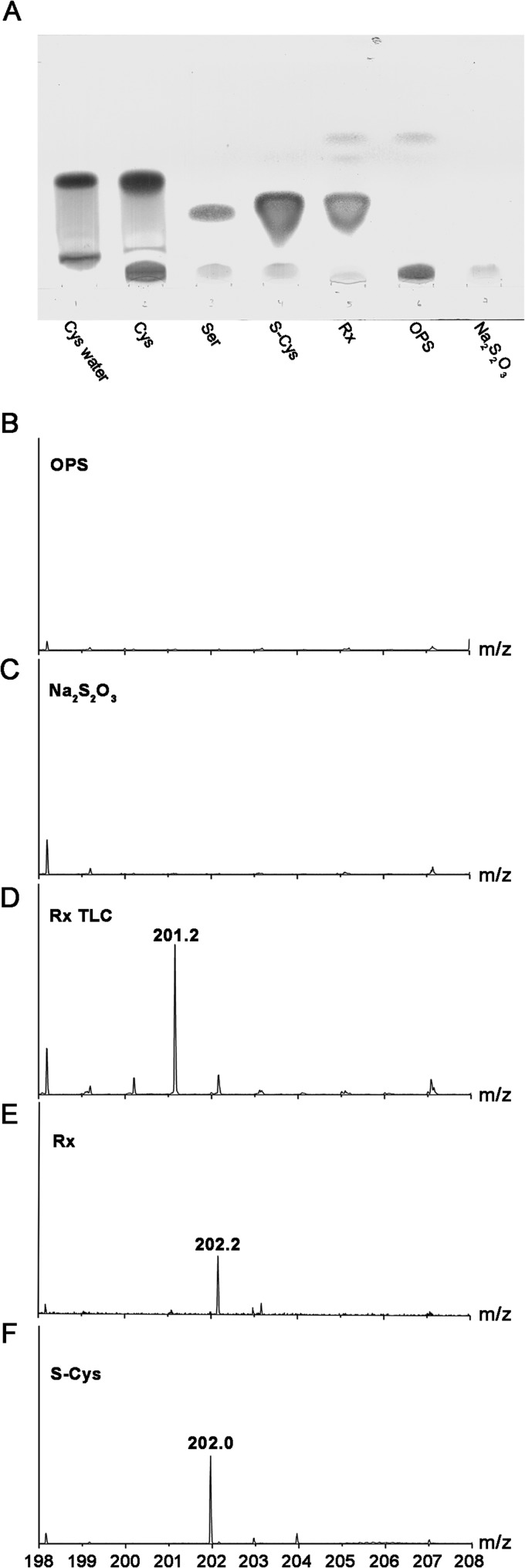
The reaction of OPS with Na2S2O3 as a sulfur donor catalyzed by CysK2 results in the formation of S-sulfocysteine as confirmed by TLC (Fig. 6A) and subsequent ESI mass spectrometry (Fig. 6B to F). Thin-layer chromatography of a reaction mixture of enzyme, OPS, and thiosulfate resulted in a new band not observed in the absence of enzyme that comigrated with the S-sulfocysteine standard (Fig. 6A). Elution of this band and subsequent analysis by ESI-MS resulted in a peak with an m/z of 201.2 (Fig. 6D) corresponding to the calculated mass of S-sulfocysteine (201.2 Da). Direct ESI-MS analysis of a reaction mix also showed the expected peak for S-sulfocysteine (m/z of 202.0), thus confirming the formation of S-sulfocysteine (Fig. 6E; see also Fig. S4A in the supplemental material).
FIG 6.
Confirmation of the S-sulfocysteine product. (A) Product confirmation using thin-layer chromatography. The reaction mixture (Rx) with 6 mM OPS and 6 mM thiosulfate as the substrates and controls were analyzed using a silica-covered aluminum TLC plate with butan-2-ol–acetic acid–water (4: 2:1) as a mobile phase. The plate was developed in 1% ninhydrin in propan-2-ol at 175°C. Relevant compounds l-cysteine (in the reaction buffer), l-cysteine (dissolved in water), l-serine, S-sulfocysteine, OPS, and sodium thiosulfate are shown for comparison. Electrospray mass spectrum of OPS standard solution (B), mass spectra of Na2S2O3 standard (C), mass spectrum of the enzymatic reaction carried out in water and eluted from the TLC plate (D), and mass spectra of the enzymatic reaction (Rx) carried out in water (E) and S-sulfocysteine (S-Cys) standard as a control (F) analyzed in positive-ionization mode.
The kinetic parameters for the enzyme reaction in the presence of S2O32− as a sulfur donor were determined as kcat of 54.8 min−1, a Km(Na2S2O3) of 43.9 μM, and a Km(OPS) of 1,085.5 μM (Fig. 5F). Thiosulfate concentrations higher than 600 μM resulted in substrate inhibition, with an inhibition constant, Ki, of >1 mM.
CysK2 can react with S-sulfocysteine to form the aminoacrylate intermediate, demonstrating the reversibility of this half-reaction (Fig. 7). The pseudo-first-order rate constants of aminoacrylate formation from S-sulfocysteine and of the spontaneous decay of the intermediate using 0.2 mM S-sulfocysteine were determined as kobs of 4.2 s−1 and 0.1 s−1, respectively (Fig. 7). CysK2 reacts also with l-cysteine to form α-aminoacrylate, albeit to a lower extent (data not shown).
FIG 7.
Investigation of the reverse reaction of S-sulfocysteine and CysK2. (A) The UV-vis spectrum of CysK2 in the presence of 2 mM S-sulfocysteine results in fast enzyme-aminoacrylate intermediate formation (blue-gray). (B) The spontaneous formation and decay of the intermediate for the back reaction were monitored in the stopped-flow spectrophotometer, and pseudo-first-order rate constants (kobs) were determined from exponential fits (insets) for the formation and the decay of the reaction intermediate.
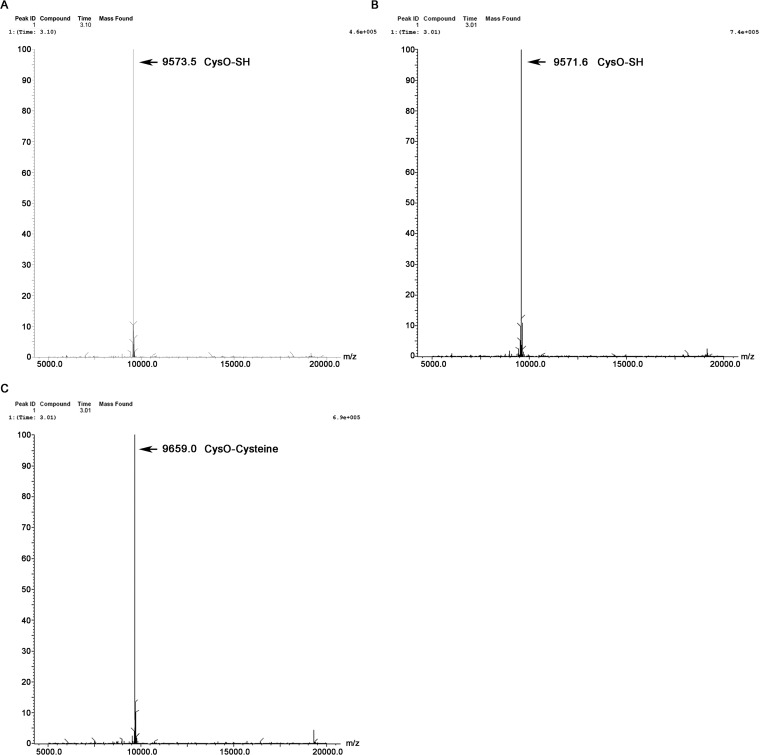
The similarity of CysK2 to CysMMtb in sequence and OPS specificity prompted us also to test if thiocarboxylated CysO (CysO-SH) is accepted as a sulfur donor, leading to a CysO-cysteine adduct as primary reaction product. The reaction of CysK2 with OPS and CysO-SH was carried out as described previously (13). However, we could not detect any CysO-cysteine adduct by ESI-MS, demonstrating that CysO-SH was not accepted by CysK2 as a sulfur donor (Fig. 8).
FIG 8.
Product identification of reactions with CysO-thiocarboxylate (CysO-SH) by electrospray mass spectrometry. (A) Mass spectrum of intact CysO-SH (9,573.5 Da). (B and C) Mass spectrum of reaction product formed by CysK2, showing intact CysO-SH mass (9,571.6 Da) (B) and of the covalent CysO-cysteine adduct (9659.0) produced by CysMMtb in the second half-reaction (C). Both enzymatic reactions were performed in the presence of 10 μg CysO-SH (106 μM) and 0.5 mM OPS.
Effect of the R243A mutation.
Amino acid sequence comparisons (Fig. 1B) suggested that R243 of CysK2 might be equivalent to the position of R220 in CysMMtb, a residue that is crucial for OPS specificity in this enzyme (13). R243 in CysK2 was therefore replaced by alanine using site-directed mutagenesis. The purified CysK2R243A mutant was folded (see Fig. S2A in the supplemental material) and showed an absorption spectrum identical to that of the wild-type enzyme. The specific activity of the R243A mutant based on phosphate release from OPS decreased to 14% of wild-type enzyme (Fig. 5D). A similar drop in enzyme activity was found based on the spectrophotometric determination of pseudo-first-order rate constant kobs (5 mM OPS), which decreased to 7.2% in comparison to the wild-type enzyme (Fig. 5B). The Michaelis-Menten parameters for the reaction with thiosulfate and OPS showed a 10-fold drop in the specificity constant kcat/Km for thiosulfate compared to wild-type enzyme but little change in the specificity constant for the substrate OPS (Table 1).
The N-terminal extension is required for efficient catalysis.
Bioinformatics analysis revealed CysK2 homologs with ≥75% sequence identity in 12 mycobacterial species. All of these homologues contain additional amino acid sequence extensions at the N terminus in comparison to CysMMtb and CysK1Mtb. For instance CysK2 carries a 20-amino-acid-long sequence stretch on its N terminus (Fig. 1B). In order to investigate the role of this additional segment, a truncated construct was expressed comprising the residue range T20 to Ser372. The removal of 19 amino acids from the N terminus had no influence on the absorption spectra, folding integrity (see Fig. S2A in the supplemental material), or the thermal stability of the protein. However, truncation of these amino acids resulted in a decrease of the first-order rate constant for aminoacrylate formation to 0.656 s−1, corresponding to 16.5% of the rate of the wild-type enzyme.
DISCUSSION
In this study, CysK2 has been identified as an S-sulfocysteine synthase embedded in the metabolic network of two previously characterized independent pathways of l-cysteine biosynthesis in M. tuberculosis (Fig. 1A). Overall, the reaction follows the scheme of PLP-dependent enzymes (Fig. 2) with the aminoacrylate intermediate as the product of the first half-reaction. CysK2 shows a pronounced preference for the acceptor substrate OPS and does not react with OAS. Formation of the aminoacrylate intermediate is coupled to phosphate release from OPS as detected by the malachite green assay. The apparent second-order rate constant for the formation of the aminoacrylate intermediate, (3.97 × 103) ± 619 M−1 s−1, is comparable to the rate of the corresponding step in CysMMtb, (1.34 × 103) ± 48.2 (13).
CysK2 accepts sulfide and thiosulfate as sulfur donors, but notably not thiocarboxylated CysO-SH, which sets it apart from the OPS-dependent CysMMtb. The reaction with OPS and sulfide leads to the formation of l-cysteine in a manner similar to that seen with the cysteine synthase CysK1Mtb, with the latter enzyme, however, using OAS and not OPS as an acceptor substrate (12). CysK2 can also catalyze the conversion of OPS and thiosulfate to S-sulfocysteine as shown by TLC and mass spectrometry. Additional evidence for S-sulfocysteine as a reaction product is the ability of the enzyme to catalyze the reverse reaction, i.e., fast formation of the aminoacrylate from S-sulfocysteine. The enzyme shows a clear preference for thiosulfate as a sulfur donor, with a 40-fold-higher specificity constant for thiosulfate compared to sulfide as the substrate, suggesting an annotation of CysK2 as an S-sulfocysteine synthase.
The OPS-dependent cysteine synthases from the archaeon Aeropyrum pernix (28) and CysMMtb (13) contain an arginine residue in the OPS binding site that is responsible for recognition of the phosphate group of OPS and thus substrate selectivity. For instance, replacement of this residue, R220, by alanine in CysMMtb resulted in a >600-fold decrease in the enzymatic rate with OPS. It is noteworthy that this amino acid replacement did not affect the reaction with the sulfur donor CysO-SH, emphasizing the role of this arginine residue in recognition of the phosphate group of OPS in CysMMtb. Amino acid sequence comparisons suggested that also the OPS-dependent CysK2 contains an arginine residue, R243, which might be involved in substrate selectivity (Fig. 1B). Replacement of this amino acid in CysK2 by alanine is, however, much less deleterious than the amino acid replacement in CysMMtb. It is noteworthy that there is no decrease in the kcat/Km value for OPS in the reaction with thiosulfate in the R243A mutant, suggesting that the contribution of this residue to the recognition of the phosphate group of OPS is not essential. This finding is consistent with a homology model of the structure of CysK2, obtained from the Swiss Model server (http://swissmodel.expasy.org/) using the coordinates of CysK1Mtb. Comparison of this model with the structure of CysMMtb shows that R243 of CysK2 is not located in the OPS binding site, with a distance of 11 Å between the guanidinium groups of the two arginine residues.
Amino acid G93 is part of the sequence stretch T92-G93-G94-T95, located in the vicinity of R243. This motif forms a tight turn in the active-site region of the enzyme and is involved in binding of the carboxylate group of the acceptor substrate. G94 is an invariant residue in CysK2 and also in the related cysteine synthases CysK1Mtb and CysMMtb (Fig. 1B), probably due to structural reasons. G93 is not strictly conserved and is sometimes replaced by serine in sequences of CysK2 from other M. tuberculosis strains. In CysMMtb and CysK1Mtb, this glycine residue is also replaced by serine (Fig. 1B). In the last two enzymes, this replacement is compatible with catalytic activity and structure, suggesting that the corresponding change in CysK2, observed in several M. tuberculosis strains, does not interfere with the activity of these CysK2 variants.
The cysK2 gene of M. tuberculosis is overexpressed during hypoxia and oxidative stress, conditions prevailing during dormancy of the pathogen in infected macrophages (10). Bacterial defense against the reactive oxygen and nitrogen species generated by macrophages requires an increased supply of l-cysteine, reflected in the upregulation of a number of mycobacterial genes related to the sulfur reduction pathway and cysteine biosynthesis (29). In this context, CysK2 could fulfill several roles. Due to its unique combination of OPS and sulfide as the substrates, it provides an additional pathway to l-cysteine during dormancy (Fig. 1). However, the higher affinity for thiosulfate suggests additional functions of this enzyme. This raises the questions of how thiosulfate is formed in M. tuberculosis under these metabolic conditions and what the role of S-sulfocysteine might be.
Thiosulfate could be formed by interference of the reactive oxygen species, generated by macrophages, with the sulfate reduction pathway, as has been suggested to occur under the light-induced oxidative stress in plants (30). Formation of S-sulfocysteine from thiosulfate would thus provide a salvage pathway whereby some of the sulfur could be rescued by incorporation into l-cysteine. S-sulfocysteine, generated from thiosulfate by S-sulfocysteine synthases, has been postulated as an intermediate of an alternative route to l-cysteine biosynthesis in Rhodospirillum tenue (31), Salmonella enterica serovar Typhimurium (32), and E. coli (33). In the last case, it was shown that thioredoxin and/or NrdH is involved in the reduction of S-sulfocysteine to cysteine and sulfite. It remains, however, to be shown whether this route to l-cysteine biosynthesis is sustainable under the oxidative stress conditions in macrophages.
If the formation of thiosulfate is due to the action of reactive oxygen species in M. tuberculosis, the formation of S-sulfocysteine could provide a signal of oxidative stress. There is some evidence that S-sulfocysteine, formed by the action of the S-sulfocysteine synthase CS26 upon light-induced oxidative stress in Arabidopsis thaliana chloroplasts, acts as a signal molecule initiating adequate responses in redox defense (34). In a similar manner, this metabolite, produced by CysK2, could act as a sensor of oxidative stress in M. tuberculosis during intracellular survival or dormancy.
In conclusion, we have shown that mycobacterial CysK2, which is upregulated under conditions simulating dormancy, is an S-sulfocysteine synthase. The enzyme catalyzes the formation of S-sulfocysteine from OPS and thiosulfate in a PLP-dependent reaction but is also able, although at lower catalytic efficiency, to convert OPS and sulfide to l-cysteine. CysK2 thus provides a third biosynthetic pathway in M. tuberculosis to l-cysteine, either directly or via the formation of S-sulfocysteine. Hypothetically, S-sulfocysteine might also be a signal molecule for the detection of oxidative stress in dormant M. tuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mahavir Singh (Lionex, Braunschweig, Germany) for the cysK2 DNA fragment and Peter Brzezinski (Stockholm University) for the access to stopped-flow instrumentation.
This work was supported by the Swedish Governmental Agency for Innovation Systems (VINNOVA) and the Swedish Science Council (VR).
Footnotes
Published ahead of print 14 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01851-14.
REFERENCES
- 1.Gengenbacher M, Kaufmann SH. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36:514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchmeier NA, Newton GL, Koledin T, Fahey RC. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47:1723–1732. 10.1046/j.1365-2958.2003.03416.x [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Farhana A, Guidry L, Saini V, Hondalus M, Steyn AJ. 2011. Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev. Mol. Med. 13:e39. 10.1017/S1462399411002079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sareen D, Newton GL, Fahey RC, Buchmeier NA. 2003. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 185:6736–6740. 10.1128/JB.185.22.6736-6740.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornemann C, Jardine MA, Spies HS, Steenkamp DJ. 1997. Biosynthesis of mycothiol: elucidation of the sequence of steps in Mycobacterium smegmatis. Biochem. J. 325:623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith S. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365–374. 10.1046/j.1365-2958.2002.03005.x [DOI] [PubMed] [Google Scholar]
- 7.Hampshire T, Soneji S, Bacon J, James BW, Hinds J, Laing K, Stabler RA, Marsh PD, Butcher PD. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 84:228–238. 10.1016/j.tube.2003.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713. 10.1084/jem.20030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhave DP, Muse WB, Carroll KS. 2007. Drug targets in mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 7:140–158. 10.2174/187152607781001772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatzios SK, Bertozzi CR. 2011. The regulation of sulfur metabolism in Mycobacterium tuberculosis. PLoS Pathog. 7:e1002036. 10.1371/journal.ppat.1002036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J, Wang D, Ma Y, Jiang T, Xin Y. 2013. Identification and characterization of serine acetyltransferase encoded by the Mycobacterium tuberculosis Rv2335 gene. Int. J. Mol. Med. 31:1229–1233 [DOI] [PubMed] [Google Scholar]
- 12.Schnell R, Oehlmann W, Singh M, Schneider G. 2007. Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis crystal structures of the enzyme A-aminoacrylate intermediate and an enzyme-inhibitor complex. J. Biol. Chem. 282:23473–23481. 10.1074/jbc.M703518200 [DOI] [PubMed] [Google Scholar]
- 13.Ågren D, Schnell R, Oehlmann W, Singh M, Schneider G. 2008. Cysteine synthase (CysM) of Mycobacterium tuberculosis is an O-phosphoserine sulfhydrylase evidence for an alternative cysteine biosynthesis pathway in mycobacteria. J. Biol. Chem. 283:31567–31574. 10.1074/jbc.M804877200 [DOI] [PubMed] [Google Scholar]
- 14.O'Leary SE, Jurgenson CT, Ealick SE, Begley TP. 2008. O-phospho-L-serine and the thiocarboxylated sulfur carrier protein CysO-COSH are substrates for CysM, a cysteine synthase from Mycobacterium tuberculosis. Biochemistry 47:11606–11615. 10.1021/bi8013664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurgenson CT, Burns KE, Begley TP, Ealick SE. 2008. Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis. Biochemistry 47:10354–10364. 10.1021/bi800915j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. 2005. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 127:11602–11603. 10.1021/ja053476x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ågren D, Schnell R, Schneider G. 2009. The C-terminal of CysM from Mycobacterium tuberculosis protects the aminoacrylate intermediate and is involved in sulfur donor selectivity. FEBS Lett. 583:330–336. 10.1016/j.febslet.2008.12.019 [DOI] [PubMed] [Google Scholar]
- 18.Rabeh WR, Cook PF. 2004. Structure and mechanism of O-acetylserine sulfhydrylase. J. Biol. Chem. 279:26803–26806. 10.1074/jbc.R400001200 [DOI] [PubMed] [Google Scholar]
- 19.Karsten WE, Cook PF. 2002. Detection of intermediates in reactions catalyzed by PLP-dependent enzymes: O-acetylserine sulfhydrylase and serine-glyoxalate aminotransferase. Methods Enzymol. 354:223–237. 10.1016/S0076-6879(02)54019-2 [DOI] [PubMed] [Google Scholar]
- 20.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843. 10.1046/j.1365-2958.2003.03474.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chattopadhyay A, Meier M, Ivaninskii S, Burkhard P. 2007. Structure, mechanism, and conformational dynamics of O-acetylserine sulfhydrylase from Salmonella typhimurium: comparison of A and B isozymes. Biochemistry 46:8315–8330. 10.1021/bi602603c [DOI] [PubMed] [Google Scholar]
- 22.Rabeh WM, Alguindigue SS, Cook PF. 2005. Mechanism of the addition half of the O-acetylserine sulfhydrylase-A reaction. Biochemistry 44:5541–5550. 10.1021/bi047479i [DOI] [PubMed] [Google Scholar]
- 23.Baykov AA, Evtushenko OA, Avaeva SM. 1988. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171:266–270. 10.1016/0003-2697(88)90484-8 [DOI] [PubMed] [Google Scholar]
- 24.Gaitonde MK. 1967. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104:627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hensel G, Trüper H. 1981. O-Acetylserine sulfhydrylase and S-sulfocysteine activities of Chromatium vinosum. Arch. Microbiol. 130:228–233. 10.1007/BF00459524 [DOI] [PubMed] [Google Scholar]
- 26.Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. 2007. A general, robust method for the quality control of intact proteins using LC–ESI-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 852:188–194. 10.1016/j.jchromb.2007.01.011 [DOI] [PubMed] [Google Scholar]
- 27.Schnackerz KD, Ehrlich JH, Giesemann W, Reed TA. 1979. Mechanism of action of D-serine dehydratase. Identification of a transient intermediate. Biochemistry 18:3557–3563 [DOI] [PubMed] [Google Scholar]
- 28.Oda Y, Mino K, Ishikawa K, Ataka M. 2005. Three-dimensional structure of a new enzyme, O-phosphoserine sulfhydrylase, involved in L-cysteine biosynthesis by a hyperthermophilic archaeon, Aeropyrum pernix K1, at 2.0A resolution. J. Mol. Biol. 351:334–344. 10.1016/j.jmb.2005.05.064 [DOI] [PubMed] [Google Scholar]
- 29.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. 2011. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2:105. 10.3389/fmicb.2011.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotor C, Romero LC. 2013. S-sulfocysteine synthase function in sensing chloroplast redox status. Plant Signal. Behav. 8:e23313. 10.4161/psb.23313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensel G, Trüper HG. 1983. O-Acetylserine sulfhydrylase and S-sulfocysteine synthase activities of Rhodospirillum tenue. Arch. Microbiol. 134:227–232. 10.1007/BF00407763 [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, Iwahashi H, Eguchi Y. 1984. Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella typhimurium. J. Bacteriol. 158:1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatani T, Ohtsu I, Nonaka G, Wiriyathanawudhiwong N, Morigasaki S, Takagi H. 2012. Enhancement of thioredoxin/glutaredoxin-mediated L-cysteine synthesis from S-sulfocysteine increases L-cysteine production in Escherichia coli. Microb. Cell Fact. 11:62. 10.1186/1475-2859-11-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bermúdez MA, Paez-Ochoa MA, Gotor C, Romero LC. 2010. Arabidopsis S-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22:403–416. 10.1105/tpc.109.071985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323. 10.1093/nar/gkg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.