Abstract
SUMMARY
The Picornaviridae represent a large family of small plus-strand RNA viruses that cause a bewildering array of important human and animal diseases. Morphogenesis is the least-understood step in the life cycle of these viruses, and this process is difficult to study because encapsidation is tightly coupled to genome translation and RNA replication. Although the basic steps of assembly have been known for some time, very few details are available about the mechanism and factors that regulate this process. Most of the information available has been derived from studies of enteroviruses, in particular poliovirus, where recent evidence has shown that, surprisingly, the specificity of encapsidation is governed by a viral protein-protein interaction that does not involve an RNA packaging signal. In this review, we make an attempt to summarize what is currently known about the following topics: (i) encapsidation intermediates, (ii) the specificity of encapsidation (iii), viral and cellular factors that are required for encapsidation, (iv) inhibitors of encapsidation, and (v) a model of enterovirus encapsidation. Finally, we compare some features of picornavirus morphogenesis with those of other plus-strand RNA viruses.
INTRODUCTION
The Picornaviridae represent a large family of small plus-strand RNA viruses that cause a bewildering array of human and animal diseases ranging from severe (poliomyelitis, encephalitis, meningitis, and hepatitis) to mild (common cold). This family, which is still growing rapidly, consists of 46 species grouped into 26 genera, the best known of which are the genera Enterovirus (e.g., poliovirus [PV], rhinovirus, coxsackievirus, and echovirus), Aphthovirus (foot-and-mouth disease virus [FMDV]), Cardiovirus (encephalomyocarditis virus [EMCV] and Theiler's virus), and Hepatovirus (hepatitis A virus [HAV]) (1).
Picornaviruses are nonenveloped particles (27 to 30 nm in diameter) that consist of a capsid with icosahedral symmetry containing a tightly packaged, nonsegmented, single-stranded, positive-sense genomic RNA (∼7,500 nucleotides [nt]). The X-ray structures of human rhinovirus 14 (HRV14) (2) and poliovirus type 1 (3) were already published in 1985. They were the first known structures of picornaviruses, but currently, the structures of many picornaviruses, most recently that of hepatitis A virus (4), have been described. Although the basic architectures of picornavirions are similar, there are many differences in their building blocks (e.g., the nature of the processed capsid precursors) and surface properties.
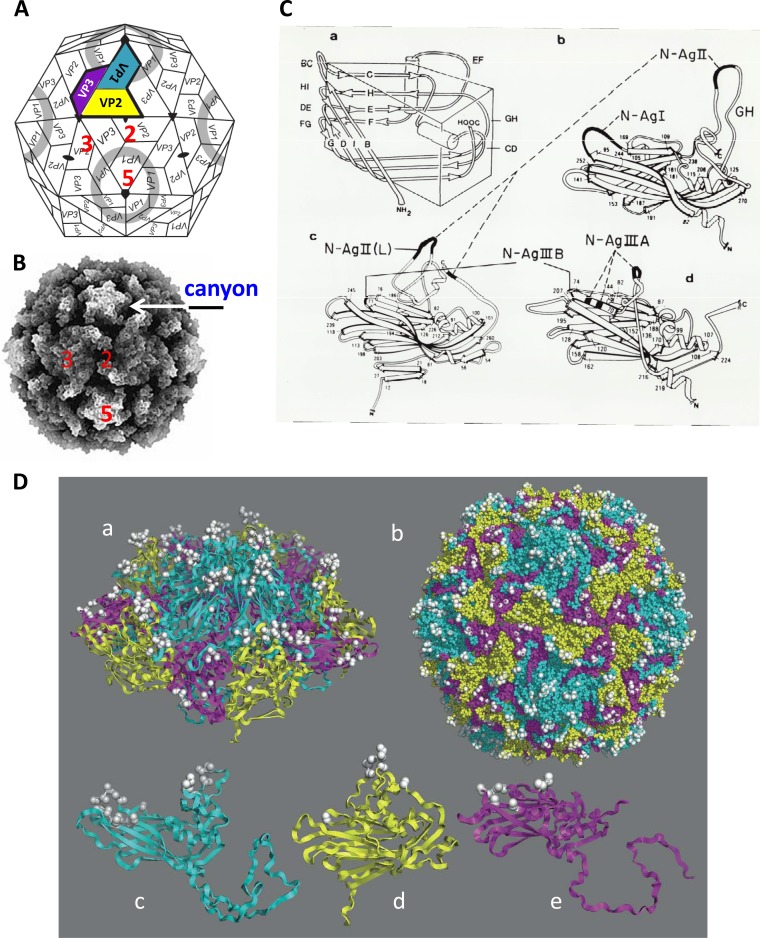
Enterovirus capsids are composed of 60 copies each of four viral polypeptides known as VP1 to VP4. They form capsids (Fig. 1A and B) that display 2-, 3-, and 5-fold symmetry axes (5). VP1, VP2, and VP3, the building blocks of the outer shell of poliovirus type 1 (Mahoney), can be presented as wedge-like structures (Fig. 1C) with major “neutralization antigenic sites” (N-Ags) (binding sites for neutralizing antibodies) that are displayed on the surface of the assembled poliovirions (discussed in Steps in Picornavirus Morphogenesis, below) (5). The small VP4 molecules reside inside the virion, but they can “breathe” into the virion surface even at physiological temperatures (6).
FIG 1.
Structure of poliovirus. (A) Schematic diagram of the structure of poliovirus with icosahedral symmetry (3, 219). The 5-fold, 3-fold, and 2-fold axes of symmetry are indicated. The capsid proteins VP1 (blue), VP2 (yellow), and VP3 (magenta) make up the outer surface of the particle, whereas VP4 is located internally. The structure shown in color is the processed protomer of which VP0 has already been cleaved into VP4 and VP2. The canyon around the 5-fold axis of symmetry is indicated with a ring. (Modified from reference 171 with permission of the publisher. Copyright 1989 Annual Reviews.) (B) Computer model of poliovirus. The 5-fold, 3-fold, and 2-fold axes of symmetry and the canyon are visible on the structure. (Reprinted from reference 172 with permission.) (C) Schematic representation of the three large poliovirus capsid proteins, each of which forms an eight-stranded, wedge-like, antiparallel β-barrel core (a) (3, 219). The antiparallel strands are connected by loops (BC, HI, DE, FG, GH, and CD). In panels b to d, the large capsid proteins are represented with ribbon diagrams (219). The four major neutralization antigenic sites (N-Ags) of poliovirus (type 1) map to surface loop extensions, as shown. N-AgI is a linear antigenic site that maps to the BC loop (amino acids 95 to 105) of VP1. All other major sites are discontinuous in nature: N-AgII (dotted line) spans VP1 and VP2 (amino acids 221 to 226 of VP1 and amino acids 164 to 172 of VP2). N-AgIII presents as two independent sites: N-AgIIIA consists of amino acids 58 to 60 and 71 to 73 of VP3 (dotted line), whereas N-AgIIIB contains amino acids 72 of VP2 and 76 to 79 of VP3. (Adapted from reference 3 with permission of AAAS.) (D) Localization of all major neutralization antigenic sites on the poliovirion indicating the density of possible neutralizing antibody-binding sites. (a) Band diagram of a pentamer containing the apex of the 5-fold symmetry axis. N-Ags are shown as white balls surrounding the mesa. Binding of antibodies to N-AgI, located on the rim of the canyon, leads to the neutralization of the virus by preventing attachment of the virus to the cellular receptor. (b) Dense distribution of N-Ags throughout the poliovirus capsid. (c to e) Band diagrams and neutralization antigenic sites of the capsid proteins VP1, VP2, and VP3. (Reprinted from reference 173 with permission of the publisher.)
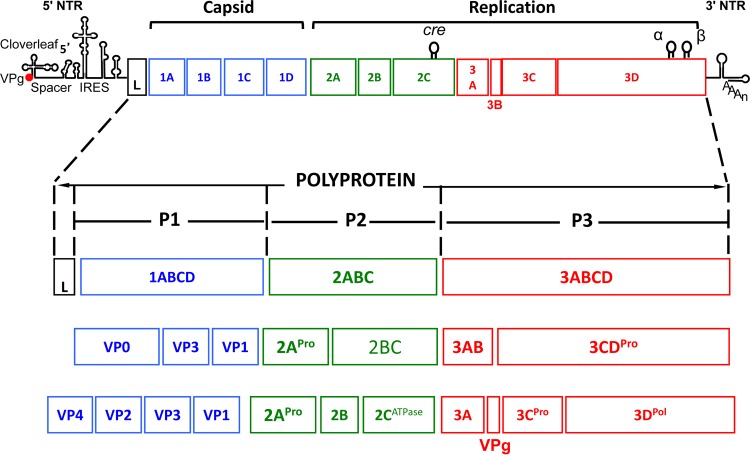
The naked protein shell of picornaviruses harbors the RNA genomes (∼7.2 to 8.4 kb), which are covalently linked at their 5′ ends through a phosphodiester bond (Tyr-p-U) to the small viral peptide VPg (22 to 25 amino acids) (7). VPg is followed by a long (500- to 1,200-nt) 5′ nontranslated region (NTR) containing replication signals (the “cloverleaf”) and the “internal ribosomal entry site” (IRES), a single large open reading frame (ORF) encoding the polyprotein, and, finally, a short (30- to 650-nt) 3′ NTR with a poly(A) tail (Fig. 2) (7). The genome has been organized by consensus into 1ABCD-2ABC-3ABCD units, where the numbers indicate three different functional domains and the letters identify proteins mapping to one of the three domains (e.g., 3Dpol is the RNA polymerase, with the superscript indicating a known enzymatic function). Members of some genera (e.g., Cardiovirus and Aphthovirus) also contain a “leader protein” (L) that is fused to the N terminus of the polyprotein (Fig. 2) (8).
FIG 2.
Genome structure of picornavirus and polyprotein processing. The picornavirus genome is a linear single-strand RNA ranging from 7.1 to 8.9 kb in length. The RNA has a covalently linked viral protein (VPg) at its 5′ end and poly(A) at its 3′ end. The ORF is flanked by a long 5′ nontranslated region (NTR) and a short 3′ NTR. The long 5′ NTR contains an internal ribosome entry site (IRES) that directs polyprotein translation. Besides the highly structured 5′ NTR and 3′ NTR, other essential RNA secondary structures have been identified in the open reading frame. For poliovirus, these structures are the cis replication element (cre) located in the 2CATPase coding sequence (106) and two RNA elements, α and β, located in the 3Dpol coding region (110). All known picornavirus genomes contain a cre element, but these structures may map to different locations in the genomes of different viruses. The first cre discovered maps to the P1 coding region of human rhinovirus 14 (HRV14) (174). No analyses of α and β elements in picornavirus genomes other than poliovirus have been published. The single open reading frame is organized as 1ABCD-2ABC-3ABCD, with the numbers indicating the three different domains and each letter representing a protein. The P1 region encodes the capsid structural polypeptides. The P2 and P3 regions encode the nonstructural proteins associated with replication. The polyproteins of member viruses of a large number of picornavirus genera (Aphthovirus, Erbovirus, Kobuvirus, Cardiovirus, Teschovirus, Sapelovirus, and Senecavirus) other than Enterovirus or Hepatovirus have an additional protein, the L protein, attached to the N terminus (8).
The life cycle of picornaviruses starts with the binding of the virus to a cell surface receptor (9). This interaction leads to virus internalization and destabilization of the capsid, which allows the release of the genome from the endosome into the cytoplasm (9, 10). After release, the RNA serves as mRNA for the IRES-mediated translation of the polyprotein (7) that is cotranslationally processed by the viral proteinases 2Apro and 3CDpro into precursor and mature proteins (11). Since poliovirus (and presumably all picornaviruses) can replicate in enucleated cells (see reference 12 and references therein), the major target of viral genetic armor is aimed at the cytoplasm, which in 2 to 3 h is remodeled for their benefit (13). All nonstructural proteins participate in this task, but, as discussed below, 2CATPase and its precursor 2BCATPase are involved in viral replication and in modifying the cell architecture to a greater extent than any other viral protein. Particularly startling is the rearrangement of cellular membranes into structures required for the formation of the RNA replication complex (14, 15). Typical for single-stranded RNA viruses of plus polarity, virus-encoded proteins (RNA polymerase 3Dpol, 3AB, VPg, 2CATPase, and more) catalyze the synthesis of minus strands, which in turn are the templates for the synthesis of progeny plus strands. This involves VPg-dependent initiation of RNA synthesis by 3Dpol (16), a process now known to function in many RNA virus systems. Notably, the functions of the IRES in controlling both translation as well as genome replication are dependent upon the participation of numerous cellular proteins, the number of which seems to increase steadily. The newly made viral RNA can be used as either mRNA for translation, a template for replication, or a substrate of encapsidation into a protective coat as soon as enough genomes and capsid proteins have been synthesized (14, 15). The mature virus exits the host cell either after cell lysis or by a not yet fully understood mechanism, possibly involving autophagosomes (17, 18). Generally, the enteroviral life cycle is very fast, with the whole process being completed in about 8 h. Among the exceptions is, most notably, HAV, whose replication cycle in tissue culture cells requires at least a week (19) (see also below).
Morphogenesis is the least-understood step in the life cycle of Picornaviridae. The packaging of genomic RNA occurs in the cytoplasm of the infected cell, and it is a complex process that, just like in other viral systems, requires a high degree of precision and specificity to regulate the interactions between participating macromolecules. Indeed, no RNA other than genomic RNA is generally encapsidated into enterovirus virions. Although the basic steps of assembly, the formation of capsid precursors followed by the capture of genomic RNA, have been known for some time, very few details about the mechanism providing specificity and the factors that regulate this process are available. Most of the available evidence about picornavirus morphogenesis is derived from studies with PV, a member of the C-cluster enteroviruses.
The recent report that the encapsidation specificity of poliovirus (and related C-cluster coxsackieviruses) is based on protein-protein interactions (2CATPase-capsid proteins) was a breakthrough in research on enterovirus morphogenesis (20, 21). Other important results of picornavirus encapsidation include (i) the observation that glutathione (GSH) is an important host factor for enterovirus morphogenesis by providing stability to capsid precursors and the mature virus (22–24); (ii) the discovery that Aichi virus, a member of the recently discovered genus Kobuvirus, uses an RNA packaging signal to confer specificity to its encapsidation (25, 26); and (iii) a report that hepatitis A virus uniquely uses its very small nonstructural protein 2A (also called X) in the formation of a precursor of the mature virus particle (27). In addition, there is an exciting report on the production of an enveloped (“membrane-cloaked”) form of hepatitis A virus (“eHAV”) that emerges from infected cells both in vitro and in vivo (28).
The aim of this review is to summarize what is currently known about the morphogenesis of picornaviruses, including steps in encapsidation, the specificity of encapsidation, viral and cellular factors required for morphogenesis, and inhibitors of morphogenesis. Emphasis is placed on the enteroviruses simply because of the greater abundance of information on these viruses. We conclude by describing an updated model of enterovirus morphogenesis, which is compared with the mechanism of particle assembly of other human and animal plus-strand RNA viruses. Unfortunately, due to the limited scope of this article, it is not possible to discuss every publication that deals with this subject in animal RNA virology or to give proper credit to everyone who has worked on this extremely complex process. However, we hope that the topics that we have selected for this review will provide the reader with an overview of the achievements and unsolved problems of picornavirus morphogenesis research that will stimulate further studies on this fascinating topic.
STEPS IN PICORNAVIRUS MORPHOGENESIS
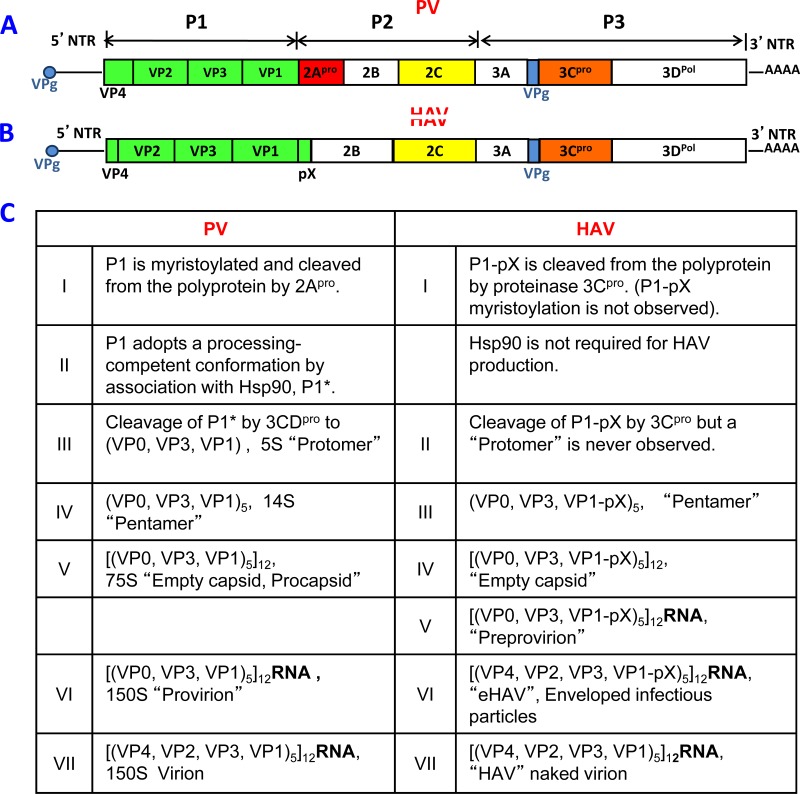
Many of the individual steps in picornavirus morphogenesis were first studied with PV and are summarized in Fig. 3. Below, we describe them briefly and note differences that exist among different picornavirus species.
FIG 3.
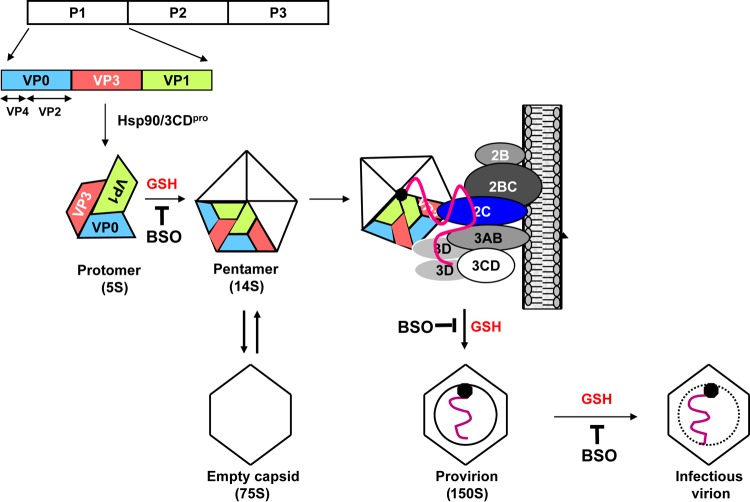
Genome organization of poliovirus (PV) and hepatitis A virus (HAV) and steps in virion formation. The general PV/HAV genome organization is the same: a VPg-linked genome; a long 5′ NTR harboring replication signals and the IRES; a single ORF divided into P1 (capsid region [green]) and P2 plus P3, encoding nonstructural proteins; and 3′-terminal RNA structures terminated by poly(A). (A) PV encodes two proteinases (red and orange), of which 3Cpro functions mostly as a processing precursor of 3CDpro. The proteinase 2Apro cleaves P1 from P2-P3 in statu nascendi. In addition, it cleaves numerous cellular proteins to the advantage of viral proliferation. The highly complex 2CATPase (yellow) is involved in numerous steps of viral replication, including morphogenesis. (B) HAV lacks 2Apro and instead retains a sequence (pX) that is involved in morphogenesis. HAV uses 3Cpro to catalyze the release of P1-pX (green) from the polyprotein. (C, left) Individual steps in PV morphogenesis. We speculate that pentamers engage in genome packaging via interactions between capsid proteins and 2CATPase. (Right) HAV morphogenesis is different from that of PV in many aspects. Nonmyristoylated HAV P1-pX (green) is cleaved and processed by 3Cpro (VP0, VP3, and VP1-pX). Hsp90 is not involved in HAV morphogenesis. We speculate that pentamers interact with genomic RNA to form “preprovirions.” The events following the formation of the preprovirion are extraordinarily different from those of all other picornaviruses: they involve “cloaking of the particles” in host cell membranes (28), which predominantly leads to infectious “eHAV” particles still carrying VP1-pX. The site and mechanism of the switch to naked virions lacking pX and VP4 are still under investigation (28). For further details, see the text.
Polyprotein Processing, Myristoylation, and Interaction of P1 with Heat Shock Protein Hsp90
Enteroviruses.
The capsid precursor domain (P1) of poliovirus is separated by the proteinase 2Apro from the rest of the polyprotein by cotranslational cleavage at a Y/G site (29) (Fig. 3C, PV step I). The N-terminal glycine of the newly formed poliovirus P1 capsid precursor is rapidly modified (perhaps also cotranslationally) by covalent linkage to myristic acid (30–32), after which it becomes a client of the cellular chaperone Hsp90 to attain a processing-competent conformation (referred to as P1* in Fig. 3C, step II) (33, 34). It is not known whether myristoylation of PV P1 is a prerequisite for its interaction with the chaperone Hsp90. However, only P1* can be effectively cleaved by the proteinase 3CDpro into VP0, VP3, and VP1 (Fig. 3C, PV step III) (33, 35). After proteolytic cleavage by 3CDpro, the capsid proteins dissociate from Hsp90, an observation which indicates that subsequent steps of morphogenesis are independent of the chaperone. Instead of separating and dispersing in the cytoplasm, VP0, VP3, and VP1 immediately assemble into the 5S protomer, which functions as the fundamental building block of the capsid (see below). Notably, myristoylation of the N-terminal domain of the polyprotein is not conserved among all picornaviruses; in particular, it is absent in hepato- and parechoviruses (36, 37).
Hepatovirus.
Unlike enteroviruses, cardio-, aphtho-, and hepatoviruses follow a different cascade for the processing of their polyproteins. In the case of HAV, the primary cleavage at the 2A/2B junction by 3Cpro yields the capsid precursor P1-2A, also referred to as P1-pX (Fig. 3C, HAV step I) (38). The small HAV 2A protein (pX) (8 kDa) has no enzymatic activity, but it is instead an essential component of the capsid precursor P1 for morphogenesis (38). HAV P1-pX is further processed by 3Cpro into VP0, VP3, and VP1-pX (Fig. 3C, HAV steps II and III). Neither HAV P1 nor VP0 is myristoylated (36), and Hsp90 is not required for HAV production (39). During later steps in HAV morphogenesis, pX is finally cleaved off from VP1 by an unidentified proteinase (see also below). Cardio- and aphthoviruses, in turn, evolved a very unusual processing mechanisms leading to the severance of P1 from the polyprotein (11). Briefly, it occurs by autocatalytic cleavage that does not involve a proteinase. Instead, it is catalyzed at a conserved motif (DxExNPG∧P) by a “CHYSEL” (cis-acting hydrolyase element) mechanism. For cardioviruses, the resulting P1 capsid precursor is linked to a 2A polypeptide lacking enzymatic activity; for aphthoviruses, it is linked to a minute “2A peptide” (only 18 amino acids long). Both of these appendices are later removed from the P1 precursors by the corresponding viral 3Cpro proteinases (11).
Formation of Small Capsid Precursors: the 5S Protomer (VP0, VP1, VP3) and the 14S Pentamer (VP0, VP1, VP3)5
After the processing of the capsid precursor P1, the capsid proteins VP0, VP3, and VP1 of all known picornaviruses remain associated and immediately form the “protomer” (VP0, VP3, VP1) (Fig. 1A and 3C, PV step III). Five protomers then assemble to form a 14S “pentamer,” (VP0, VP3,VP1)5 (Fig. 1D and 3C, PV step IV). As discussed below (see “Myristic Acid”), the myristate moiety at the N terminus of the PV P1 precursor (VP4) enhances the assembly of protomers into pentamers (40–42). It should be noted that the temperature-sensitive (ts) growth phenotype of the PV type 3 Sabin vaccine strain is related to a defect at the nonpermissive temperature in the early assembly process, specifically at the protomer/pentamer step (43). In the case of HAV, 5S particles are not observed in infected cells, presumably because of the highly efficient association of protomers into pentamers (Fig. 3C, HAV steps II and III) (44), which is enhanced by the presence of pX (2A) in VP1-pX (45).
The exact role of pentamers in the assembly of the mature virus particle is still controversial. According to the currently accepted model, pentamers condense around the viral RNA during the assembly process to form a provirion. The alternate model proposes that pentamers first assemble into an empty capsid, into which the viral RNA is inserted to yield the provirion. Several lines of evidence favor the first model: (i) only 14S pentamers but not empty capsids display RNA-binding activity in vitro, and 14S pentamers undergo a conformational change upon RNA binding (46); (ii) during in vitro cell-free synthesis of mature poliovirus, only 14S pentamers interact with the newly made viral RNA to form mature virus (47); (iii) 14S pentamers can be found in cells infected with all picornaviruses (48, 49); (iv)14S pentamers can be made to accumulate in infected cells by using a temperature block, and after removal of the block, they can be chased into mature viruses (50); and (v) 14S pentamers are associated with the replication complex in infected cells and can be cross-linked to viral RNA but not to the replicative intermediate (RI) (51). Our recent studies with l-buthionine sulfoximine (BSO), an inhibitor of GSH synthesis, also suggest that 14S pentamers are critical for the formation of mature virus particles and that the 75S empty capsids may not be functional intermediates in virus morphogenesis (22) (see below).
Formation of 75S Empty Capsids: [(VP0, VP1, VP3)5]12
Empty capsids, also known as procapsids, contain 12 pentamers but no viral RNA and sediment at 75S in sucrose gradients (Fig. 3C, PV step V). They have been known for decades to be formed during poliovirus infection (52, 53) but are not present in cells infected with mengovirus, a species of Cardiovirus (49). Early studies suggested the existence of two functional states of procapsids in PV-infected cells, one active and one inactive (48, 54, 55). The active procapsid can be disassembled into pentamers and can be reassembled into stable procapsids. Two different forms of procapsids, similar to those found in vivo, can also be observed during in vitro assembly of 14S particles (48, 56). It was proposed that the active particle exists in vivo, while the inactive particle is only a by-product of isolation procedures or conditions used during in vitro assembly. Putnak and Phillips (48) reported that the conformation of empty capsids made from 14S particles is enhanced by a “morphopoietic factor” present in infected cell extracts, but the identity of such a putative factor, if it exists, has never been elucidated.
As noted above, the role of the procapsid in viral morphogenesis is not yet clear. Some early studies suggested the possibility that it plays a role as a precursor. The addition of 1 to 3 mM guanidine hydrochloride (GnHCl) at midinfection, which inhibits RNA synthesis, caused the accumulation of procapsids, which could be chased into virus particles (57, 58). From our studies with BSO, we found that 75S particles, although they are made in normal amounts during infection in GSH-depleted cells, do not progress into forming mature virus, an observation which suggests that 75S may not be a functional capsid intermediate (22).
The Provirion: [(VP0, VP1, VP3)5]12RNA
In poliovirus morphogenesis, the provirion contains viral RNA in an “immature shell” composed of VP0, VP1, and VP3 (58, 59) (Fig. 3C, PV step VI) and sediments at 150S in sucrose gradients (60). The RNA in provirions is resistant to degradation by pancreatic RNase. However, in contrast to mature poliovirus, treatment of the particles with EDTA or 1% SDS results in the release of the viral RNA, yielding an empty capsid (60). The encapsidation of the viral RNA requires compact condensation of the RNA molecule and neutralization of the negatively charged phosphate groups by cations (61).
Using BSO, the inhibitor of poliovirus morphogenesis, we observed that GSH depletion leads to the accumulation of “provirion-like” particles, which migrate slightly faster on sucrose gradients than the 150S particles made under normal growth conditions and contain essentially no infectious virions, suggesting that GSH is required for the final maturation of viral particles (22).
In HAV, there is yet another intermediate, the “preprovirion,” [(VP0, VP3, VP1-pX)5]12RNA, that still contains polypeptide X linked to VP1 (Fig. 3C, HAV step V) (62).
Mature Virions: [(VP4, VP2, VP3, VP1)5]12RNA
With most picornaviruses, the final step of morphogenesis is accomplished by the cleavage of VP0 into VP2 and VP4 (Fig. 3C, PV step VII). This triggers a rearrangement of the capsid proteins and confers stability to the resulting icosahedral particle (3). There is no evidence that the RNA is structured within the mature virus particle (63). In contrast to provirions, mature poliovirus particles are resistant to SDS or EDTA treatment and are not permeable to Cs+ ions (64). It should be noted that the mature virus particles of Aichi virus (65) and echovirus 22 (66) contain VP0 but no VP4.
The maturation cleavage of PV VP0 during the last step of virion formation is efficient, yet 1 or 2 VP0 molecules out of 60 remain uncleaved (67). The mechanism of VP0 cleavage is most likely autocatalytic and dependent on RNA encapsidation (3, 67). An asparagine and a serine form the VP4/VP2 cleavage sites (N∧S) of most picornaviruses (68). It was originally speculated that a serine (VP2 S10) near the N terminus of VP2 was activated during virion maturation to catalyze VP0 cleavage (69). However, mutation of this conserved serine to either cysteine (S10C) or alanine (S10A) yielded no maturation phenotypes of the respective virus variants (68). In contrast, a histidine (H195) in VP2, conserved in all picornaviruses, has been identified as being essential for efficient cleavage at the VP4/VP2 junction (70). An H195T or H195R mutation in PV VP2 resulted in the assembly of highly unstable 150S particles that contain uncleaved VP0 (70). It was suggested that the histidine activates local water molecules and initiates a nucleophilic attack on the scissile bond.
There are several amino acids in the capsid proteins that appear to directly affect morphogenesis, uncoating, or both. For example, two determinants of mouse adaptation of PV, the VP1 T22I and VP2 S32T mutations, affect both uncoating and particle assembly (71). Similarly, a deletion of residues 8 and 9 in VP1 (mutant 101) affects RNA release, while a deletion of residues 1 to 4 in VP1 (mutant 102) leads to defects in both encapsidation and uncoating (72). Finally, a ts mutant of PV (VP2-103), containing a single amino acid change, Q76R, accumulates provirions but no mature virus at the nonpermissive temperature (73).
Structural analyses have indicated that there is a major difference between empty capsids and mature viruses in the network formed by the N-terminal extensions of capsid proteins on the inner surface of the shell (3). On the inner surface of the mature capsid, the NH2-terminal strands of VP1, VP2, and VP3 form an extensive network, which links the five protomers together to form a pentamer (3). The NH2 termini of five VP3 molecules intertwine around the 5-fold axis and form a twisted tube, which was proposed to contribute to the stability of pentamers. The small capsid protein VP4 has a more extended structure than the other three capsid proteins, and it is similar to the NH2-terminal strands of VP1 and VP3 in its position (3).
Other factors also affect virus maturation. As noted above, myristoylation of VP4 is important for virus maturation (36). As discussed below (see “3CD and VPg”), virus maturation is also enhanced by the viral protein 3CD in an in vitro translation/RNA replication system (74, 75). Moreover, we have recently shown that GSH is required for the production of infectious 150S particles (22). Finally, Richards and Jackson (18) proposed that virion maturation occurs in a cellular compartment known as the acidic autophagosome, but whether these structures are essential for poliovirus replication remains uncertain. Those authors observed that inhibitors of vesicle acidification have no effect on the production of 150S virus particles but inhibit the cleavage of VP0 to VP2 and VP4 and the accompanying formation of infectious virions. Nevertheless, it should be remembered that poliovirus RNA can translate→replicate→encapsidate in the cell-free environment of a cytoplasmic extract of HeLa cells (76), and although membranous components are absolutely required (77), it is unlikely that highly organized membrane structures would survive the preparation of the cell extract.
With the maturation of the viral particle, there is a mesa formed at the top of the 5-fold axis of the poliovirion that is surrounded by a cleft that has been termed “canyon” (Fig. 1A and B) (2). All enteroviruses (including rhinoviruses) have this canyon (78). Although canyons of several enteroviruses function as the receptacle of their respective receptors (e.g., PV, HRV14, and coxsackie B virus 3 [CBV3]), a large number of enteroviruses use structures other than this cleft to attach to the cell (HRV2, some echoviruses, and coxsackie B viruses) (9, 79–82).
Of those picornaviruses analyzed, VP1, the largest capsid protein, forms at the bottom of canyons a peculiar hydrophobic pouch or “pocket,” first observed by Hogle and colleagues (3). VP1 pockets found in a large number of picornavirions are occupied by a “pocket factor” of cellular origin whose identity and function are still ill defined. In general, they have been described as fatty acid-related, hydrophobic compounds, but some enteroviruses (HRV14 and HRV3) and some cardioviruses (EMCV and mengovirus) do not appear to carry pocket factors altogether (78, 82). Although there is no proof that pockets and pocket factors are involved in morphogenesis or uncoating, it is highly unlikely that their existence is just an accident.
The so-called WIN compounds are among the best-known inhibitors of picornavirus replication. They invade and lodge inside the hydrophobic pocket of enteroviruses beneath the canyon floor of the capsid (78, 83, 84). Because the binding sites of the viral receptor and the pocket factor overlap, only one WIN compound fits into the space. As was found for human rhinoviruses, binding of these compounds can have two distinct consequences: they may inhibit either receptor binding, as in the case of HRV14 (85, 86), or uncoating, as with HRV1A (87).
Mature poliovirions display on their surface an impressive web of neutralization antigenic sites (N-Ags), which are defined as structures (linear or nonlinear) to which neutralizing antibodies can bind. The positions of the N-Ags on the individual large capsid proteins are shown in Fig. 1C and Dc to e. The entire web of N-Ags on virions is shown in Fig. 1Db. With the use of specific monoclonal antibodies, these epitopes offer convenient ways to identify mature virions in tissue culture cells or by light microscope imaging experiments. Generally, poliovirions express major (N-AgIA, N-AgIIA, N-AgIIIA, and N-AgIIIB) and minor (N-AgIB and N-AgIIB) neutralization antigenic sites. Altogether, polioviruses express three unique sets (in sequence and/or structure) of all these sites; hence, poliovirus exists as three serotypes. We note that if poliovirus should attempt to evolve a fourth serotype, it would have to change sequences and/or structures of all N-Ags, thereby generating an entirely new set. Fortunately, no new poliovirus serotypes have been discovered since the three serotypes were originally identified in 1951 (88). Thus, changing of an entire set of N-Ags appears to be extremely difficult, even over a time of, say, 1 century. In contrast, human rhinoviruses, whose crystal structures are very similar to those of the polioviruses, have evolved dozens of unique sets of N-Ags; hence, human rhinoviruses exist as dozens of serotypes.
Special Case of a Picornavirus: the Naked and Membrane-Cloaked Particles of Hepatitis A Virus
A very special case among the picornaviruses is HAV. HAV is released from tissue culture cells and its target tissue in vivo (in infected humans) in not only in a nonenveloped (naked) form (Fig. 3C, step VII) but also in an enveloped form (eHAV) (Fig. 3C, step VI) (28). HAV released from cells is cloaked in host-derived membranes, which protect the virus from neutralization by antibody. Using a pathway involving the endosomal sorting complexes, the preprovirions envelop themselves into membranes (1, 2, or even 3 particles per vesicle) and egress cells as enveloped particles called eHAV (Fig. 3C, step VI) (28). During this process, VP0 undergoes maturation cleavage, processing VP0 into VP2 and VP4 (23 amino acids long), presumably by RNA-induced autocatalysis. pX is retained as a component of eHAV (60 copies of VP1-pX) until the eHAVs shed the membranes prior to fecal egress from a patient, a step still poorly understood (Fig. 3C, step VII) (28) but leading to the maturation of naked virions in stool. During the last step of morphogenesis, pX is cleaved off from all VP1 proteins of the particle by an unknown cellular proteinase (19).
DO PICORNAVIRUSES USE RNA PACKAGING SIGNALS IN MORPHOGENESIS?
Morphogenesis of progeny viral genomes is highly specific, and RNA viruses have evolved different elaborate mechanisms to discriminate against nucleic acids as substrates other than their own. The best-known mechanism, used by many RNA viruses, is an RNA packaging signal that is recognized by one or more capsid proteins to provide specificity to the encapsidation process (89–91). Another mechanism has been suggested (without experimental proof), that specific interactions between capsid proteins and the nonstructural proteins of the replication complex are involved (92). This would place the progeny RNA near the site of assembly. As we will show, picornaviruses have evolved to use both strategies: encapsidation specificity by protein-protein interactions (enteroviruses) and encapsidation by recognition of packaging signals (Aichi virus of the genus Kobuvirus).
All Searches for RNA Packaging Signals in Enterovirus Genomes Have Failed
As outlined below, numerous studies aimed at identifying RNA-based encapsidation signals in the 5′ and 3′ NTRs of enteroviral genomes have been unsuccessful. These searches generally included the construction of chimeric genomes in which various segments were exchanged between genomes of different enteroviruses, followed by assaying for the nature of the progeny virus (if there was one). Alternatively, the polyprotein of enterovirus was radically recoded by changing codon pairs without altering the codon usage or the amino acid sequence of the target protein. By the latter strategy, it was assumed that RNA packaging signals would be destroyed without interference with genome replication, provided that essential replication signals, like cre(2C), were not modified.
The 5′ NTR.
The first evidence against the 5′ NTR carrying an RNA packaging signal was provided by Semler and colleagues, who exchanged nearly the entire 5′ NTR of PV with that of CBV3 (nt 1 to 627) without losing the PV-specific replication-and-encapsidation phenotype (93, 94). Subsequently, numerous PVs with exchanges of IRESs from different viruses (including that from hepatitis C virus [HCV]) were constructed and analyzed; none of the chimeras showed significant translation/replication/morphogenesis phenotypes (7, 95). Similarly, the cloverleaf at the 5′ end of the PV genome was exchanged with that of the quite distantly related human rhinovirus 2 (HRV2). This chimera also expressed an adequate proliferation phenotype (96). Combined, these data strongly suggest that the PV genome, and perhaps all enterovirus genomes, does not contain a packaging signal in the 5′ NTR.
RNA sequences in the polyprotein-encoding region.
(i) The P1 capsid domain.
PV proliferation can lead to the evolution of defective interfering (DI) particles, in which a portion (of various lengths) of the P1 domain encoding the capsid proteins has been deleted (97, 98). DI particles recruit their capsid proteins in trans from coinfecting wild-type (wt) PV, thereby reducing (interfering with) the replication of the wt virus. Kuge and colleagues subsequently made the important observation that the deletions in the P1 domain of DI particles must be in frame with the rest of the polyprotein, or else there is no PV genome replication and encapsidation (99); they concluded correctly that translation of the ORF of the polyprotein (regardless of whether it contains P1 sequences or not) is required in cis for genome replication. Novak and Kirkegaard later provided strong support for this hypothesis (100) (see below). Kajigaya et al. proposed that the P1 region is unlikely to harbor an RNA packaging signal since the genomes of DI particles can be encapsidated in trans (101).
A number of investigators then replaced part or all of the capsid coding sequence in P1 with foreign genes, thereby generating replicons suitable for studies of PV replication and encapsidation, even for gene therapy. Porter and colleagues expressed a multitude of different genes in PV replicons, with firefly luciferase being the most suitable for studies of encapsidation (102). Barclay and colleagues used chloramphenicol acetyltransferase (CAT) inserted into the P1 domain as a foreign marker gene (103). These replicons carrying foreign genes could be encapsidated in trans by coinfection with either a vaccinia virus expressing the poliovirus capsid proteins (104) or PV (105). The exquisite specificity of encapsidation was revealed when enteroviruses other than PV were used in coinfections: the PV replicons could not be packaged in trans into capsids of bovine enterovirus, C-cluster coxsackievirus A21, B-cluster coxsackieviruses B3 and B4, rhinovirus 14, or enterovirus 70 (102, 103).
(ii) The P2-P3 nonstructural domains.
A computational analysis of conserved RNA structural elements in genomes of PV and related C-cluster coxsackie A viruses (C-CAVs) identified an RNA element mapping to the coding region of PV 2CATPase, termed cre(2C) (106). cre(2C) plays an essential role in genome replication; equivalent cre elements have been found in the genomes of all picornaviruses (107, 108). Using all enterovirus genome sequences known in 2001, Witwer et al. identified a highly conserved stem-loop structure in the C-terminal coding region of the PV RNA polymerase 3Dpol (109), subsequently called the beta stem-loop (110). However, no evidence has been uncovered to suggest that either cre(2C) or the beta stem-loop is involved in morphogenesis (107, 110).
Recently, we used a genome-wide scan involving large-scale recoding of the ORF of the PV polyprotein in combination with synthetic biology. This allowed us to search for functional RNA elements in enterovirus RNAs that had escaped previous discovery (111, 112). A computer algorithm, called “Scrambled” (SD), shuffled synonymous codons in different domains of the PV polyprotein, thereby introducing hundreds of silent mutations into the ORF without altering the amino acid sequence or codon usage. Scrambling of the P1 capsid-encoding region had no effect on PV proliferation, although 934 mutations were introduced into the 2,642-nt P1 coding domain (112). Scrambling of the P2 domain plus a small segment of the P3 domain (701 mutations in 2,214 nt), however, killed the virus, as expected, because cre(2C) in P2 was destroyed. The virus was revived when cre(2C) was rebuilt into the otherwise scrambled sequence (110). Genetic analyses provided no evidence for cre as a packaging signal (113). Similarly, the scrambled sequence (on average, every fourth nucleotide was changed) is not expected to support morphogenesis. Scrambling of P3 (625 mutations in 1,764 nt) revealed the existence of 2 closely spaced, functionally redundant RNA elements (α and β) in the close vicinity of the C terminus of the 3Dpol-encoding region (Fig. 2), which we found are required for RNA replication but not for encapsidation (110). This leads us to the conclusion that it is highly improbable that there are RNA sequence elements essential for particle assembly in the coding region of the PV polyprotein.
The 3′ NTR.
Several studies have shown the importance of the PV 3′ NTR for genome replication. For example, Pilipenko et al. provided genetic evidence that “kissing” between stem-loops X and Y in the 3′ NTR was required for adequate genome synthesis (114). Similarly, an intact PV 3′ NTR was essential for efficient PV RNA synthesis followed by encapsidation (115). While there is no controversy about these data (116), the following surprising observations clearly show that the PV 3′ NTR is not required for PV replication and morphogenesis. First, the entire 3′ NTR can be replaced with structurally very different 3′ NTRs from HRV14 or B-cluster CBV4 without significant impairment of PV replication and PV encapsidation (117). Second, Todd et al. reported the astounding observation that the entire PV 3′ NTR can be deleted [except for poly(A)], yet the resulting variant replicated and packaged albeit with reduced efficiency (118).
Encapsidation of enterovirus negative-strand RNAs under certain circumstances.
Under normal conditions of infection, only plus-strand RNAs are encapsidated. As an exception, the encapsidation of minus-strand RNAs has also been observed in noncytopathogenic CVB3 that was isolated from persistently infected murine hearts (119). The RNA genomes of these viruses contain 5′-terminal deletions in the cloverleaf, and when replicated, the amount of progeny minus strand present is much increased relative to the amount of plus strand. Since minus and plus strands have complementary rather than identical sequences, these results argue against an RNA signal determining the specificity of encapsidation (see Model of Enterovirus Morphogenesis, below).
Together, we conclude that there is no evidence that the genome of enterovirus harbors an RNA packaging signal essential for morphogenesis.
RNA Packaging Signal in the Genome of Aichi Virus
The Picornaviridae are a huge family of viruses that have evolved specialties in different stages of their replication, although generally, they follow a common proliferation plan. It therefore may not be surprising that morphogenesis of Aichi virus of the genus Kobuvirus involves an RNA packaging signal. The 5′ NTR of Aichi virus contains a hairpin at the very 5′ end that has been reported to play a role in particle assembly (25). Mutations constructed into the stem of the hairpin lead to the production of mostly empty capsids and very few mature virus particles. In addition to this hairpin, the leader (L) protein of Aichi virus harbors a possible function in virus assembly (26).
SPECIFICITY OF ENTEROVIRUS ENCAPSIDATION IS PROVIDED BY A SPECIFIC INTERACTION BETWEEN CAPSID PROTEINS AND NONSTRUCTURAL PROTEIN 2CATPase
During the life cycle of picornaviruses, the plus-stranded full-length genomic RNA serves three main functions: (i) as mRNA for translation, (ii) as the template for RNA replication, and (iii) as the genome for encapsidation. As first suggested by K. Kirkegaard, the three fundamental processes of translation, replication, and morphogenesis in picornavirus proliferation are so tightly linked that they serve as a “higher-order” proofreading mechanism: without adequate translation, there is no genome replication, and without genome replication, there is no encapsidation (92, 95). It follows that ready-made genomic RNA (VPg-linked genomic RNA, infectious mRNA, or genomic transcripts) present in the cytoplasm has no chance to serve as the substrate for the assembly of virions, as was observed in studies of a cell-free replication system developed by Molla et al. (76). Thus, a positive identification of an encapsidation signal is possible only if modifying that signal has no detrimental effect on RNA translation or RNA replication (20). This observation and the lack of apparent RNA packaging signals in enterovirus genomes raised the possibility that the specificity of encapsidation of poliovirus, and perhaps of all enteroviruses, is provided solely by an interaction between capsid and nonstructural proteins, a mechanism that would be quite unique in virology.
Multiple Roles of Enterovirus 2CATPase in the Virus Replication Cycle
2CATPase is a complex nonstructural polypeptide that expresses numerous functions: (i) it has been proven to be crucial for RNA replication and binding (120–124); (ii) it contains a nucleoside triphosphate (NTP)-binding domain (125) and possesses ATPase activity (discovered in vitro), which is inhibited by guanidine hydrochloride (GnHCl) (126); (iii) it has been implicated (weakly) in virus uncoating (127); (iv) together with its processing precursor 2BCATPase, it is involved in cellular membrane rearrangements and the formation of the cytoplasmic vesicle “web” typical of all enterovirus replication (13, 128–131); and (v) drug (hydantoin) inhibition and genetic studies have provided convincing genetic evidence that 2CATPase is involved in encapsidation (20, 21, 132).
Analyses of the linkage map of nonstructural proteins revealed that 2CATPase has binding affinity for 3AB (133, 134) or 3Cpro (135). For enterovirus 71 infections, it has been reported that 2CATPase binds the host protein reticulon-3 (136), but this was not found in a yeast two-hybrid scan with poliovirus 2CATPase (C. Wang and E. Wimmer, unpublished data). In a recent study, the interaction of 2BCATPase/2CATPase with VCP/p97, a cellular ATPase, was demonstrated, which was strongly enhanced by GnHCl (137). The significance of this observation, however, has not been elucidated. Considering the multitude of 2CATPase functions in various phases of enterovirus replication combined with the functional linkage between translation, replication, and morphogenesis, deciphering a role of 2CATPase in morphogenesis seemed hopeless.
Unique Assay Separating Replication from Morphogenesis in Tissue Culture Experiments
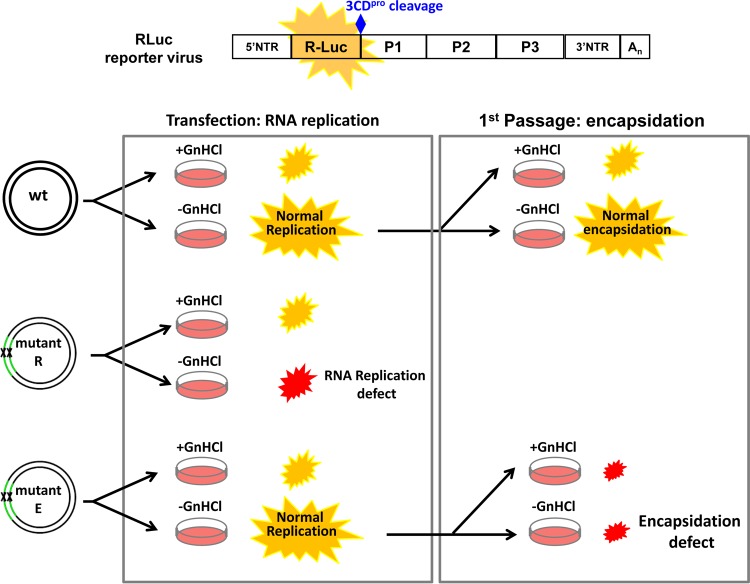
A key for these studies was a novel reporter virus that has allowed us to distinguish the function of 2CATPase in RNA replication from that in encapsidation in a two-step experiment (20) (Fig. 4). The reporter virus genome contains the renilla luciferase (RLuc) gene fused to the N terminus of the polyprotein that is cleaved off from the polyprotein by 3CDpro after translation. RNA replication and encapsidation are measured by luciferase activity after (i) transfection into HeLa cells (assay for replication) and (ii) the first passage of virus (if morphogenesis took place) into new naive HeLa cells (assay for morphogenesis). Specifically, RNA transcripts of the reporter cDNA are transfected into HeLa cells, in the absence and presence of GnHCl, a potent inhibitor of RNA replication (124, 138, 139). The luciferase activity produced in the absence of the drug is a measure of both translation and RNA replication of genomes (and, hence, a measurement of replication), while in the presence of the drug, only protein translation is assayed. In the second step, lysates of cells transfected in the absence of the drug are used to infect fresh HeLa cells. For the wt reporter virus, which is normal in both RNA replication and encapsidation, luciferase activity is produced during both transfection and infection after passage in new cells (Fig. 4). With a mutant defective in RNA replication (“mutant R”), only luciferase activity from protein translation of the transfected viral RNA is produced (Fig. 4). However, encapsidation-defective mutants (“mutant E”), even though luciferase activity can be observed from RNA replication in transfected cells, does not produce any significant levels of luciferase activity after passage into fresh HeLa cells (Fig. 4). This is because no intact infectious virus capable of infecting fresh cells upon passage is produced after transfection. Using this strategy, we have discovered that at least one of the functions of 2CATPase in assembly is to interact with one of the capsid proteins (VP3 and/or VP1) (20, 21).
FIG 4.
Use of renilla luciferase (RLuc) reporter virus to distinguish a defect in RNA replication from a defect in encapsidation. The genome of the RLuc reporter virus contains an RLuc gene fused to the N terminus of the PV polyprotein coding sequence. The RLuc reporter virus can distinguish a defect in RNA replication (mutant R) from that in encapsidation (mutant E) in a two-step experiment. In the first step, the reporter transcripts are transfected into HeLa cells in the absence and presence of guanidine hydrochloride (GnHCl), a potent inhibitor of RNA replication. After transfection, luciferase activity produced in the absence of the drug is a measure of RNA replication, while in the presence of the drug, only translation of the transfecting RNA is measured. In the second step, lysates of cells transfected in the absence of the drug are used to infect fresh HeLa cells. Luciferase activity in the absence of the drug is a measure of encapsidation in the first HeLa cells that were transfected. Mutant R has a defect in genome replication; hence, only a small Luc signal derived from translation of the transfected RNA is observed. Mutant E has a defect in encapsidation. It produces a robust Luc signal after transfection (first HeLa cell monolayer), but no Luc signal is detected in infected HeLa cells because no infectious progenies are formed during transfection.
Analyses of Chimeras between C-Cluster Coxsackie Viruses and Poliovirus Suggest That 2CATPase-Capsid Protein Interaction Provides Specificity of Morphogenesis
In the course of the global eradication of poliovirus with the live, oral Sabin vaccine (OPV), the observation was made that OPV can readily recombine in the field with closely related C-CAVs (140). The recombinants evolved rapidly into viruses that were as neurovirulent as the wt polioviruses targeted for eradication, and they caused small epidemics of poliomyelitis in many parts of the world. All neurovirulent recombinants were found to have capsid domain P1 of PV (and, hence, the virulence identity of PV) and various parts of downstream domains P2 and P3 of C-CAV. We abbreviate the recombinants “P-C-C,” indicating the three-domain polyproteins from two different viruses. In our studies of this highly important phenomenon for polio eradication, we generated P-C-C viruses in the laboratory and compared them with their C-P-P counterparts, e.g., consisting of the capsid domain of C-CAV and the two nonstructural domains of PV. C-P-P has the identity of a coxsackievirus because of its interaction with ICAM-1 as a receptor and pathogenesis resembling that of the common cold. An unexpected observation was made, that all P-C-C constructs grew very well, whereas most of the C-P-P constructs did not grow at all (141). Importantly, the genomes of C-P-P translated and genome replicated with wt kinetics, but as Luc chimeras, they did not produce virions in our two-step assay. However, blind passage of a C-P-P variant on HeLa cells generated individually two poorly growing small-plaque suppressor mutants, with the first having a single amino acid substitution in C-CAV VP3 (C*-P-P) and the second having a single amino acid exchange in PV 2CATPase (C-P*-P). The poor-growth phenotype of C*-P-P or C-P*-P in tissue culture cells changed to a variant growing with wt kinetics and a large-plaque phenotype when both mutations were engineered into the chimeric genome, yielding the C*-P*-P genotype (20). These results suggested a direct interaction between 2CATPase and the capsid protein VP3, which was subsequently proven by coimmunoprecipitation assays.
Alanine Scanning Analysis of 2CATPase Confirms the Linkage between the Nonstructural Protein and Capsid Proteins in Morphogenesis
The protein-protein mechanism for encapsidation specificity has been further supported by alanine scanning mutagenesis of 2CATPase, a strategy involving the sequential replacement of clusters of charged amino acids in a polypeptide with alanine residues (21). Of many mutants that were generated, one was “quasi-infectious” (qi), a phenotype with greatly debilitated replication such that progeny virions could not be isolated. However, blind passage of the qi variant yielded second-site suppressor mutations leading to new variants that could be grown and plaque titrated. In the case of the 2CATPase qi mutant, the second-site suppressor mutations mapped to capsid proteins, a result supporting rescue by regeneration of the 2CATPase-capsid protein interaction. Surprisingly, in two independently rescued variants, the second-site suppressor mutations each mapped to either VP3 or VP1. These genetic results are strong evidence that 2CATPase and capsid proteins must communicate for morphogenesis to occur, most likely by direct binding (20).
Other Nonstructural Proteins Involved in Picornavirus Morphogenesis
3CD and VPg.
The PV proteinase 3CDpro, a multifunctional polypeptide, is the precursor of the RNA polymerase 3Dpol (lacking RNA polymerase activity) as well as of the proteinase 3Cpro. The 3CDpro protein, an important proteinase itself (Fig. 3C, PV step III), is also an important RNA-binding protein during RNA replication. It interacts with both the 5′ cloverleaf structure of the viral RNA, in conjunction with 3AB or PCBP2 (142, 143), and the cre(2C) RNA element during the protein-priming step of RNA synthesis (144, 145). The best-known function of 3CDpro in encapsidation is the proteolytic processing of P1. It appears that 3CDpro can cleave all designated Q∧G scissile bonds in the PV polyprotein, whereas 3Cpro cannot: the latter is insufficient for processing P1 (146). Thus, 3CDpro is for as-yet-unknown reasons exclusively responsible for the cleavage of the PV capsid precursor P1 into VP0, VP1, and VP3.
Another role for the 3CD polypeptide in encapsidation, which is independent of its protease activity, was discovered with studies of an in vitro translation/RNA replication system that produces viable poliovirus (76). It was observed that proteolytically inactive 3CD (designated 3CD instead of 3CDpro) strongly stimulated mature virus production in the in vitro system (74, 75). Stimulation was dependent on the RNA-binding activity of the protein, residing in the 3C domain, and the integrity of “interface I” in the 3D domain. Interestingly, 3CD only stimulates the encapsidation of a VPg-linked viral RNA template (74, 75). A T7 RNA polymerase-generated transcript RNA could not replace VPg-linked RNA in the stimulation reaction, even if the two 5′-terminal G's were removed by ribozyme. Thus, it appears that 3CD has an affinity for the VPg-linked 5′ end of the poliovirus genome that may aid in encapsidation, an observation that also suggests a role for VPg in encapsidation.
2A of hepatitis A virus.
In contrast to most members of the Picornaviridae, HAV expresses only one proteinase (3Cpro) for the proteolytic cleavage of its polyprotein (147). The initial cleavage by HAV 3Cpro is between 2A and 2B, thereby producing a capsid precursor, P1-2A, which is subsequently cleaved into VP0, VP3, and VP1-2A. The 2A protein of HAV, also called pX, is a C-terminal extension of the structural protein VP1, and it lacks proteinase activity. Interestingly, pX (2A) functions in both particle assembly, by promoting the formation of pentamers, and the subsequent particle maturation step (38, 62, 147). Using genetic analyses, Morace and colleagues (27) showed that the C terminus of the 2A protein is important for the liberation of VP1-pX from the polyprotein, while a basic residue at the C terminus of VP1 is required for efficient particle assembly. The pX protein is removed from VP1-pX by an unknown host proteinase(s) so that mature nonenveloped HAV particles lack pX (28). However, in eHAVs, which are the dominant forms of virus released from infected cells, pX is still attached to VP1, but VP0 is already cleaved into VP2 and VP4 (28).
Leader protein of Aichi virus.
The Aichi virus genome encodes a 170-amino-acid-long L protein upstream of the capsid-encoding region (Fig. 2). This polypeptide exhibits no sequence similarity to the L proteins of other picornaviruses, such as that of aphthovirus, cardiovirus, or teschovirus (26). A deletion analysis of the Aichi virus L protein revealed that this polypeptide is important for both RNA replication and encapsidation. Specifically, a deletion of the C-terminal 50 amino acids of the L protein resulted in efficient RNA replication, but the virus yield was strongly reduced compared to that of the wt virus. A sedimentation analysis of the mutant virus showed that it has a severe defect in the formation of mature virions but not in that of empty capsids (26), indicating that L protein is involved in virus morphogenesis.
CELLULAR FACTORS INVOLVED IN PV MORPHOGENESIS
Heat Shock Proteins Hsp70 and Hsp90
Heat shock proteins are a group of functionally related proteins that aid in the folding or unfolding of proteins. Their expression levels are increased when cells are exposed to elevated temperatures or other stresses. An interaction between the PV P1 capsid precursor and Hsp70 was first observed by Macejak and Sarnow (34). Subsequently, the involvement of the cellular chaperone Hsp90 in picornavirus assembly was discovered with the use of geldanamycin, a specific inhibitor of Hsp90, to inhibit poliovirus growth in tissue culture cells (33). In uninfected cells, Hsp70 delivers proteins to Hsp90 in a partially folded state (148, 149). It was proposed that Hsp90 aids in the proper folding of the PV P1 capsid precursor, maintains it in a processing-competent conformation, and protects it from proteasomal degradation (33). A recent report suggests that the role of Hsp90 in FMDV morphogenesis is to stimulate the formation of pentamers from protomers rather than to promote P1 processing (150).
Myristic Acid
Myristic acid, also called tetradecanoic acid, is a saturated fatty acid cotranslationally linked to the N terminus of the PV P1 precursor (30–32). The myristic acid moiety functions in particle assembly, possibly both early and late in the process (151, 152). Two possible functions of myristoylation in the early particle assembly process have been proposed. First, myristoylated VP0 targets P1 to the membranous replication complex (42). Second, the myristic moiety facilitates protomer-protomer interactions for pentamer assembly (40, 41). Similar functions were proposed for myristoylation during pentamer formation of FMDV (36). VP0 myristoylation also appears to be important in late stages of PV assembly during virus maturation (40, 151, 152).
Glutathione
Glutathione (GSH) (γ-l-glutamyl-l-cysteinylglycine) is an important reducing agent in human and animal cells, which helps maintain the redox potential of cells and is important for immune function. It also has additional functions in processes such as signal transduction, apoptosis, gene expression, and regulation of protein function (153). The thiol group of GSH is responsible for its biological activity. GSH exists in cells either in a reduced form (GSH) or, to a lesser extent, in an oxidized state, glutathione disulfide (GSSG).
Glutathione synthesis in cells involves two consecutive steps. In the first step, glutamate and cysteine are linked to form a dipeptide by the enzyme glutamylcysteine synthase. In the second step, glutathione synthase catalyzes the addition of glycine to the dipeptide. l-Buthionine sulfoximine (BSO) is a potent inhibitor of glutamylcysteine synthase and of GSH biosynthesis (154). Depletion of GSH by BSO completely inhibits the growth of enteroviruses at the stage of virus morphogenesis (22, 24), but it has no effect on translation or genome replication. Moreover, PV or C-CAV20 variants resistant to depletion of GSH can readily be isolated, with the mutations mapping exclusively to viral capsid polypeptides (VP3 and VP1), an observation suggesting that GSH is needed for steps in morphogenesis downstream of RNA synthesis. We have recently shown that GSH depletion inhibits the accumulation of pentamers in PV-infected cells and that GSH directly interacts in vitro with capsid precursors and mature virus (22). Using a different inhibitor, TP219, Thibaut and colleagues also made a similar observation that GSH is required for the morphogenesis of coxsackie B virus (23). Indeed, GSH pulldown assays have shown that the compound interacts directly with 14S, 75S, provirions, and mature virus particles (22). In separate experiments, we have found that GSH protects mature virus from heat inactivation in vitro (22). We propose that the function of GSH in enterovirus morphogenesis is to stabilize the pentamers and the mature virus during virus assembly.
Cellular Membranes
The rearrangement of cellular membranes into specific structures associated with RNA replication/encapsidation is a characteristic of the plus-strand RNA virus life cycle in eukaryotic cells. Several cellular pathways, such as the secretory pathway, autophagy, and lipid biosynthesis, are involved in the formation of replication organelles (155–158). The PV-induced structures appear as vesicles of different sizes (50 nm to 400 nm in diameter) that are associated with the endoplasmic reticulum (ER) and later are clustered in the perinuclear region (159, 160). RNA replication of enteroviruses occurs in association with tubular structures, where the replication proteins are localized on external membranous surfaces facing the cytoplasm (158, 159). Early in infection, the tubular structures have a single membrane, but a very large fraction of them are later converted into double-membrane vesicles (155, 158, 161).
Relatively little is known about the requirement for membranes in morphogenesis. However, it was shown that membrane-associated PV replication complexes synthesizing viral RNA contain 5S protomers and 14S pentamers (162). These subviral particles and each of the capsid proteins (VP0, VP1, and VP3) can be UV cross-linked to progeny viral RNA. It was proposed that 14S pentamers associate with the viral RNA as the first step in encapsidation (51). A recent study suggested that blocking of autophagosome formation reduced PV RNA replication, while vesicle acidification was required only for the final maturation cleavage of the virus particles (163, 164). Specifically, the ratio of uncleaved VP0 to VP3 in 150S sucrose gradient peaks of PV-infected cell lysates was found to be increased about 2-fold in cells treated with vesicle acidification inhibitors such as ammonium chloride or bafilomycin A1. So far, the available evidence suggests that autophagosomes are supportive of but not required for PV replication and maturation.
INHIBITORS OF MORPHOGENESIS
Hydantoin
The small organic compound hydantoin [5(3,4-dichlorophenyl)methylhydantoin] strongly inhibits enterovirus morphogenesis in tissue culture by aborting maturation at an 110S intermediate (132). The effect of hydantoin is reversible since the 110S particles can be chased into mature virions after removal of the drug. PVs resistant to hydantoin were readily isolated, and the mutations were found to map to the 2CATPase protein. In subsequent studies with hydantoin in a cell-free translation/RNA replication system (76), inhibition of protein processing was observed, but no 110S particles were found (47).
Guanidine Hydrochloride
GnHCl reversibly inhibits viral RNA synthesis and encapsidation when used at concentrations where cellular macromolecular synthesis is not affected. When GnHCl is added to cells halfway through PV infection, the newly made RNA is trapped in 80S particles, called the guanidons, which accumulate (57). After the removal of the drug, the amount of 80S particles decreases, along with increases in the amounts of provirions and virions. Subsequent studies showed that GnHCl inhibits the ATPase activity of purified 2CATPase (126). Whether or not the ATPase activity of the 2CATPase protein is directly involved in encapsidation is not yet clear.
l-Buthionine Sulfoximine
As noted above (see “Glutathione”), BSO is a drug that inhibits glutamylcysteine synthase during the biosynthesis of the cellular reducing agent glutathione (24, 154). GSH depletion by BSO treatment has no effect on protein translation or RNA replication of enteroviruses. In particular, BSO inhibits the accumulation of 14S particles and finally affects the production of mature virus in infected cells. Our recent studies with GSH pulldown assays have shown that GSH interacts directly with 14S, 75S, and mature virus particles. Drug-resistant mutants, which contained mutations in VP1 and VP3 at protomer-protomer interfaces, evolved during passaging, suggesting that the role of GSH during enterovirus morphogenesis is to stabilize the pentamers and mature virus particles both during and after the viral assembly.
Py-11
The nucleoside analogue Py-11 (2-amino-4,6-dichloropyrimidine) is a potent inhibitor of PV morphogenesis at the stage of cleavage of the capsid precursor P1, thus preventing the assembly of capsid proteins into pentamers (165). The effect of the drug can be antagonized by the joint addition of glutamine and cysteine during infection. The target of the drug has not yet been identified.
Geldanamycin
Geldanamycin is a benzoquinone ansamycin antibiotic, originally discovered in Streptomyces hygroscopicus, which binds to the ATP/ADP-binding pocket of Hsp90 and thereby inhibits its function (166, 167). Hsp90, a cellular chaperone, plays an essential role in the folding and function of protein kinases, steroid hormone receptors, and numerous cellular proteins involved in controlling the cell cycle and apoptosis. In 2007, Geller and coworkers discovered that geldanamycin inhibited the replication of three different picornaviruses in tissue culture, PV, HRV, and coxsackievirus (33). Those authors showed that the effect of geldanamycin is due to an inhibition of Hsp90-aided folding of its client, the viral capsid precursor P1. This folding, in turn, is essential for the processing of P1 by the proteinase 3CDpro (see “Polyprotein Processing, Myristoylation, and Interaction of P1 with Heat Shock Protein Hsp90,” above). Interestingly, attempts to isolate PV mutants resistant to geldanamycin inhibition failed.
COMPARISON WITH OTHER HUMAN AND ANIMAL PLUS-STRAND RNA VIRUSES
Besides picornaviruses, morphogenesis has been studied in any detail only with member viruses of three other families of enveloped human and animal plus-strand RNA viruses: Flaviviridae, Togaviridae, and Coronaviridae. These families encompass enveloped viruses with a nucleocapsid consisting of multiple copies of capsid protein complexed with the viral RNA. We summarize what is known about some important aspects of the morphogenesis of these viruses and compare them to those of picornaviruses in Table 1. Members of all four families exhibit a linkage of encapsidation to RNA replication, and they all use membranous structures for the site of morphogenesis. The majority of Picornaviridae and Flaviviridae do not possess any RNA-based encapsidation signal, or at least, none has been discovered so far. The exceptions are Aichi virus of the Picornaviridae and dengue virus (DENV) of the Flaviviridae, which have been grouped separately from the other members of the families in Table 1. On the other hand, for Togaviridae and Coronaviridae, there is strong evidence for RNA-based encapsidation signals for all members of the family examined so far. Stable capsid intermediates (except for empty capsids) have been observed only for the Picornaviridae. Picornaviridae and Flaviviridae are similar in that there are interactions between capsid and nonstructural proteins that are important for morphogenesis (20, 168). Based on genetic and biochemical experiments, we have recently proposed for PV and other enteroviruses that protein-protein interactions are sufficient to provide specificity to the encapsidation process (20). We suggest that the same mechanism might also apply to HCV and other viruses that do not possess specific RNA encapsidation signals.
TABLE 1.
Comparison of morphogenesis of Picornaviridae to those of other plus-strand RNA virus families
| Morphogenesis trait | Presence of trait in virus family (reference[s])a |
|||||
|---|---|---|---|---|---|---|
|
Picornaviridae |
Flaviviridae |
Togaviridae (SINV, SFV, rubella virus, Mayaro virus, VEEV) | Coronaviridae (MHV, SARS-CoV) | |||
| PV, CAV, CBV, FMDV | Aichi virus | HCV, YFV, WNV | DENV | |||
| RNA replication and encapsidation linked | + | + (25) | + (168, 175–181) | + (182) | + (183) | NA |
| Assembly associated with membrane | + (51, 162) | NA | + (184–187) | +(182) | + | + (188–190) |
| Interaction between capsid and NS protein required | + (20, 21) | NA | + (191–199) | NA | + (200) | NA |
| RNA encapsidation signal | − | + (25, 26) | − | + (201) | + (202, 203) | + (204–206) |
| Stable capsid intermediates | + (63) | NA | − | − | − | − |
| Involvement of host factors | + (22–24, 30, 32, 33) | NA | + (207–215) | NA | + (216–218) | NA |
PV, poliovirus; CAV, coxsackie A virus; CBV, coxsackie B virus; FMDV, foot-and-mouth disease virus; HCV, hepatitis C virus; YFV, yellow fever virus; WNV, West Nile virus; DENV, dengue virus; SINV, Sindbis virus; SFV, Semliki Forest virus; VEEV, Venezuelan equine encephalitis virus; MHV, mouse hepatitis virus; SARS-CoV, severe acute respiratory syndrome coronavirus; NA, not available.
MODEL OF ENTEROVIRUS MORPHOGENESIS
Any model of enterovirus morphogenesis has to take into account that only newly replicated RNAs are encapsidated, which indicates a tight linkage between RNA replication and encapsidation (76, 92). In Fig. 5, we propose a model of morphogenesis based on our studies of enteroviruses. Except for Aichi virus or HAV, it is possible that this model may be applicable to most picornaviruses, but the Picornaviridae are such a large family (169) that there may be different pathways of morphogenesis for the proliferation of member viruses of some genera.
FIG 5.
Model for enterovirus morphogenesis. After the newly made capsid precursor P1 is released from the polyprotein, it interacts with the chaperone Hsp90 to assume a conformation competent for cleavage by 3CDpro. After proteolytic processing, a protomer is formed spontaneously, consisting of one copy each of VP0, VP1, and VP3. In the presence of GSH, five protomers assemble to generate a pentamer, which, by interactions between VP3 and 2CATPase or between VP1 and 2CATPase, is recruited to the replication complex to associate with the newly made VPg-linked plus-strand viral RNA. Subsequently, 12 pentamers condense around the RNA to produce a noninfectious provirion. The last step is the maturation cleavage of VP0, which is autocatalytic and RNA dependent and yields a stable particle containing 60 copies of VP1 and VP3, 59 copies of VP2 and VP4, and 1 copy of VP0. The steps at which GSH is required, as shown by inhibition with BSO, are marked.
As detailed in Fig. 3C and 5, we propose that the newly made capsid precursor P1 that is released from the polyprotein and myristoylated (Fig. 3C, PV stage I) interacts with the chaperone Hsp90 to achieve a conformation (PV stage II) competent for cleavage by 3CDpro (33, 34). After proteolytic processing, assembly of the cleavage products occurs in a stepwise manner. Notably, processing of P1 does not result in the release of “free” polypeptides. Instead, a “protomer” is formed directly from P1, consisting of one copy each of VP0, VP1, and VP3 (PV stage III). Five protomers then assemble to generate a pentamer (PV stage IV). Based on the interaction between 2CATPase and the capsid protein VP3 or VP1 (20, 21), we suggest that 2CATPase, a component of the membrane-associated replication complex, recruits the 14S capsid precursors to the site of particle assembly. Whether the 5′-terminal VPg that is likely to emerge first from the replication complex interacts with capsid proteins is not yet known. However, at this stage, 12 pentamers are likely to condense around the RNA to produce a noninfectious provirion (PV stage VI). Whether or not 2CATPase is also involved in the condensation of the viral RNA (via ATP hydrolysis) during packaging is not yet known. The last step is the maturation cleavage of VP0, which is autocatalytic and RNA dependent (67, 70, 170) and yields a stable particle containing VP1, VP2, VP3, and VP4 (PV stage VII). The role of the procapsids (PV stage V) in morphogenesis is still debated. Most likely, it is a by-product capable of reversible dissociation into pentamers. However, our evidence suggests that procapsids are not direct precursors of mature poliovirus (22). We propose that GSH is required for the stability of pentamers and mature virions during the early and late stages of assembly (22, 23).
In wt enterovirus infections, only plus-strand RNAs that emerge from the replication complex are encapsidated. This is most likely due to the fact that minus strands are normally sequestered from the replication complex through associations with plus stands in either the replicative form (RF) or replicative intermediate (RI) structures and not released. As pointed out above, the replication complex of special variants of coxsackie B virus can produce free minus-stranded RNAs, and in turn, those can be encapsidated (119). As noted above, the last steps of morphogenesis of HAV differ from those of enteroviruses (Fig. 3C, HAV steps I to VII).
CONCLUSIONS
Since the discovery of the basic steps of assembly more than 3 decades ago, progress in the field of picornavirus encapsidation has been slow. This may be the reason why the last full review on picornavirus morphogenesis was published in 1981 (49). Here, we have attempted to summarize older data collected decades ago, and we have detailed new discoveries made during last few years (20–23, 26, 28, 141). Comparison with other human and animal plus-strand RNA viruses reveals certain differences but also some unifying principles that link aspects of morphogenesis.
Among the many unsolved mysteries in picornavirus morphogenesis, questions that remain to be answered are as follows. (i) What is the precise mechanism by which provirions are formed—do pentamers condense around genomic RNA, or are genomes actively “inserted” into the empty capsids? (ii) What is the role of 2CATPase in the delivery of newly synthesized RNA from the replication complex to the capsid? (iii) What is the role of 3CD in enhancing virus maturation? (iv) What is the role of VPg in viral assembly? (v) What are the roles of host factors in encapsidation? Future studies using a combination of genetics and high-power microscopic imaging will hopefully extend and refine our knowledge of the mechanism of picornavirus morphogenesis and enhance our ability for drug design to limit the toll of picornavirus diseases.
ACKNOWLEDGMENT
This work was supported by NIH grant AI 15122.
REFERENCES
- 1.Knowles E, Delwart E, Gorbalenya AE, Hovi T, Hyypia T, King AMQ, LinBerg AM, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. 2014. Picornaviridae: 26 genera, 46 species and growing, p 98 Abstr. Europic 18th Int. Picornavirus Meet., Blankenberge, Belgium [Google Scholar]
- 2.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Rueckert RR, Sherry B, Vriend G. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. 10.1038/317145a0 [DOI] [PubMed] [Google Scholar]
- 3.Hogle JM, Chow M, Filman DJ. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358–1365. 10.1126/science.2994218 [DOI] [PubMed] [Google Scholar]
- 4.Stuart DWX, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands D, Yin W, Wang J, Rao Y, Fry E. 2014. Structural analysis of Hepatitis A virus, p P30 Abstr. Europic 18th Int. Picornavirus Meet., Blankenberge, Belgium [Google Scholar]
- 5.Fry EE, Stuart DI. 2010. Virion structure, p 59–72 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 6.Li Q, Yafal AG, Lee YM, Hogle J, Chow M. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 68:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wimmer E, Hellen CU, Cao X. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353–436. 10.1146/annurev.ge.27.120193.002033 [DOI] [PubMed] [Google Scholar]
- 8.Palmenberg A, Neubauer D, Skern T. 2010. Genome organization and encoded proteins, p 3–18 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 9.Bergelson JM. 2010. Receptors, p 73–86 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 10.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Cell entry: a biochemical and structural perspective, p 87–104 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 11.Martinez-Salaz E, Ryan MD. 2010. Translation and protein processing, p 141–161 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 12.Detjen BM, Lucas J, Wimmer E. 1978. Poliovirus single-stranded RNA and double-stranded RNA: differential infectivity in enucleate cells. J. Virol. 27:582–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Kuppeveld F, Belov G, Ehrenfeld E. 2010. Remodeling cellular membranes, p 181–193 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 14.Kirkegaard K, Semler BL. 2010. Genome replication II: the process, p 127–140 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 15.Rozovics JM, Semler BL. 2010. Genome replication I: the players, p 107–126 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 16.Paul AV, van Boom JH, Filippov D, Wimmer E. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280–284. 10.1038/30529 [DOI] [PubMed] [Google Scholar]
- 17.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. 10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards AL, Jackson WT. 2013. How positive-strand RNA viruses benefit from autophagosome maturation. J. Virol. 87:9966–9972. 10.1128/JVI.00460-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Z, Lemon SM. 2010. Hepatitis A virus, p 383–396 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 20.Liu Y, Wang C, Mueller S, Paul AV, Wimmer E, Jiang P. 2010. Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 6:e1001066. 10.1371/journal.ppat.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Jiang P, Sand C, Paul AV, Wimmer E. 2012. Alanine scanning of poliovirus 2CATPase reveals new genetic evidence that capsid protein/2CATPase interactions are essential for morphogenesis. J. Virol. 86:9964–9975. 10.1128/JVI.00914-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma HC, Liu Y, Wang C, Strauss M, Rehage N, Chen YH, Altan-Bonnet N, Hogle J, Wimmer E, Mueller S, Paul AV, Jiang P. 2014. An interaction between glutathione and the capsid is required for the morphogenesis of C-cluster enteroviruses. PLoS Pathog. 10:e1004052. 10.1371/journal.ppat.1004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thibaut HJ, van der Linden L, Jiang P, Thys B, Canela MD, Aguado L, Rombaut B, Wimmer E, Paul A, Perez-Perez MJ, van Kuppeveld FJ, Neyts J. 2014. Binding of glutathione to enterovirus capsids is essential for virion morphogenesis. PLoS Pathog. 10:e1004039. 10.1371/journal.ppat.1004039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AD, Dawson H. 2006. Glutathione is required for efficient production of infectious picornavirus virions. Virology 353:258–267. 10.1016/j.virol.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 25.Sasaki J, Taniguchi K. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542–3548. 10.1128/JVI.77.6.3542-3548.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki J, Nagashima S, Taniguchi K. 2003. Aichi virus leader protein is involved in viral RNA replication and encapsidation. J. Virol. 77:10799–10807. 10.1128/JVI.77.20.10799-10807.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morace G, Kusov Y, Dzagurov G, Beneduce F, Gauss-Muller V. 2008. The unique role of domain 2A of the hepatitis A virus precursor polypeptide P1-2A in viral morphogenesis. BMB Rep. 41:678–683. 10.5483/BMBRep.2008.41.9.678 [DOI] [PubMed] [Google Scholar]
- 28.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. 10.1038/nature12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyoda H, Nicklin MJ, Murray MG, Anderson CW, Dunn JJ, Studier FW, Wimmer E. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761–770. 10.1016/0092-8674(86)90790-7 [DOI] [PubMed] [Google Scholar]
- 30.Paul AV, Schultz A, Pincus SE, Oroszlan S, Wimmer E. 1987. Capsid protein VP4 of poliovirus is N-myristoylated. Proc. Natl. Acad. Sci. U. S. A. 84:7827–7831. 10.1073/pnas.84.22.7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow M, Moscufo N. 1995. Myristoyl modification of viral proteins: assays to assess functional roles. Methods Enzymol. 250:495–509. 10.1016/0076-6879(95)50093-6 [DOI] [PubMed] [Google Scholar]
- 32.Chow M, Newman JF, Filman D, Hogle JM, Rowlands DJ, Brown F. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482–486. 10.1038/327482a0 [DOI] [PubMed] [Google Scholar]
- 33.Geller R, Vignuzzi M, Andino R, Frydman J. 2007. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21:195–205. 10.1101/gad.1505307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macejak DG, Sarnow P. 1992. Association of heat shock protein 70 with enterovirus capsid precursor P1 in infected human cells. J. Virol. 66:1520–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ypma-Wong MF, Semler BL. 1987. Processing determinants required for in vitro cleavage of the poliovirus P1 precursor to capsid proteins. J. Virol. 61:3181–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin S, Tuthill TJ, Arias A, Killington RA, Rowlands DJ. 2009. Foot-and-mouth disease virus assembly: processing of recombinant capsid precursor by exogenous protease induces self-assembly of pentamers in vitro in a myristoylation-dependent manner. J. Virol. 83:11275–11282. 10.1128/JVI.01263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tesar M, Jia XY, Summers DF, Ehrenfeld E. 1993. Analysis of a potential myristoylation site in hepatitis A virus capsid protein VP4. Virology 194:616–626. 10.1006/viro.1993.1301 [DOI] [PubMed] [Google Scholar]
- 38.Cohen L, Benichou D, Martin A. 2002. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J. Virol. 76:7495–7505. 10.1128/JVI.76.15.7495-7505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aragones L, Guix S, Ribes E, Bosch A, Pinto RM. 2010. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 6:e1000797. 10.1371/journal.ppat.1000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ansardi DC, Porter DC, Morrow CD. 1992. Myristylation of poliovirus capsid precursor P1 is required for assembly of subviral particles. J. Virol. 66:4556–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YM, Chow M. 1992. Myristate modification does not function as a membrane association signal during poliovirus capsid assembly. Virology 187:814–820. 10.1016/0042-6822(92)90485-8 [DOI] [PubMed] [Google Scholar]
- 42.Martin-Belmonte F, Lopez-Guerrero JA, Carrasco L, Alonso MA. 2000. The amino-terminal nine amino acid sequence of poliovirus capsid VP4 protein is sufficient to confer N-myristoylation and targeting to detergent-insoluble membranes. Biochemistry 39:1083–1090. 10.1021/bi992132e [DOI] [PubMed] [Google Scholar]
- 43.Macadam AJ, Ferguson G, Arnold C, Minor PD. 1991. An assembly defect as a result of an attenuating mutation in the capsid proteins of the poliovirus type 3 vaccine strain. J. Virol. 65:5225–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borovec SV, Anderson DA. 1993. Synthesis and assembly of hepatitis A virus-specific proteins in BS-C-1 cells. J. Virol. 67:3095–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Probst C, Jecht M, Gauss-Muller V. 1999. Intrinsic signals for the assembly of hepatitis A virus particles. Role of structural proteins VP4 and 2A. J. Biol. Chem. 274:4527–4531 [DOI] [PubMed] [Google Scholar]
- 46.Nugent CI, Kirkegaard K. 1995. RNA binding properties of poliovirus subviral particles. J. Virol. 69:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verlinden Y, Cuconati A, Wimmer E, Rombaut B. 2000. Cell-free synthesis of poliovirus: 14S subunits are the key intermediates in the encapsidation of poliovirus RNA. J. Gen. Virol. 81:2751–2754. 10.1099/vir.0.17247-0 [DOI] [PubMed] [Google Scholar]
- 48.Putnak JR, Phillips BA. 1981. Differences between poliovirus empty capsids formed in vivo and those formed in vitro: a role for the morphopoietic factor. J. Virol. 40:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putnak JR, Phillips BA. 1981. Picornaviral structure and assembly. Microbiol. Rev. 45:287–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rombaut B, Vrijsen R, Boeye A. 1990. New evidence for the precursor role of 14 S subunits in poliovirus morphogenesis. Virology 177:411–414. 10.1016/0042-6822(90)90502-I [DOI] [PubMed] [Google Scholar]
- 51.Pfister T, Egger D, Bienz K. 1995. Poliovirus subviral particles associated with progeny RNA in the replication complex. J. Gen. Virol. 76(Part 1):63–71 [DOI] [PubMed] [Google Scholar]
- 52.Hummeler K, Anderson TF, Brown RA. 1962. Identification of poliovirus particles of different antigenicity by specific agglutination as seen in the electron microscope. Virology 16:84–90. 10.1016/0042-6822(62)90205-2 [DOI] [PubMed] [Google Scholar]
- 53.Scharff MD, Levintow L. 1963. Quantitative study of the formation of poliovirus antigens in infected HeLa cells. Virology 19:491–500. 10.1016/0042-6822(63)90043-6 [DOI] [PubMed] [Google Scholar]
- 54.Marongiu ME, Pani A, Corrias MV, Sau M, La Colla P. 1981. Poliovirus morphogenesis. I. Identification of 80S dissociable particles and evidence for the artifactual production of procapsids. J. Virol. 39:341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rombaut B, Vrijsen R, Brioen P, Boeye A. 1982. A pH-dependent antigenic conversion of empty capsids of poliovirus studied with the aid of monoclonal antibodies to N and H antigen. Virology 122:215–218. 10.1016/0042-6822(82)90393-2 [DOI] [PubMed] [Google Scholar]
- 56.Putnak JR, Phillips BA. 1982. Poliovirus empty capsid morphogenesis: evidence for conformational differences between self- and extract-assembled empty capsids. J. Virol. 41:792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baltimore D. 1969. The replication of picornaviruses, p 103–176 In Levy HB. (ed), The biochemistry of viruses. Marcel Dekker, New York, NY [Google Scholar]
- 58.Fernandez-Tomas CB, Baltimore D. 1973. Morphogenesis of poliovirus. II. Demonstration of a new intermediate, the proviron. J. Virol. 12:1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Tomas CB, Guttman N, Baltimore D. 1973. Morphogenesis of poliovirus. III. Formation of provirion in cell-free extracts. J. Virol. 12:1181–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guttman N, Baltimore D. 1977. Morphogenesis of poliovirus. IV. Existence of particles sedimenting at 150S and having the properties of provirion. J. Virol. 23:363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koch F, Koch G. 1985. The molecular biology of poliovirus, p 463 Springer-Verlag, New York, NY [Google Scholar]
- 62.Anderson DA, Ross BC. 1990. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J. Virol. 64:5284–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossmann MG. 2002. Picornavirus structure overview, p 27–68 In Semler BL, Wimmer E. (ed), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 64.Koch F, Koch G. 1985. The molecular biology of poliovirus, p 94 Springer-Verlag, New York, NY [Google Scholar]
- 65.Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Miyamura T, Yamazaki S. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyypia T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. U. S. A. 89:8847–8851. 10.1073/pnas.89.18.8847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basavappa R, Syed R, Flore O, Icenogle JP, Filman DJ, Hogle JM. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651–1669. 10.1002/pro.5560031005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harber JJ, Bradley J, Anderson CW, Wimmer E. 1991. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J. Virol. 65:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold E, Luo M, Vriend G, Rossmann MG, Palmenberg AC, Parks GD, Nicklin MJ, Wimmer E. 1987. Implications of the picornavirus capsid structure for polyprotein processing. Proc. Natl. Acad. Sci. U. S. A. 84:21–25. 10.1073/pnas.84.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hindiyeh M, Li QH, Basavappa R, Hogle JM, Chow M. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73:9072–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Couderc T, Delpeyroux F, Le Blay H, Blondel B. 1996. Mouse adaptation determinants of poliovirus type 1 enhance viral uncoating. J. Virol. 70:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirkegaard K. 1990. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J. Virol. 64:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Compton SR, Nelsen B, Kirkegaard K. 1990. Temperature-sensitive poliovirus mutant fails to cleave VP0 and accumulates provirions. J. Virol. 64:4067–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. 2005. Stimulation of poliovirus synthesis in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. J. Virol. 79:6358–6367. 10.1128/JVI.79.10.6358-6367.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. 2005. Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. Virol. J. 2:86. 10.1186/1743-422X-2-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molla A, Paul AV, Wimmer E. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647–1651. 10.1126/science.1661029 [DOI] [PubMed] [Google Scholar]
- 77.Molla A, Paul AV, Wimmer E. 1993. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 67:5932–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rossmann MG, He Y, Kuhn RJ. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324–331. 10.1016/S0966-842X(02)02383-1 [DOI] [PubMed] [Google Scholar]
- 79.Bergelson JM, Chan M, Solomon KR, St John NF, Lin H, Finberg RW. 1994. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. U. S. A. 91:6245–6248. 10.1073/pnas.91.13.6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bergelson JM, Mohanty JG, Crowell RL, St John NF, Lublin DM, Finberg RW. 1995. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69:1903–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. U. S. A. 91:1839–1842. 10.1073/pnas.91.5.1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo M, Vriend G, Kamer G, Minor I, Arnold E, Rossmann MG, Boege U, Scraba DG, Duke GM, Palmenberg AC. 1987. The atomic structure of Mengo virus at 3.0 A resolution. Science 235:182–191. 10.1126/science.3026048 [DOI] [PubMed] [Google Scholar]
- 83.Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286–1293. 10.1126/science.3018924 [DOI] [PubMed] [Google Scholar]
- 84.Rossmann MG. 1989. The structure of antiviral agents that inhibit uncoating when complexed with viral capsids. Antiviral Res. 11:3–13. 10.1016/0166-3542(89)90016-8 [DOI] [PubMed] [Google Scholar]
- 85.Pevear DC, Tull TM, Seipel ME, Groarke JM. 1999. Activity of pleconaril against enteroviruses. Antimicrob. Agents Chemother. 43:2109–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diana GD, Pevear DC, Otto MJ, McKinlay MA, Rossmann MG, Smith T, Badger J. 1989. Inhibitors of viral uncoating. Pharmacol. Ther. 42:289–305. 10.1016/0163-7258(89)90028-4 [DOI] [PubMed] [Google Scholar]
- 87.Kim KH, Willingmann P, Gong ZX, Kremer MJ, Chapman MS, Minor I, Oliveira MA, Rossmann MG, Andries K, Diana GD, Dutko FJ, McKinlay MA, Pevear DC. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206–227. 10.1006/jmbi.1993.1137 [DOI] [PubMed] [Google Scholar]
- 88.Salk JE. 1951. Committee on Typing of the National Foundation of Infantile Paralysis. Am. J. Hyg. 54:191–27414877799 [Google Scholar]
- 89.McBride MS, Schwartz MD, Panganiban AT. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartenschlager R, Schaller H. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frolova E, Frolov I, Schlesinger S. 1997. Packaging signals in alphaviruses. J. Virol. 71:248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nugent CI, Johnson KL, Sarnow P, Kirkegaard K. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Semler BL, Johnson VH, Tracy S. 1986. A chimeric plasmid from cDNA clones of poliovirus and coxsackievirus produces a recombinant virus that is temperature-sensitive. Proc. Natl. Acad. Sci. U. S. A. 83:1777–1781. 10.1073/pnas.83.6.1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson VH, Semler BL. 1988. Defined recombinants of poliovirus and coxsackievirus: sequence-specific deletions and functional substitutions in the 5′-noncoding regions of viral RNAs. Virology 162:47–57. 10.1016/0042-6822(88)90393-5 [DOI] [PubMed] [Google Scholar]
- 95.Wimmer E, Paul AV. 2010. The making of a picornavirus, p 33–55 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 96.Rieder E, Xiang W, Paul A, Wimmer E. 2003. Analysis of the cloverleaf element in a human rhinovirus type 14/poliovirus chimera: correlation of subdomain D structure, ternary protein complex formation and virus replication. J. Gen. Virol. 84:2203–2216. 10.1099/vir.0.19013-0 [DOI] [PubMed] [Google Scholar]
- 97.Cole CN, Smoler D, Wimmer E, Baltimore D. 1971. Defective interfering particles of poliovirus. I. Isolation and physical properties. J. Virol. 7:478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nomoto A, Jacobson A, Lee YF, Dunn J, Wimmer E. 1979. Defective interfering particles of poliovirus: mapping of the deletion and evidence that the deletions in the genomes of DI(1), (2) and (3) are located in the same region. J. Mol. Biol. 128:179–196. 10.1016/0022-2836(79)90125-6 [DOI] [PubMed] [Google Scholar]
- 99.Kuge S, Saito I, Nomoto A. 1986. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J. Mol. Biol. 192:473–487. 10.1016/0022-2836(86)90270-6 [DOI] [PubMed] [Google Scholar]
- 100.Novak JE, Kirkegaard K. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726–1737. 10.1101/gad.8.14.1726 [DOI] [PubMed] [Google Scholar]
- 101.Kajigaya S, Arakawa H, Kuge S, Koi T, Imura N, Nomoto A. 1985. Isolation and characterization of defective-interfering particles of poliovirus Sabin 1 strain. Virology 142:307–316. 10.1016/0042-6822(85)90339-3 [DOI] [PubMed] [Google Scholar]
- 102.Porter DC, Ansardi DC, Wang J, McPherson S, Moldoveanu Z, Morrow CD. 1998. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology 243:1–11. 10.1006/viro.1998.9046 [DOI] [PubMed] [Google Scholar]
- 103.Barclay W, Li Q, Hutchinson G, Moon D, Richardson A, Percy N, Almond JW, Evans DJ. 1998. Encapsidation studies of poliovirus subgenomic replicons. J. Gen. Virol. 79(Part 7):1725–1734 [DOI] [PubMed] [Google Scholar]
- 104.Porter DC, Ansardi DC, Morrow CD. 1995. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J. Virol. 69:1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Percy N, Barclay WS, Sullivan M, Almond JW. 1992. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J. Virol. 66:5040–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, Evans DJ. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590–4600. 10.1128/JVI.74.10.4590-4600.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paul AV, Yin J, Mugavero J, Rieder E, Liu Y, Wimmer E. 2003. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J. Biol. Chem. 278:43951–43960. 10.1074/jbc.M307441200 [DOI] [PubMed] [Google Scholar]
- 108.Paul AV. 2002. Possible unifying mechanism of picornavirus genome replication, p 227–246 In Semler BL, Wimmer E. (ed), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 109.Witwer C, Rauscher S, Hofacker IL, Stadler PF. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079–5089. 10.1093/nar/29.24.5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song Y, Liu Y, Ward CB, Mueller S, Futcher B, Skiena S, Paul AV, Wimmer E. 2012. Identification of two functionally redundant RNA elements in the coding sequence of poliovirus using computer-generated design. Proc. Natl. Acad. Sci. U. S. A. 109:14301–14307. 10.1073/pnas.1211484109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. 2006. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 80:9687–9696. 10.1128/JVI.00738-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. 10.1126/science.1155761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371–10380. 10.1128/JVI.74.22.10371-10380.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pilipenko EV, Poperechny KV, Maslova SV, Melchers WJ, Slot HJ, Agol VI. 1996. cis-element, oriR, involved in the initiation of (-) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing') interactions. EMBO J. 15:5428–5436 [PMC free article] [PubMed] [Google Scholar]
- 115.Brown DM, Cornell CT, Tran GP, Nguyen JH, Semler BL. 2005. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J. Virol. 79:11962–11973. 10.1128/JVI.79.18.11962-11973.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Ooij MJ, Polacek C, Glaudemans DH, Kuijpers J, van Kuppeveld FJ, Andino R, Agol VI, Melchers WJ. 2006. Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element. Nucleic Acids Res. 34:2953–2965. 10.1093/nar/gkl349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rohll JB, Moon DH, Evans DJ, Almond JW. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Todd S, Towner JS, Brown DM, Semler BL. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868–8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim KS, Tracy S, Tapprich W, Bailey J, Lee CK, Kim K, Barry WH, Chapman NM. 2005. 5′-terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 79:7024–7041. 10.1128/JVI.79.11.7024-7041.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodriguez PL, Carrasco L. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105–10112. 10.1074/jbc.270.17.10105 [DOI] [PubMed] [Google Scholar]
- 121.Paul AV, Molla A, Wimmer E. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199:188–199. 10.1006/viro.1994.1111 [DOI] [PubMed] [Google Scholar]
- 122.Li JP, Baltimore D. 1988. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J. Virol. 62:4016–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pfister T, Jones KW, Wimmer E. 2000. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334–343. 10.1128/JVI.74.1.334-343.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pincus SE, Diamond DC, Emini EA, Wimmer E. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mirzayan C, Wimmer E. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547–555. 10.1016/0042-6822(92)90578-D [DOI] [PubMed] [Google Scholar]
- 126.Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001. 10.1074/jbc.274.11.6992 [DOI] [PubMed] [Google Scholar]
- 127.Li JP, Baltimore D. 1990. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 64:1102–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Teterina NL, Bienz K, Egger D, Gorbalenya AE, Ehrenfeld E. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66–77. 10.1006/viro.1997.8775 [DOI] [PubMed] [Google Scholar]
- 129.Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129–145. 10.1006/viro.1994.1329 [DOI] [PubMed] [Google Scholar]
- 131.Aldabe R, Barco A, Carrasco L. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134–23137. 10.1074/jbc.271.38.23134 [DOI] [PubMed] [Google Scholar]
- 132.Vance LM, Moscufo N, Chow M, Heinz BA. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yin J, Liu Y, Wimmer E, Paul AV. 2007. Complete protein linkage map between the P2 and P3 non-structural proteins of poliovirus. J. Gen. Virol. 88:2259–2267. 10.1099/vir.0.82795-0 [DOI] [PubMed] [Google Scholar]
- 134.Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E. 2011. Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J. Virol. 85:4284–4296. 10.1128/JVI.02398-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Banerjee R, Weidman MK, Echeverri A, Kundu P, Dasgupta A. 2004. Regulation of poliovirus 3C protease by the 2C polypeptide. J. Virol. 78:9243–9256. 10.1128/JVI.78.17.9243-9256.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tang WF, Yang SY, Wu BW, Jheng JR, Chen YL, Shih CH, Lin KH, Lai HC, Tang P, Horng JT. 2007. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 282:5888–5898. 10.1074/jbc.M611145200 [DOI] [PubMed] [Google Scholar]
- 137.Arita M. 2014. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol. Immunol. 58:239–256. 10.1111/1348-0421.12144 [DOI] [PubMed] [Google Scholar]
- 138.Pincus SE, Wimmer E. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pincus SE, Rohl H, Wimmer E. 1987. Guanidine-dependent mutants of poliovirus: identification of three classes with different growth requirements. Virology 157:83–88. 10.1016/0042-6822(87)90316-3 [DOI] [PubMed] [Google Scholar]
- 140.Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. 2005. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59:587–635. 10.1146/annurev.micro.58.030603.123625 [DOI] [PubMed] [Google Scholar]
- 141.Jiang P, Faase JA, Toyoda H, Paul A, Wimmer E, Gorbalenya AE. 2007. Evidence for emergence of diverse polioviruses from C-cluster coxsackie A viruses and implications for global poliovirus eradication. Proc. Natl. Acad. Sci. U. S. A. 104:9457–9462. 10.1073/pnas.0700451104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xiang W, Cuconati A, Paul AV, Cao X, Wimmer E. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892–904 [PMC free article] [PubMed] [Google Scholar]
- 143.Gamarnik AV, Andino R. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219–2226. 10.1128/JVI.74.5.2219-2226.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359–10370. 10.1128/JVI.74.22.10359-10370.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shen M, Reitman ZJ, Zhao Y, Moustafa I, Wang Q, Arnold JJ, Pathak HB, Cameron CE. 2008. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J. Biol. Chem. 283:875–888. 10.1074/jbc.M707907200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265–270. 10.1016/0042-6822(88)90172-9 [DOI] [PubMed] [Google Scholar]
- 147.Schultheiss T, Kusov YY, Gauss-Muller V. 1994. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology 198:275–281. 10.1006/viro.1994.1030 [DOI] [PubMed] [Google Scholar]
- 148.Riggs DL, Cox MB, Cheung-Flynn J, Prapapanich V, Carrigan PE, Smith DF. 2004. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 39:279–295. 10.1080/10409230490892513 [DOI] [PubMed] [Google Scholar]
- 149.Pratt WB, Toft DO. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228:111–133 [DOI] [PubMed] [Google Scholar]
- 150.Newman J, Jackson T, Curry S, Tuthill T. 2014. The cellular chaperone heat shock protein 90 is required for the assembly of FMDV cpasid pentamers, p 38 Abstr. Europic 18th Int. Picornavirus Meet., Blankenberge, Belgium [Google Scholar]
- 151.Marc D, Masson G, Girard M, van der Werf S. 1990. Lack of myristoylation of poliovirus capsid polypeptide VP0 prevents the formation of virions or results in the assembly of noninfectious virus particles. J. Virol. 64:4099–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Moscufo N, Simons J, Chow M. 1991. Myristoylation is important at multiple stages in poliovirus assembly. J. Virol. 65:2372–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pompella A, Visvikis A, Paolicchi A, De Tata V, Casini AF. 2003. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 66:1499–1503. 10.1016/S0006-2952(03)00504-5 [DOI] [PubMed] [Google Scholar]
- 154.Meister A, Griffith OW, Novogrodsky A, Tate SS. 1979. New aspects of glutathione metabolism and translocation in mammals. Ciba Found. Symp. 72:135–161 [DOI] [PubMed] [Google Scholar]
- 155.Belov GA, van Kuppeveld FJ. 2012. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr. Opin. Virol. 2:740–747. 10.1016/j.coviro.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kirkegaard K, Jackson WT. 2005. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy 1:182–184. 10.4161/auto.1.3.2065 [DOI] [PubMed] [Google Scholar]
- 158.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. 10.1371/journal.ppat.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bienz K, Egger D, Pfister T, Troxler M. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Egger D, Bienz K. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707–718. 10.1099/vir.0.80442-0 [DOI] [PubMed] [Google Scholar]
- 161.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Barcena M. 2011. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio 2(5):00166-11. 10.1128/mBio.00166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pfister T, Pasamontes L, Troxler M, Egger D, Bienz K. 1992. Immunocytochemical localization of capsid-related particles in subcellular fractions of poliovirus-infected cells. Virology 188:676–684. 10.1016/0042-6822(92)90522-Q [DOI] [PubMed] [Google Scholar]
- 163.Richards AL, Jackson WT. 2013. Behind closed membranes: the secret lives of picornaviruses? PLoS Pathog. 9:e1003262. 10.1371/journal.ppat.1003262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Richards AL, Jackson WT. 2012. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 8:e1003046. 10.1371/journal.ppat.1003046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.La Colla P, Corrias MV, Marongiu ME, Pani A. 1981. Dichloropyrimidines: inhibitors of viral morphogenesis, p 21–35 In Gauri KK. (ed), Antiviral chemotherapy: design of inhibitors of viral functions. Academic Press, New York, NY [Google Scholar]
- 166.He W, Wu L, Gao Q, Du Y, Wang Y. 2006. Identification of AHBA biosynthetic genes related to geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Curr. Microbiol. 52:197–203. 10.1007/s00284-005-0203-y [DOI] [PubMed] [Google Scholar]
- 167.Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D, Neckers LM. 1998. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3:100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6:e1001233. 10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Knowles NJ, Hovi T, King MQ, Stanway G. 2010. Overview of taxonomy, p 19–32 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 170.Curry S, Fry E, Blakemore W, Abu-Ghazaleh R, Jackson T, King A, Lea S, Newman J, Stuart D. 1997. Dissecting the roles of VP0 cleavage and RNA packaging in picornavirus capsid stabilization: the structure of empty capsids of foot-and-mouth disease virus. J. Virol. 71:9743–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rossmann MG, Johnson JE. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533–573. 10.1146/annurev.bi.58.070189.002533 [DOI] [PubMed] [Google Scholar]
- 172.Hogle JM, Rancaniello VR. 2002. Poliovirus receptors and cell entry, p 71–84 In Semler BL, Wimmer E. (ed), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 173.Hogle JM, Filman DJ. 1989. The antigenic structure of poliovirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 323:467–478. 10.1098/rstb.1989.0024 [DOI] [PubMed] [Google Scholar]
- 174.McKnight KL, Lemon SM. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569–1584. 10.1017/S1355838298981006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jirasko V, Montserret R, Appel N, Janvier A, Eustachi L, Brohm C, Steinmann E, Pietschmann T, Penin F, Bartenschlager R. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562. 10.1074/jbc.M803981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383. 10.1128/JVI.00690-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Moradpour D, Penin F. 2013. Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 369:113–142. 10.1007/978-3-642-27340-7_5 [DOI] [PubMed] [Google Scholar]
- 178.Phan T, Kohlway A, Dimberu P, Pyle AM, Lindenbach BD. 2011. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. J. Virol. 85:1193–1204. 10.1128/JVI.01889-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Phan T, Beran RK, Peters C, Lorenz IC, Lindenbach BD. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379–8395. 10.1128/JVI.00891-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639. 10.1128/JVI.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633–4640. 10.1128/JVI.75.10.4633-4640.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lee JY, Marshall JA, Bowden DS. 1992. Replication complexes associated with the morphogenesis of rubella virus. Arch. Virol. 122:95–106. 10.1007/BF01321120 [DOI] [PubMed] [Google Scholar]
- 184.Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. 2011. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology 141:1057–1066. 10.1053/j.gastro.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 185.Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. 2012. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 8:e1002466. 10.1371/journal.ppat.1002466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Op De Beeck A, Molenkamp R, Caron M, Ben Younes A, Bredenbeek P, Dubuisson J. 2003. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 77:813–820. 10.1128/JVI.77.2.813-820.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Op De Beeck A, Rouille Y, Caron M, Duvet S, Dubuisson J. 2004. The transmembrane domains of the prM and E proteins of yellow fever virus are endoplasmic reticulum localization signals. J. Virol. 78:12591–12602. 10.1128/JVI.78.22.12591-12602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ruch TR, Machamer CE. 2012. The coronavirus E protein: assembly and beyond. Viruses 4:363–382. 10.3390/v4030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klumperman J, Locker JK, Meijer A, Horzinek MC, Geuze HJ, Rottier PJ. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tooze J, Tooze S, Warren G. 1984. Replication of coronavirus MHV-A59 in sac- cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33:281–293 [PubMed] [Google Scholar]
- 191.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. 10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976. 10.1128/JVI.00826-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 11:688–700. 10.1038/nrmicro3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jones DM, Atoom AM, Zhang X, Kottilil S, Russell RS. 2011. A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J. Virol. 85:12351–12361. 10.1128/JVI.05313-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mousseau G, Kota S, Takahashi V, Frick DN, Strosberg AD. 2011. Dimerization-driven interaction of hepatitis C virus core protein with NS3 helicase. J. Gen. Virol. 92:101–111. 10.1099/vir.0.023325-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342–3352. 10.1128/JVI.02447-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773–4784. 10.1128/JVI.76.10.4773-4784.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 82:4731–4741. 10.1128/JVI.00002-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Heliot L, Rouille Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7:e1001278. 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kim DY, Atasheva S, Frolova EI, Frolov I. 2013. Venezuelan equine encephalitis virus nsP2 protein regulates packaging of the viral genome into infectious virions. J. Virol. 87:4202–4213. 10.1128/JVI.03142-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. 2012. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology 432:511–526. 10.1016/j.virol.2012.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I. 2011. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J. Virol. 85:8022–8036. 10.1128/JVI.00644-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu Z, Yang D, Qiu Z, Lim KT, Chong P, Gillam S. 1996. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J. Virol. 70:2184–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kuo L, Masters PS. 2013. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 87:5182–5192. 10.1128/JVI.00100-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qin L, Xiong B, Luo C, Guo ZM, Hao P, Su J, Nan P, Feng Y, Shi YX, Yu XJ, Luo XM, Chen KX, Shen X, Shen JH, Zou JP, Zhao GP, Shi TL, He WZ, Zhong Y, Jiang HL, Li YX. 2003. Identification of probable genomic packaging signal sequence from SARS-CoV genome by bioinformatics analysis. Acta Pharmacol. Sin. 24:489–496 [PubMed] [Google Scholar]
- 206.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79:13848–13855. 10.1128/JVI.79.22.13848-13855.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789. 10.1128/JVI.02343-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945. 10.4161/auto.5.7.9243 [DOI] [PubMed] [Google Scholar]
- 209.Parent R, Qu X, Petit MA, Beretta L. 2009. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology 49:1798–1809. 10.1002/hep.22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. 2012. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 8:e1002829. 10.1371/journal.ppat.1002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. 2012. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 8:e1002845. 10.1371/journal.ppat.1002845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari FV. 2012. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 86:11043–11056. 10.1128/JVI.07101-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hirsch AJ, Medigeshi GR, Meyers HL, DeFilippis V, Fruh K, Briese T, Lipkin WI, Nelson JA. 2005. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79:11943–11951. 10.1128/JVI.79.18.11943-11951.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hunt TA, Urbanowski MD, Kakani K, Law LM, Brinton MA, Hobman TC. 2007. Interactions between the West Nile virus capsid protein and the host cell-encoded phosphatase inhibitor, I2PP2A. Cell. Microbiol. 9:2756–2766. 10.1111/j.1462-5822.2007.01046.x [DOI] [PubMed] [Google Scholar]
- 215.Carpp LN, Galler R, Bonaldo MC. 2011. Interaction between the yellow fever virus nonstructural protein NS3 and the host protein Alix contributes to the release of infectious particles. Microbes Infect. 13:85–95. 10.1016/j.micinf.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 216.Sousa IP, Jr, Carvalho CA, Ferreira DF, Weissmuller G, Rocha GM, Silva JL, Gomes AM. 2011. Envelope lipid-packing as a critical factor for the biological activity and stability of alphavirus particles isolated from mammalian and mosquito cells. J. Biol. Chem. 286:1730–1736. 10.1074/jbc.M110.198002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Richardson CD, Vance DE. 1978. Chemical cross-linking of proteins of Semliki Forest virus: virus particles and plasma membranes from BHK-21 cells treated with colchicine or dibucaine. J. Virol. 28:193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Richardson CD, Vance DE. 1978. The effect of colchicine and dibucaine on the morphogenesis of Semliki Forest virus. J. Biol. Chem. 253:4584–4589 [PubMed] [Google Scholar]
- 219.Pfister T, Mirzayan C, Wimmer E. 1999. Polioviruses (Picornaviridae): molecular biology, p 1330–1348 In Granoff WA. (ed), The encyclopedia of virology, 2nd ed. Academic Press, New York, NY [Google Scholar]