Abstract
Shigella flexneri O-antigen is an important and highly variable cell component presented on the outer leaflet of the outer membrane. Most Shigella flexneri bacteria share an O-antigen backbone composed of →2)-α-l-RhapIII-(1→2)-α-l-RhapII-(1→3)-α-l-RhapI-(1→3)-β-d-GlcpNAc-(1→ repeats, which can be modified by adding various chemical groups to different sugars, giving rise to diverse O-antigen structures and, correspondingly, to various serotypes. The known modifications include glucosylation on various sugar residues, O-acetylation on RhaI or/and RhaIII, and phosphorylation with phosphoethanolamine on RhaII or/and RhaIII. Recently, a new O-antigen modification, namely, O-acetylation at position 6 of N-acetylglucosamine (GlcNAc), has been identified in S. flexneri serotypes 2a, 3a, Y, and Yv. In this study, the genetic basis of the 6-O-acetylation of GlcNAc in S. flexneri was elucidated. An O-acyltransferase gene designated oacD was found to be responsible for this modification. The oacD gene is carried on serotype-converting bacteriophage SfII, which is integrated into the host chromosome by lysogeny to form a prophage responsible for the evolvement of serotype 2 of S. flexneri. The OacD-mediated 6-O-acetylation also occurs in some other S. flexneri serotypes that carry a cryptic SfII prophage with a dysfunctional gtr locus for type II glucosylation. The 6-O-acetylation on GlcNAc confers to the host a novel O-antigen epitope, provisionally named O-factor 10. These findings enhance our understanding of the mechanisms of the O-antigen variation and enable further studies to understand the contribution of the O-acetylation to the antigenicity and pathogenicity of S. flexneri.
INTRODUCTION
Shigella flexneri is the major pathogen mainly responsible for shigellosis, or bacterial dysentery, in developing countries. About 125 million shigellosis cases are estimated to occur annually in Asia, with 14,000 deaths, the majority of which are those of children under 5 years old (1). The O-polysaccharide chain of the lipopolysaccharide (LPS) called O-antigen plays an important role in the pathogenesis of S. flexneri; it protects the bacterium from the lytic action of serum complement and also enhances the adherence and internalization of the bacterium to intestinal epithelial cells (2–4). The fine structure of the O-antigen provides the chemical basis for serotyping of S. flexneri. All serotypes except for serotype 6 share a polysaccharide backbone, having a tetrasaccharide repeat (O-unit) of three l-rhamnose residues (RhaI to RhaIII) and one N-acetylglucosamine (GlcNAc) residue: →2)-α-l-RhapIII-(1→2)-α-l-RhapII-(1→3)-α-l-RhapI-(1→3)-β-d-GlcpNAc-(1→ (5).
The basic O-antigen is referred to as serotype Y, and adding various chemical groups to different sugars gives rise to diverse O-antigen structures and, correspondingly, to various serotypes (6). The host immune response to S. flexneri infection is serotype specific, and antigenic diversity provided by O-antigen modifications enhances the survival of the pathogens as it helps them to escape the host defense (6). Furthermore, some modifications, such as glucosylation on GlcNAc, RhaI, and RhaII, promote bacterial invasion into host cells mediated by the type III secretion system (2). Therefore, elucidation of the O-antigen modification mechanisms is important for understanding the antigenicity and pathogenicity of S. flexneri.
The well-known O-antigen modification types in S. flexneri are 2-O-acetylation and glucosylation (6). 2-O-acetylation occurs on RhaI in serotypes 1b, 3a, 3b, 4b, and 7b, conferring to the host group 6 and (in serotypes 3a and 3b) type III antigenic determinants (O-factors) (7, 8). An acetyltransferase protein encoded by the oac gene which is carried on prophage Sf6 mediates the 2-O-acetylation (9, 10). Glucosylation has been identified at different positions on one or two of the four sugar residues in the O-unit; it defines type I, IC, II, IV, and V as well as group 7,8 antigenic determinants in various serotypes (6, 11). Three genes, gtrA, gtrB, and gtr (type specific), are responsible for the glucosylation in S. flexneri; they are carried on six prophages or cryptic prophages (SfI, SfIC, SfII, SfIV, SfV, and SfX) and are arranged in a single operon known as the gtr locus (10–17). The first two genes are highly conserved and interchangeable between serotypes, whereas the third gene, gtr (type), encodes a serotype-specific glucosyltransferase responsible for the addition of a glucosyl group to a certain sugar residue in the O-unit (6). The Sf6 and SfIC prophages are located on the bacterial chromosome at the tRNA-argW site adjacent to the conserved yfdC gene and at the site adjacent to the yejO locus, respectively (11, 18), whereas the other prophages are integrated into the tRNA-thrW gene between the conserved proA and adrA genes (6, 19).
Recently, a third type of S. flexneri O-antigen modification has been identified, namely, that corresponding to the addition of phosphoethanolamine (PEtN) to position 3 of RhaIII or RhaII or both, which confers to the host the MASF IV-1 (E1037) epitope called the “variant” (v) factor in newly proposed serotypes Xv, 4av, and Yv (20–23). An O-antigen PEtN transferase protein (Opt) encoded by a single dimorphic opt gene carried on a 6.8-kb plasmid (pSFxv_2 or pSFyv_2) mediates the PEtN modification (20, 21, 23). The pSFxv_2 plasmid can be transferred among S. flexneri serotypes by conjugation in the laboratory (24), demonstrating the potential for dissemination of this modification in nature.
O-acetylation sites other than position 2 of RhaI had been overlooked in early studies of S. flexneri O-antigen structures and have been identified only recently (5, 25–28). These are position 3 or 4 of RhaIII (3/4-O-acetylation) in serotypes 1a, 1b, 2a, 5a, Y, and 6 and position 6 of GlcNAc (6-O-acetylation) in serotypes 2a, 3a, Y, and Yv. The 3/4-O-acetylation on RhaIII interferes with the glucosylation (group O-factor 7,8) and PEtN phosphorylation (group O-factor IV-1) at the same sugar residue, thus abolishing the 7,8 determinant and reducing the IV-1 determinant manifestation (24). Further studies have indicated that an acyltransferase protein (OacB) which is encoded by gene oacB carried on a transposon-like element of the chromosome mediates the 3/4-O-acetylation in serotypes 1a, 1b, 2a, 5a, and Y (28), whereas an OacB homolog named OacC encoded by a phage-like structure is responsible for the 3/4-O-acetylation in serotype 6 (see the supplemental material). The oacB and oacC genes are widespread in serotypes 1a, 1b, 2a, 5a, Y, and 6 and confer to the host a novel antigenic determinant called group O-factor 9 (29). oacB and oacC are not involved with the 6-O-acetylation on GlcNAc as they could not be detected in strains carrying only this modification and their inactivation in strains carrying both 3/4-O-acetylation and 6-O-acetylation did not affect the 6-O-acetylation on GlcNAc (28) (see the supplemental material). Therefore, there is another mechanism involved with the 6-O-acetylation of S. flexneri O-antigens.
In this study, we identified yet another oac homolog, designated oacD, and demonstrated that this gene is responsible for the 6-O-acetylation on GlcNAc in S. flexneri O-antigens. oacD was carried on the serotype-converting bacteriophage SfII genome, which was integrated into the host chromosome by lysogeny to form a prophage present in serotype 2 strains. Some other serotypes of S. flexneri (3a, Y, and Yv) were found to possess a cryptic SfII prophage with a dysfunctional gtr locus. It was also shown that the OacD-mediated 6-O-acetylation confers to the host a novel antigenic determinant provisionally named O-factor 10.
MATERIALS AND METHODS
Ethics statement.
This study was reviewed and approved by the ethics committee of National Institute for Communicable Disease Control and Prevention, China Center for Disease Control (CDC).
Bacterial strains, plasmids, and culturing conditions.
Strains and plasmids used in this study are listed in Table 1. S. flexneri strain Sf301 (serotype 2a) (28) with the 6-O-acetylated GlcNAc in the O-antigen was used as the reference strain for oacD gene deletion and complementation analysis. S. flexneri 51571 (serotype 1a) and 51577 (serotype 4b) (24) were employed as hosts in function analysis of the oacD gene. Escherichia coli JM109 (TaKaRa, Japan) was used for plasmid propagation. pMD20T vector (TaKaRa, Japan) was used for oacD gene cloning and DNA sequencing analysis. Plasmid pRS551 was used for kanamycin resistance gene amplification. pKOBEG encoding a homologous recombination system was used in the oacD gene deletion analysis. Thirty-one S. flexneri strains with known O-antigen structures (listed in Table 2) were used for oacD PCR detection analyses. A total of 641 more S. flexneri isolates representing 18 serotypes were tested using the oacD gene PCR detection and antiserum 10 agglutination assay (Table 3). These strains were isolated from diarrheal patients in a surveillance program performed by China CDC during 2000 to 2012 or were purchased from the National Collection of Type Cultures (NCTC) or were kindly donated by B. Liu (Nankai University, Tianjin, China). Twelve strains of Shigella dysenteriae (one each of serotypes 1 to 12), 18 strains of Shigella boydii (one each of serotypes 1 to 18), 14 strains of Shigella sonnei (2 phase I and 12 phase II), and 10 strains of E. coli (one each of serotypes O6, O8, O13, O44, O71, O78, O127, O128, O157, and O159) were used for the oacD gene PCR detection and antiserum 10 specificity evaluation (see Table S1 in the supplemental material). Strains were grown in a 37°C incubator or orbital shaker in Luria-Bertani broth (LB) supplemented with ampicillin (100 μg ml−1), kanamycin (40 μg ml−1), or chloramphenicol (50 μg ml−1) when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| S. flexneri strains | ||
| Sf301 | Serotype 2a with 6-O-acetylation on GlcNAc used for oacD gene cloning and inactivation analysis, Aps Kms | 28 |
| Sf301ΔoacD | Sf301 with the oacD gene replaced by the kanamycin resistance gene (kan) from pSR551, Kmr Aps | This work |
| Sf301ΔoacD_pSQZ6 | Sf301ΔoacD transformed by plasmid pSQZ6 | This work |
| 51571 | Serotype 1a with 4-glucosylation on GlcNAc used for oacD function analysis, Aps | 24 |
| 51571_pSQZ6 | 51571 transformed by plasmid pSQZ6 | This work |
| 51577 | Serotype 4b with 6-glucosylation on GlcNAc used for oacD function analysis, Aps | 24 |
| 51577_pSQZ6 | 51577 transformed by plasmid pSQZ6 | This work |
| E. coli JM109 | E. coli strain used for plasmid propagation and gene cloning | TaKaRa |
| Plasmids | ||
| pMD20T | T-A vector, Apr | TaKaRa |
| pSR551 | Kmr, used for amplification of the kan gene | 30 |
| pKOBEG | A thermosensitive replicon that carries the λ phage redγβα operon expressed under the control of the arabinose-inducible pBAD promoter | 31 |
| pSQZ6 | pMD20T carrying the whole sequence of the oacD gene from strain Sf301, Apr | This work |
Ap, ampicillin.
TABLE 2.
Correlation between the 6-O-acetylation on GlcNAc, the presence of the oacD gene, and the reactivity with grouping antiserum 10 of S. flexneri strains
| Strain | Serotype | 6-O-acetylation on GlcNAc | oacD gene PCR amplification | Antiserum 10 reactivity | Reference for the O-antigen structure |
|---|---|---|---|---|---|
| 51571 | 1a | − | − | − | 24 |
| G1661 | 1a | − | − | − | 26 |
| 51572 | 1b | − | − | − | 24 |
| G1662 | 1b | − | − | − | 26 |
| X6 | 1c | − | − | − | 24 |
| HN153 | 1d | − | − | − | 32 |
| 51250 | 2a | + | + | + | 24 |
| G1663 | 2a | + | + | + | 26 |
| Sf301 | 2a | + | + | + | 28 |
| 51251a | 2b | − | + | − | 24 |
| 51575 | 3a | + | + | + | 24 |
| G1665 | 3a | + | + | + | 5 |
| G1666 | 3b | − | − | − | 5 |
| NCTC9725 | 4a | − | − | − | 20 |
| G1668 | 4av | − | − | − | 22 |
| 51577 | 4b | − | − | − | 24 |
| G1669 | 4b | − | − | − | 5 |
| 51247 | 5a | − | − | − | 24 |
| G1036 | 5a | − | − | − | 25 |
| 51580 | X | − | − | − | 20 |
| G1039 | X | − | − | − | 5 |
| 2002017 | Xv | − | − | − | 20 |
| 2003055 | Xv | − | − | − | 20 |
| 51581b | Y | + | + | + | 24 |
| G1040b | Y | + | + | + | 5 |
| 036 | Y | − | − | − | 23 |
| HN006b | Yv | + | + | + | 23 |
| AH012b | Yv | + | + | + | 23 |
| HN011b | Yv | + | + | + | 23 |
| 51579 | 6 | − | − | − | 24 |
| G1038 | 6 | − | − | − | 5 |
| G1671 | 6 | − | − | − | 5 |
The oacD gene in serotype 2b strain 51251 has a one-base deletion (A) at position 191, which resulted in a stop codon at amino acid 64, rendering the gene defective.
Strains HN006, AH012, and HN011 (all serotype Yv) and strains 51581 and G1040 (both serotype Y) carry a SfII prophage genome with a mutational gtrII gene.
TABLE 3.
Distribution of the oacD gene in S. flexneri strains and their cross-reactivity with grouping antiserum 10
| Serotype | No. of strains tested | No. of oacD-positive strains | No. of antiserum 10-reactive strains |
|---|---|---|---|
| 1a | 76 | 0 | 0 |
| 1b | 22 | 0 | 0 |
| 1c | 1 | 0 | 0 |
| 1d | 5 | 0 | 0 |
| 2a | 154 | 154 | 154 |
| 2ba | 29 | 29 | 23 |
| 3ab | 7 | 2 | 2 |
| 3b | 2 | 0 | 0 |
| 4a | 3 | 0 | 0 |
| 4av | 3 | 0 | 0 |
| 4b | 3 | 0 | 0 |
| 5a | 2 | 0 | 0 |
| Xc | 39 | 1 | 1 |
| Xvd | 189 | 1 | 0 |
| Ye | 37 | 14 | 13 |
| Yvf | 21 | 13 | 13 |
| 6 | 76 | 0 | 0 |
| 7b | 2 | 0 | 0 |
| Total | 672 | 214 | 206 |
Six serotype 2b strains (51251, 05BJ13, 08GS74, 08SX28, 2005049, and 2005001) are antiserum 10 negative. All carry a defective oacD gene, with strain 2005001 having a one-base (T) insertion at position 177 and the others having a one-base (A) deletion at position 191.
Two serotype 3a strains (51575 and G1665) carry a cryptic SfII prophage lacking gtrII.
One serotype X isolate (2001006) carries a cryptic SfII prophage with a defective gtrII gene having a single-base (T) deletion at base 1,031 resulting in a stop codon at amino acid 394.
One serotype Xv strain (2003005) carries a cryptic SfII prophage with a defective gtrII gene having a single-base (T) deletion at base 1,291 resulting in a stop codon at amino acid 444; the oacD gene of 2003005 has a one-base (A) deletion at position 191 resulting in a stop codon at amino acid 64.
Ten serotype Y strains carry a cryptic SfII prophage with a defective gtrII gene, with 5 strains (2000025, 2000026, 03HL08, 10HN064, and HN078) having a single-base (T) deletion at base 1,031 of gtrII, 2 strains (51581 and G1040) a two-base insertion (AT) at base 1,181, 2 strains (HN126 and XZ014) a four-base deletion (ACAT) at base 1,197 to base 1,282, and one isolate (XZ002) a one-base substitution (G to A) at base 87, all resulting in a stop codon at amino acid 28 or 344 or 394 or 445 in GtrII. Four serotype Y strains (HB06, 03HL32, AH104, and HN114) have one nonsynonymous mutation (A to C) in gene gtrB at position 560, resulting in an amino acid change (Q to P) at position 187; 06AH104 carries a dysfunctional oacD having a one-base (A) deletion at position 191, resulting in a stop codon at amino acid 64.
Thirteen serotype Yv strains (HN006, AH012, HN068, HN069, AH028, HN171, HN182, HN049, AH029, HN011, HN033, HN116, and HN106) carry a dysfunctional gtrII locus (either one or both of the mutations in gtrII at position 1,222 and in gtrB at position 560).
Bioinformatics analysis.
The protein sequences of Oac of Sf6 (GenBank accession no. NP_958191.1), OacB of Sf301 (NP_706267.1), and OacC of CDC796-83 (WP_005054336.1) were compared to the protein database sequence of S. flexneri strain Sf301 (NC_004337.1) using the BLASTP web server (http://www.ncbi.nlm.nih.gov/BLAST). The candidates were further searched against the genomes of S. flexneri strains 2002017 (CP001383) and 036 (CP004056) (both lacking the 6-O-acetylation on GlcNAc) (20, 23) and 51581 (AZPG00000000) and HN006 (CP004057) (both with 6-O-acetylation) (23) using BLASTn. Searches for homologs to the OacD protein in the NCBI database were carried out using the BLASTP search engine.
DNA techniques.
Genomic DNA and plasmid DNA were extracted using a DNA extraction kit according to the manufacturer's instructions (Qiagen, Germany). Primers used in this study were listed in Table 4. The oacD-1 primer pair was used for oacD gene detection. The oacD-2 primer pair was used for oacD gene function analysis. The oacD-3 primer pair was used to amplify regions flanking oacD in S. flexneri strains 51575 and G1665 (both serotype 3a). The gtrII-1 primer pair was used to amplify SfII-specific gene gtrII for prophage SfII genome detection. The gtrII-2 primer pair, whose product spans the whole gtr locus (gtrA, gtrB, and gtrII), was used for sequencing analysis. The PCR products of the oacD-2 primer pair were purified and cloned into the TA-pMD20T vector to generate the pSQZ6 expression plasmid. The recombinant plasmid was first transformed into commercial E. coli JM109 competent cells and then into S. flexneri strains, using a standard chemical protocol (33). The transformants were selected on LB plates supplemented with ampicillin (100 μg ml−1) and confirmed by PCR amplification of the oacD gene. Oligonucleotide primers were synthesized by Sangon Biotech (Shanghai, China). PCR amplifications were performed using a TaKaRa PCR amplification kit (TaKaRa, Japan) following a standard protocol.
TABLE 4.
Primers used in this study
| Primer | Primer sequence (5′–3′) | Length of the PCR fragment (bp) | Target gene(s) or reference sequence |
|---|---|---|---|
| oacD-1 | F: GGGGCTGGCAAATTGTATCC | 822 | oacD |
| R: GCCTATAATTGCTAAAGCCATAGG | |||
| oacD-2 | F: GACCATGGTGCGAGAGTGGCAGG | 1,935 | oacD |
| R: CTGGGCGAAGCATCAGGAAGGC | |||
| kan | F: GGTCACCTTGGGTTGGGGGCTGGCAAATTGTATCCTTTTGTTTTTAGTTATCACGTCCCACTATTCTTTTTCGCTGCCACGTTGTGTCTCAAAATCT | 1,130 | kan and oacD |
| R: CAGGTATATATGATTGCGCAGATAGGATTTGGAATAGTCACGCTGAAGCCTATAATTGCTAAAGCCATAGGGCGTCCCGTCAAGTCAGCGTA | |||
| oacD-3 | F: GGATACAATTTGCCAGCCCCC | 3,775 | oacD, gtrB |
| R: CCCAGTACGCGGGTATCCCTCCC | |||
| gtrII-1 | F: ATTTATTGTTATTGGGGGTGGTTG | 1,268 | gtrII |
| R: ATTTGTTCTTTATTTGCTGGTT | |||
| gtrII-2 | F: TGAAAATTTTCTGGGGATCCCTCAG | 3,383 | NC_004337 |
| R: ATGGTGCCGATAATAGGAGTCGAAC |
oacD gene functional deletion and complementation analysis.
Deletion of the oacD gene was performed on S. flexneri strain Sf301 using a one-step method as described previously (28, 34). The aminoglycoside 3′-phosphotransferase gene encoding kanamycin resistance (Kmr) was PCR amplified from plasmid pRS551 using the kan primer pair (Table 4). The PCR products were electroporated into strain Sf301 carrying plasmid pKOBEG and selected on an LB plate with chloramphenicol (50 μg ml−1) and kanamycin (40 μg ml−1). oacD gene deletion mutant Sf301ΔoacD was further confirmed by PCR amplification of oacD using the oacD-1 and oacD-2 primer pairs. Plasmid pSQZ6 was transformed into Sf301ΔoacD, 51571 (1a), and 51577 (4b), giving rise to oacD-complemented strains Sf301ΔoacD_pSQZ6, 51571_pSQZ6, and 51577_pSQZ6, respectively.
Preparation of specific antiserum 10 against a 6-O-acetylated GlcNAc-linked epitope.
Immunization and preparation of antisera were performed as described previously (29, 35). Briefly, three New Zealand White rabbits (female, 1.5 to 2 kg body weight) were immunized intravenously with heat-killed cells of S. flexneri strain Sf301 twice a week at increasing doses (1 × 109, 2 × 109, 4 × 109, 8 × 109, 16 × 109, and 16 × 109 CFU). The serum was separated 1 week after the last immunization and mixed with heat-killed cells of S. flexneri isolate Sf301ΔoacD to absorb nonspecific antibodies that cross-react with other O-antigenic epitopes as described previously (35). The antiserum that agglutinated strain Sf301 but not Sf301ΔoacD is referred to as antiserum 10 below.
Serotyping analysis.
The serological features of S. flexneri strains were revealed by a slide agglutination test using commercially available Shigella monovalent antisera (Denka Seiken, Japan) and monoclonal antibody (Reagensia AB, Sweden), as well as 3/4-O-acetylated RhaIII-specific antiserum 9 prepared previously (29). Antiserum 10 prepared in this study was used for detection of the 6-O-acetylated GlcNAc-linked epitope by slide agglutination, and agglutination apparent to the naked eye within 20 s was recorded as representing a positive result.
Western blot analysis.
LPSs were prepared using an LPS extraction kit (iNtRON, South Korea) according to the manufacturer's instructions. The LPSs were electrophoresed on 15% polyacrylamide gels and detected by silver staining as described previously (36, 37). A Western blot assay of the LPS was performed as described previously (29). The LPS separated by SDS-PAGE was transferred onto a polyvinylidene difluoride (PVDF) membrane and incubated with antiserum 10. After washing, the membrane was incubated with anti-rabbit antibody labeled with fluorescent IRDye 800 (Rockland), and the fluorescence was detected using an Odyssey infrared imaging system (Li-COR).
O-polysaccharide isolation and structure analysis.
Lipopolysaccharides of wild-type strains, mutants, and transformants were isolated by phenol-water extraction of bacterial cells (38). The crude extract without separation of the layers was dialyzed against tap water to remove phenol, nucleic acids and proteins were removed by adding aqueous 50% CCl3CO2H to the dialysis retentate with stirring at 4°C for 5 min to reach pH 2, the precipitate was discarded, and the LPS-containing supernatant was dialyzed against distilled water with two water changes over the period of 48 h and freeze-dried to yield purified LPS preparations in yields of 7.6% to 9.6% of dry bacterial mass. Each LPS was degraded with aqueous 2% acetic acid (HOAc) at 100°C, and the O-polysaccharides were isolated in yields of 18% to 51% of the LPS mass by gel permeation chromatography on Sephadex G-50 Superfine medium (Amersham Biosciences, Sweden)–0.05 M pyridinium acetate buffer, pH 4.5, monitored with a differential refractometer (Knauer, Germany).
Structures of the O-polysaccharides were elucidated using two-dimensional nuclear magnetic resonance (NMR) spectroscopy essentially as described previously (23). Assignment of the 1H and 13C NMR spectra was performed using correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and 1H,13C heteronuclear single-quantum coherence (HSQC) experiments. The assigned 1H and 13C NMR chemical shifts were essentially identical to published data for the correspondingly substituted Rha and GlcNAc residues (5, 24, 26, 28). Positions of O-acetyl groups were determined by characteristic downfield displacements of the NMR signals for 1H and 13C atoms at the O-acetylation sites (see Table S2 in the supplemental material) compared with the corresponding O-polysaccharides lacking the O-acetylation (5, 24). In particular, the signals for H-3 and C-3 of RhaIII were shifted from δH 3.86 and δC 71.2 to δH 5.07 to 5.08 and δC 73.6 to 74.2, H-4 and C-4 of RhaIII from δH 3.33 to 3.35 and δC 73.8 to 73.9 to δH 4.79 to 4.82 and δC 75.3 to 75.6, and H-6a,6b and C-6 of GlcNAc from δH 3.77 to 3.98 and δC 61.9 to 62.6 to δH 4.30 to 4.42 and δC 64.1 to 64.5, respectively. The degree of 6-O-acetylation was determined by the relative integral intensities of the 1H NMR signals of the O-acetylated and non-O-acetylated GlcNAc residues.
Nucleotide sequence accession numbers.
The genome sequences of S. flexneri strains 036 and HN006 have been deposited in GenBank under accession numbers CP004056 and CP004057, respectively.
RESULTS
Identification of an O-acyltransferase gene, oacD, that is present only in S. flexneri strains with 6-O-acetylation on GlcNAc.
Until now, three acyltransferase-encoding genes responsible for O-acetylation of S. flexneri O-antigens had been identified, including oac (which we have renamed oacA [28]) for the 2-O-acetylation of RhaI (9, 10) and oacB and oacC for the 3/4-O-acetylation of RhaIII (28, 29) (see the supplemental material). We hypothesized that the 6-O-acetylation of GlcNAc in serotypes 2a, 3a, Y, and Yv is mediated by an acyltransferase that is encoded by an oac homolog. To identify it, the OacA, OacB, and OacC protein sequences were searched against the genome of S. flexneri Sf301 (serotype 2a), which possesses the 6-O-acetylation on GlcNAc, using BLASTp and tBLASTn searches. Several proteins homologous to the Oac proteins (see Table S3 in the supplemental material) were retrieved, and further searches of the genomes of S. flexneri strains HN006 and 51581 (both having the 6-O-acetylation on GlcNAc) and strains 2002017 and 036 (both lacking the 6-O-acetlyation) were performed. As a result, a hypothetical protein encoded by a chromosomal gene, SF0309, was found in strains Sf301, 51581, and HN006 but not in strains 2002017 and 036. The predicted protein showed homology to OacC, with 27% identity and 45% similarity in the amino terminal domain (amino acids 13 to 93), suggesting that it is responsible for the 6-O-acetylation of GlcNAc.
The protein encoded by SF0309 contained 349 amino acids and possessed conserved domains of the acyltransferase or acetyltransferase family (Acyl_transf_3 or NoIL). A BLAST search also revealed the presence of this protein in completely or partially sequenced S. flexneri strains 2457T (NP_706261.1), SFL124 (AY900451.1), K-1770 (WP_000282634.1), and 2747-71 (WP_000613535.1), serotype-converting bacteriophage SfII (YP_008318503.1), and E. coli strains KTE33 (WP_016159269.1), 1-176-05_S3_C2 (EYD87781.1), KTE-18 (WP_001579928.1), and UMEA 3703-1 (WP_001579928.1) with 98% to 100% identity at the protein and DNA levels. It also showed 24% to 41% identity to predicted acyltransferase family proteins of Pantoea ananatis PA13 (YP_005993804.1), Clostridium cellulovorans 743Bv (YP_003844488.1), Leptospira kmetyi (WP_020986752.1), and some other species. The suspected SF0309-encoded acetyltransferase was named OacD and the corresponding gene oacD, following the designations for OacA to OacC (28, 29) (see the supplemental material) mediating the 2-O- and 3/4-O-acetylations.
PCR screening was performed on 31 other S. flexneri strains with known O-antigen structures (Table 2), using the oacD-1 primer pair. The expected PCR product (822 bp) was amplified from strains 51250 and G1663 (both serotype 2a), 51251 (serotype 2b), 51575 and G1665 (both serotype 3a), 51581 and G1040 (both serotype Y), and HN006, AH012, and HN011 (all serotype Yv); except for strain 51251, all possessed 6-O-acetylation (5, 23, 24). In contrast, the other strains tested that lacked 6-O-acetylation were oacD negative (Table 2). The whole oacD gene in 10 oacD-positive strains was PCR amplified and sequenced using the oacD-2 primer pair. It was found that the oacD gene in the strains tested was identical to that of Sf301, except for strain 51251, whose oacD carried a one-base (A) deletion at position 191, resulting in a stop codon at amino acid 64.
The oacD gene is responsible for the 6-O-acetylation of GlcNAc in S. flexneri O-antigens.
To confirm the function of oacD, a deletion and complementation analysis was performed on strain Sf301. The oacD gene from 168 to 902 bp from the start site of translation was replaced with the aminoglycoside 3′-phosphotransferase-encoding gene (kan) sequence, resulting in a dysfunctional oacD gene in the deletion mutant.
To construct the oacD expression vector, the entire oacD gene of 1,050 bp, together with 885-bp sequences upstream and downstream to include its potential promoter and terminator sequences, was cloned from strain Sf301 into plasmid pMD20T to construct plasmid pSQZ6. The latter was transformed into the Sf301ΔoacD deletion mutant and two wild-type S. flexneri strains lacking oacD and 6-O-acetylation but carrying 4- or 6-glucosylation on GlcNAc (serotype 1a strain 51571 and serotype 4b strain 51577; for the O-polysaccharide structures, see Table 5). A serological assay using Shigella antisera of Denka Seiken, Japan, and antiserum 9 (29) revealed no differences among the Sf301 parental strain, its Sf301ΔoacD deletion mutant, and complementation mutant Sf301ΔoacD_pSQZ6 (Table 6). Similarly, the pSQZ6 complementation mutants of the wild-type strains were serologically identical to the parental strains, except that 51577_pSQZ6 had acquired the type III antigenic determinant (Table 6).
TABLE 5.
Structures of the O-polysaccharides of S. flexneri strains studied

TABLE 6.
Serological features of oacD gene deletion and complementation mutants of S. flexneri
| Strain | Reactivity with type and group antisera of Seiken |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | 3,4 | 6 | 7,8 | 9a | 10b | |
| Sf301 (2a) | − | + | − | − | − | − | + | − | − | + | + |
| Sf301ΔoacD | − | + | − | − | − | − | + | − | − | + | − |
| Sf301ΔoacD_pSQZ6 | − | + | − | − | − | − | + | − | − | + | + |
| 51571 (1a) | + | − | − | − | − | − | + | − | − | + | − |
| 51571_pSQZ6 | + | − | − | − | − | − | + | − | − | + | + |
| 51577 (4b) | − | − | − | + | − | − | − | + | − | − | − |
| 51577_pSQZ6 | − | − | + | + | − | − | − | + | − | − | + |
Antiserum 9 was prepared previously (29).
Antiserum 10 was prepared in this work.
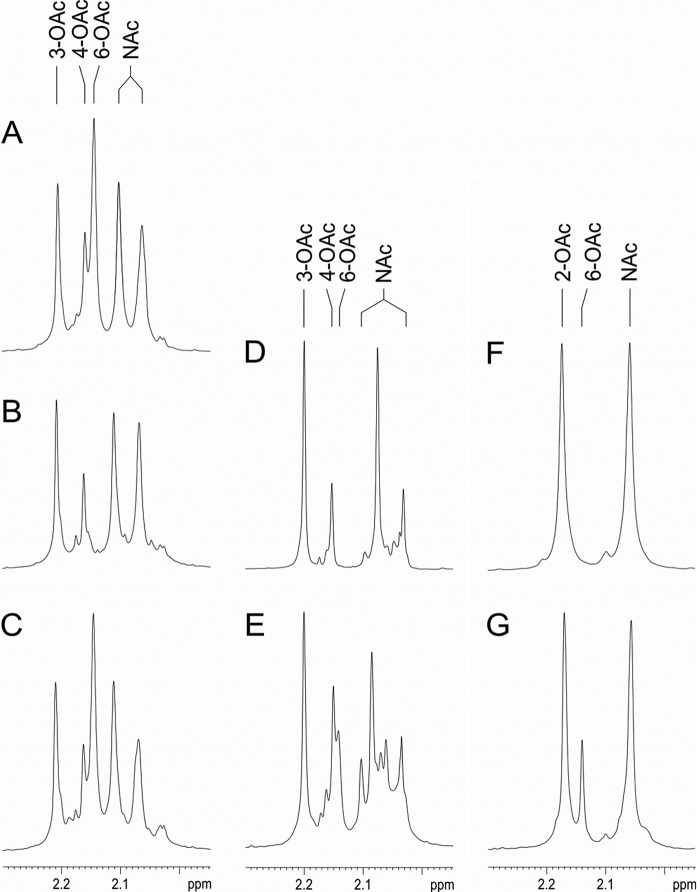
The O-polysaccharides of the deletion and complemented mutants were isolated from the LPS and analyzed using NMR spectroscopy as described in Materials and Methods. The O-polysaccharide structures thus established are shown in Table 5. The NMR spectra of the Sf301ΔoacD deletion mutant lacked signals for the 6-O-acetyl group at δH 2.14 (Fig. 1A and B) and 6-O-acetylated GlcNAc, particularly those for C-6 and H-6a and H-6b, which were present in the spectra of the parental strain Sf301 at δC 64.8 and at δH 4.21 and 4.33. The signals for the 3- and 4-O-acetyl groups at δH 2.20 and 2.16, respectively, and the correspondingly acetylated RhaIII residues were present in the spectra of the mutant and were as intense as in the parental strain (∼45% and ∼25%, respectively). Therefore, the mutant O-antigen lost the 6-O-acetylation on GlcNAc, whereas the 3/4-O-acetylation on RhaIII was unaffected. Both O-acetyl modifications were present in Sf301ΔoacD_pSQZ6 (Fig. 1C) and, hence, the 6-O-acetylation was restored by transformation of Sf301ΔoacD with the oacD-carrying pSQZ6 plasmid. As in the parental strain, the 6-O-acetylation in the transformant was nonstoichiometric (∼65%).
FIG 1.
Parts of 1H NMR spectra of the O-polysaccharides from wild-type strains and transformants showing signals for O-acetyl and N-acetyl groups. A, Sf301; B, Sf301ΔoacD; C, Sf301ΔoacD_pSQZ6; D, 51571; E, 51571_pSQZ6; F, 51577; G, 51577_pSQZ6. 2-OAc, 3-OAc, 4-OAc, and 6-OAc indicate O-acetyl groups at position 2 of RhaI, position 3 of RhaIII, position 4 of RhaIII, and position 6 of GlcNAc, respectively. The presence of multiple signals for the N-acetyl group (NAc) reflects structural heterogeneity due to nonstoichiometric O-acetylation and (in strain 51571_pSQZ6) nonstoichiometric glucosylation.
As judged by the appearance in the NMR spectra of minor signals for the 6-O-acetyl group at δH 2.14 (Fig. 1D to G), and 6-O-acetylated GlcNAc (δC 64.4 and δH 4.30 to 4.31 and 4.41 to 4.42), the pSQZ6 transformants of strains 51571 (1a) and 51577 (4b) acquired the 6-O-acetyl group on ∼25% and ∼30% GlcNAc residues, respectively (Table 5). In both cases, the degree of glucosylation on GlcNAc decreased correspondingly from 100% to ∼75% and ∼70%. In 51577_pSQZ6, the incomplete 6-glucosylation is the basis for the manifestation of the type III antigenic determinant, which is associated with the 2-O-acetylation on RhaI but is blocked by glucosylation on GlcNAc in wild-type serotype 4b strains.
The oacD gene was carried on serotype-converting bacteriophage SfII, which is present as a prophage in serotype 2 (2a and 2b) or as a cryptic prophage with a dysfunctional gtr locus in other serotypes.
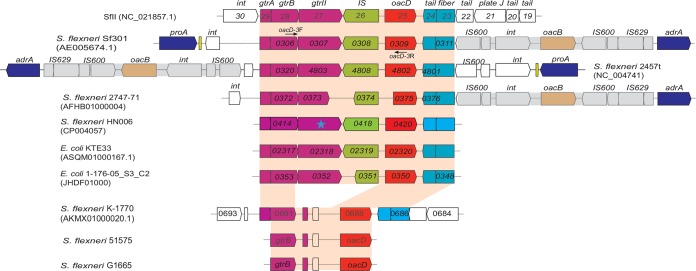
The genomic regions flanking the oacD gene in sequenced strains of S. flexneri (Sf301, 2457T, 2747-71, HN006, and 51581) and E. coli (KTE33 and 1-176-05_S3_C2) were analyzed, and their genetic organizations are shown in Fig. 2. In all cases, the oacD was located downstream of the gtr locus (gtrA, gtrB, and gtrII) of the SfII prophage. One insertion sequence (ISsf13) and two genes encoding phage tail fiber proteins were located upstream and downstream of oacD, respectively. This structure of gtr-IS-oacD-phage tail genes is identical in both genetic organization and DNA sequence to that of serotype-converting bacteriophage SfII (NC_021857.1), a free particle that has been induced from serotype 2a strain NCTC4 (39) (Fig. 2).
FIG 2.
Genetic organizations of the genomic regions of SfII, S. flexneri, and E. coli strains carrying the oacD gene. Sequences of serotype-converting phage SfII, S. flexneri strains Sf301, 2457T, 2747-71, HN006, and K-1770, and E. coli strains KTE33 and 1-176-05_S3_C2 were obtained from the NCBI database. Genomic sequences of serotype 3a strains 51575 and G1665 were obtained by PCR amplification. The open reading frames (ORFs) were annotated as submitted in NCBI or predicted using ORF Finder and are shown as thick arrows. The locus tags are shown within arrows, and the encoding proteins are listed above. The conserved genes are shown in different colors: proA and adrA, dark blue; gtr locus, pink; ISsf13, green; oacD, red; tail fiber light blue. The conserved oacD-carrying prophage genome and oacB-carrying transposon are highlighted in red and gray, respectively. The defective gtrII gene in strain HN006 is marked with a blue star.
The oacD region from serotype 3a strains 51575 and G1665 was PCR amplified and sequenced using complementation of the oacD-3 primer pair to genes gtrB and oacD (Fig. 2). The 2,295-bp PCR products obtained were shorter than the 3,775-bp PCR product from reference strain Sf301. The two PCR products were identical and differed from that of Sf301 in the middle of the sequence, with a 1,675-bp insertion between gtrB and oacD, resulting in a shorter GtrB product and complete deletion of gtrII and the region up to ISsf13 (Fig. 2). The 1,675-bp fragment encoded two putative proteins of unknown function and showed no homology to any sequence in the NCBI database. A similar genomic structure was found in partially sequenced S. flexneri strain K-1770 (Fig. 2). Therefore, a cryptic SfII prophage is present in these three strains.
The presence of the conserved gtr-ISsf13-oacD structure in both the free SfII phage particle and oacD-positive strains suggested that oacD originated from phage SfII and therefore should occur in all prophage SfII-carrying strains. To confirm this, 183 serotype 2 strains (154 of 2a and 29 of 2b) were tested by PCR amplification of the oacD gene using the oacD-1 primer pair, and all were found to be oacD positive (Table 3). Further screening of 488 S. flexneri isolates of various other serotypes showed that all were oacD negative, except for 31 strains belonging to serotypes 3a (2 strains), Xv (1), X (1), Y (14), and Yv (13) (Table 3).
Previously, we showed that all 13 oacD-positive strains of serotype Yv carry a dysfunctional gtrII locus with either one or both of the mutations in gtrII (position 1,222) and gtrB (position 560) (21). Therefore, it was concluded that these Yv strains originated from serotype 2a strains and, hence, should carry a cryptic SfII prophage. PCR detection of the SfII-specific gtrII gene using the gtrII-1 primer pair showed that 18 other oacD-positive non-serotype 2 strains were gtrII positive, except for serotype 3a strains 51575 and G1665, which had lost the gtrII gene (see above). In 10 serotype Y strains and 1 strain each of serotypes X and Xv, the gtrII had frameshift mutations due to a base deletion, insertion, or substitution (see footnote to Table 3), rendering it defective. Four gtrII-positive serotype Y strains had no defect in gtrII but did have a nonsynonymous mutation in the gtrB gene, which gave rise to an amino acid change at position 187 and might be responsible for the defective gtrII-based type II glucosylation.
Based on these data, we conclude that the oacD gene was carried on the phage SfII genome, which was integrated into the host chromosome by lysogeny to form a prophage. As a result, the oacD gene always coexists with the gtr locus in serotype 2 strains and is also present in strains of some other serotypes carrying a cryptic SfII prophage with a defective gtr locus. This conclusion was confirmed by serological studies described in the next section.
OacD-mediated 6-O-acetylation on GlcNAc confers to the host a new antigenic determinant.
Rabbit polyclonal antiserum specific to a putative new epitope(s) associated with 6-O-acetylated GlcNAc was prepared using serotype 2a strain Sf301 carrying 6-O-acetylation on GlcNAc for immunization and its 6-O-acetylation-lacking oacD deletion mutant for absorption of the crude antiserum. The absorption eliminated cross-reactive antibodies to all O-antigen-linked epitopes except for the target one. After repeated absorptions, the antiserum obtained only agglutinated strain Sf301 but not the Sf301ΔoacD deletion mutant (Table 6). It was called grouping antiserum 10.
The specificity of antiserum 10 was characterized by an immunoblotting assay performed with the LPS of Sf301, its deletion mutant Sf301ΔoacD, and complementation mutant Sf301ΔoacD_pSQZ6 as well as serotype 1a strain 51571 and its complementation mutant 51571_pSQZ6. The LPS samples were resolved by SDS-PAGE on a 15% gel and visualized by silver staining (Fig. 3A). A typical ladder-like banding pattern of an LPS with an O-antigen composed of various numbers of O-units was observed for all strains (Fig. 3A). Differences were found in the LPS profiles of the parental strains and constructed mutants, with the complementation mutants (Sf301ΔoacD_pSQZ6 and 51571_pSQZ6) having more rough- and semirough LPS than the parental strains (Fig. 3A). We do not know the exact mechanism causing this difference, and further studies are needed. In Western blot analyses, antiserum 10 reacted only with the ladder-like LPS bands of Sf301, 51571_pSQZ6, and Sf301ΔoacD_pSQZ6 carrying the 6-O-acetylation on GlcNAc but did not recognize LPS of 51571 and Sf301ΔoacD lacking this modification (Fig. 3B). This serological specificity was further confirmed by an agglutination assay performed on other oacD transformants, which indicated that antiserum 10 reacted only with oacD-carrying transformants Sf301ΔoacD_pSQZ6, 51571_pSQZ6, and 51577_pSQZ6 but not with oacD-lacking strains 51571 and 51577 (Table 6). Therefore, antiserum 10 is specific to a 6-O-acetylated GlcNAc-linked epitope(s) on the O-antigen.
FIG 3.
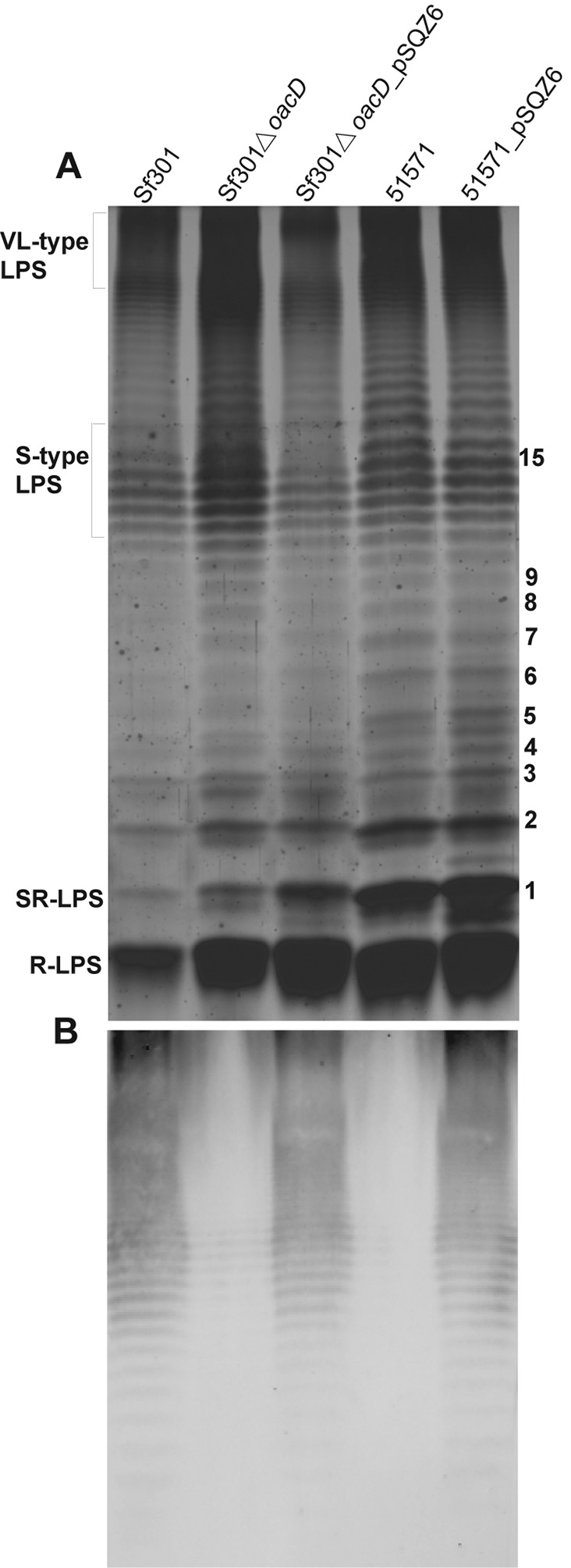
SDS-PAGE and Western blot analysis of the LPS of S. flexneri Sf301, its deletion mutant Sf301ΔoacD and complementation mutant Sf301ΔoacD_pSQZ6, and serotype 1a strain 51571 and its oacD transformant 51571_pSQZ6. (A) Silver-staining detection of LPS profiles on 15% polyacrylamide gels. The positions of LPSs of various types are indicated on the left. R-LPS, rough LPS; SR-LPS, semirough LPS; L-type LPS, long-type LPS; VL-type LPS, very-long-type LPS. The numbers of O-units are indicated on the right. (B) Immunoblotting detection of LPS with antiserum 10. The LPSs separated by SDS-PAGE were transferred onto a PVDF membrane and hybridized with grouping antiserum 10. An anti-rabbit antibody labeled with fluorescent IRDye 800 (Rockland) was used as the secondary antibody. Fluorescence was detected using an Odyssey infrared imaging system (Li-COR).
The 672 strains used for the oacD PCR detection were tested by slide agglutination, and a good correlation between the presence of the functional oacD gene and the antiserum 10 reactivity was observed. The agglutinated strains (206 in total, all oacD positive) belonged to serotypes 2a (154 strains), 2b (23), 3a (2), X (1), Y (13), and Yv (13) (Table 3). Except for 8 strains (6 of serotype 2b, 1 Xv, and 1 Y), all oacD-carrying strains were antiserum 10 positive, whereas all oacD-lacking strains were negative (Table 3). The oacD gene in the 8 aberrant strains was amplified and sequenced using the oacD-3 primer pair, and all were found to carry a dysfunctional oacD gene, with serotype 2b strain 2005001 having a one-base (T) insertion at position 177 and the others having a one-base (A) deletion at position 191; all resulted in a stop codon at amino acid 59 or 64, rendering the protein defective in these isolates.
The data obtained demonstrate that the 6-O-acetylation on GlcNAc confers to the host a previously unknown O-antigen epitope(s). Following the designations for S. flexneri group O-factors 3,4, O-factor 6, O-factors 7,8, and O-factor 9, we name the new antigenic determinant group O-factor 10 and suggest that it should be included in the current serotyping scheme of S. flexneri for antiserum 10-positive strains carrying the 6-O-acetylation on GlcNAc.
A total of 54 strains of other species were tested by slide agglutination and found to be antiserum 10 negative, except for E. coli strains 042 (serotype O44) and G1237 (serotype O13) as well as 12 strains of S. sonnei phase II (see Table S1 in the supplemental material). E. coli O13 reacted with antiserum 10, evidently because it has a S. flexneri serotype 2a-like O-antigen structure with 6-O-acetylation on GlcNAc (25). The positive oacD gene PCR amplification confirmed that a similar oacD-mediating mechanism is involved in this modification in E. coli O13. In contrast, the O-antigen of E. coli O44 is devoid of 6-O-acetylation on GlcNAc (40), and S. sonnei phase II has no O-antigen; therefore, further studies are necessary to elucidate the structural basis for their reactivity with antiserum 10.
DISCUSSION
Modifications of the O-antigen of S. flexneri give rise to the expression of enormously diverse O-antigenic determinants on the same O-polysaccharide backbone. So far, the following molecular factors involved with these modifications have been revealed: (i) serotype-converting prophages carrying a gtr locus for glucosylation of various monosaccharides or the single oac (oacA) gene for 2-O-acetylation of RhaI (6); (ii) transposon-like elements carrying oacB or oacC genes for 3/4-O-acetylation of RhaIII (28) (see the supplemental material); and (iii) plasmids carrying an optII or optIII gene for PEtN phosphorylation of RhaII and/or RhaIII (20, 23). One can speculate that the coexistence of several O-antigen modification mechanisms allows S. flexneri to change rapidly the antigenic landscape to escape the serotype-specific host immunity and to promote the spread of shigellosis in the human population.
In this work, it was found that serotype-converting bacteriophage SfII, which possesses the gtrII locus responsible for the 4-glucosylation of RhaI, also carries the oacD gene and that the corresponding prophage is responsible for the 6-O-acetylation of GlcNAc of S. flexneri. The following data elucidated the role of oacD in modification of S. flexneri O-antigens: (i) the occurrence of conserved domains of the acyltransferase family (Acyl_transf_3) in the predicted OacD protein and its similarity to acyltransferases; (ii) a clear correlation between the presence of the functional oacD gene and the 6-O-acetylation on GlcNAc in the O-antigen; (iii) deletion of oacD resulting in the loss of the 6-O-acetylation and the finding that cloned oacD mediated the 6-O-acetylation upon transformation; and (iv) the finding that naturally occurring dysfunctional mutations in oacD in some strains abolished the 6-O-acetylation.
This is the first report showing that one serotype-converting phage carries two factors involved in different types of O-antigen modifications in S. flexneri. Being located in the SfII genome, the oacD gene may be transferred, together with the gtrII locus, by the SfII infection mechanism among S. flexneri strains in nature. The coexistence of the gtrII locus and oacD in SfII might indicate that the two factors are functionally dependent, but the presence of a dysfunctional gtrII locus in 6-O-acetylation-positive strains showed that this modification can be done without the assistance of the gtr locus. This is supported by the acquisition of the 6-O-acetylation-positive phenotype upon transformation with the oacD expression vector of serotype Y strain 036, which has no known serotype-converting phage within the genome (our unpublished data).
The serotype-converting SfII bacteriophage is integrated into the host chromosome at the tRNA-thrW site located between conserved genes proA and adrA by lysogeny (6, 13). Our previous work demonstrated that the oacB-carrying transposon responsible for the 3/4-O-acetylation of RhaIII maps upstream of the adrA gene (28). Therefore, the genetic elements involved in glucosylation and two types of O-acetylation are located in the proA-adrA region in serotype 2 strains, suggesting that this site is the hot spot for mobilization of O-antigen modification factors in S. flexneri.
In various S. flexneri serotypes, the same monosaccharide may carry different chemical groups; for instance, a glucosyl, O-acetyl, or PEtN group is attached at position 3 of RhaIII in serotypes X, 1, and 4av, giving rise to O-factors 7,8, O-factor 9, and O-factor IV-1, respectively (8, 20, 29). Earlier, we demonstrated that the 3-glycosyl group on RhaIII in serotype 2b and X strains can be completely or (in serotype X) almost completely replaced with the 3- or 4-O-acetyl group (28) upon transformation with the oacB gene for the specific acetyltransferase. In this work, a similar replacement of the glucosyl group on GlcNAc, whether it occurred at position 4 or 6, with the 6-O-acetyl group was observed upon transformation of serotype 1a and 4b strains with the oacD gene. However, the replacement was only partial and the degree of 6-O-acetylation (∼25% to ∼30%) was lower than in S. flexneri strains with the natural occurrence of this modification (40% to 65%) (5, 24). Therefore, the interrelations between O-acetylation and glucosylation are distinct on different monosaccharides mediated by different acetyltransferase proteins: in the case of GlcNAc glucosylation and OacD-mediated O-acetylation, the modifications proceeded competitively, whereas on RhaIII, the OacB-mediated O-acetylation suppressed glucosylation. This competitiveness or suppression may be due to a high expression level of the acetyltransferase proteins as, in both cases, the cloned oac gene dosage is much higher (>500) than that of the chromosomal gtr locus.
Like the 3/4-O-acetylation on RhaIII, the 6-O-acetylation on GlcNAc confers to the host an additional O-antigen epitope, which had been neglected earlier owing to the absence of specific antiserum. This new epitope, which we provisionally name O-factor 10, is expressed in all serotype 2 strains as well as in strains of other serotypes that carry a cryptic SfII phage with a dysfunctional gtr locus. It should be noted that serotype 2 is one of the most predominant serotypes in China and other countries (41, 42), and the possibility is not excluded that the coexistence of 4-glucosylation on RhaI and 6-O-acetylation on GlcNAc, and coexpression of the corresponding type II and group 10 antigenic determinants, contributes to the prevalence of this serotype in nature.
Conventional Shigella serotyping tools revealed the same LPS serological pattern for each O-factor 10-positive strain and its negative counterpart; hence, the 6-O-acetylation on GlcNAc does not affect other antigenic determinants. Therefore, it would be expedient to add the group O-factor 10 into the current serotyping scheme of S. flexneri and to use antiserum 10 for detection of strains carrying the functional oacD gene and, accordingly, the 6-O-acetylation on GlcNAc.
The findings of this work enhance our understanding of the serotype conversion mechanisms in S. flexneri, which will assist epidemiological monitoring of S. flexneri and development of vaccines against shigellosis. Further studies are necessary to gain an insight into the impact of the expression of the 6-O-acetylation-linked epitope on the pathogenicity of S. flexneri.
Supplementary Material
ACKNOWLEDGMENTS
Y.A.K., S.N.S., and A.S.S. were supported by the Russian Science Foundation (no. 14-14-01042); Q.S., J.W., X.L., R.L., and J.X. were supported by the National Natural Science Foundation of China (no. 81271788 and 81290345), the National Basic Research Priorities Program (2011CB504901), and the National Key Program for Infectious Diseases of China (2013ZX10004221, 2013ZX10004216-001-002, and 2012ZX10004215).
Footnotes
Published ahead of print 11 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02009-14.
REFERENCES
- 1.Bardhan P, Faruque AS, Naheed A, Sack DA. 2010. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg. Infect. Dis. 16:1718–1723. 10.3201/eid1611.090934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West NP, Sansonetti P, Mounier J, Exley RM, Parsot C, Guadagnini S, Prevost MC, Prochnicka-Chalufour A, Delepierre M, Tanguy M, Tang CM. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313–1317. 10.1126/science.1108472 [DOI] [PubMed] [Google Scholar]
- 3.Köhler H, Rodrigues SP, McCormick BA. 2002. Shigella flexneri interactions with the basolateral membrane domain of polarized model intestinal epithelium: role of lipopolysaccharide in cell invasion and in activation of the mitogen-activated protein kinase ERK. Infect. Immun. 70:1150–1158. 10.1128/IAI.70.3.1150-1158.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong M, Payne SM. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24:779–791. 10.1046/j.1365-2958.1997.3731744.x [DOI] [PubMed] [Google Scholar]
- 5.Perepelov AV, Shekht ME, Liu B, Shevelev SD, Ledov VA, Senchenkova SN, L'Vov VL, Shashkov AS, Feng L, Aparin PG, Wang L, Knirel YA. 2012. Shigella flexneri O-antigens revisited: final elucidation of the O-acetylation profiles and a survey of the O-antigen structure diversity. FEMS Immunol. Med. Microbiol. 66:201–210. 10.1111/j.1574-695X.2012.01000.x [DOI] [PubMed] [Google Scholar]
- 6.Allison GE, Verma NK. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8:17–23. 10.1016/S0966-842X(99)01646-7 [DOI] [PubMed] [Google Scholar]
- 7.Foster RA, Carlin NI, Majcher M, Tabor H, Ng LK, Widmalm G. 2011. Structural elucidation of the O-antigen of the Shigella flexneri provisional serotype 88–893: structural and serological similarities with S. flexneri provisional serotype Y394 (1c). Carbohydr. Res. 346:872–876. 10.1016/j.carres.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 8.Kenne L, Lindberg B, Petersson K, Katzenellenbogen E, Romanowska E. 1978. Structural studies of Shigella flexneri O-antigens. Eur. J. Biochem. 91:279–284. 10.1111/j.1432-1033.1978.tb20963.x [DOI] [PubMed] [Google Scholar]
- 9.Verma NK, Brandt JM, Verma DJ, Lindberg AA. 1991. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol. Microbiol. 5:71–75. 10.1111/j.1365-2958.1991.tb01827.x [DOI] [PubMed] [Google Scholar]
- 10.Clark CA, Beltrame J, Manning PA. 1991. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene 107:43–52. 10.1016/0378-1119(91)90295-M [DOI] [PubMed] [Google Scholar]
- 11.Stagg RM, Tang SS, Carlin NI, Talukder KA, Cam PD, Verma NK. 2009. A novel glucosyltransferase involved in O-antigen modification of Shigella flexneri serotype 1c. J. Bacteriol. 191:6612–6617. 10.1128/JB.00628-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q, Lan R, Wang Y, Wang J, Li P, Du P, Xu J. 2013. Isolation and genomic characterization of SfI, a serotype-converting bacteriophage of Shigella flexneri. BMC Microbiol. 13:39. 10.1186/1471-2180-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavris M, Manning PA, Morona R. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26:939–950. 10.1046/j.1365-2958.1997.6301997.x [DOI] [PubMed] [Google Scholar]
- 14.Guan S, Bastin DA, Verma NK. 1999. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145:1263–1273. 10.1099/13500872-145-5-1263 [DOI] [PubMed] [Google Scholar]
- 15.Allison GE, Angeles D, Tran-Dinh N, Verma NK. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974–1987. 10.1128/JB.184.7.1974-1987.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adhikari P, Allison G, Whittle B, Verma NK. 1999. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexneri chromosome. J. Bacteriol. 181:4711–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams MM, Allison GE, Verma NK. 2001. Type IV O antigen modification genes in the genome of Shigella flexneri NCTC 8296. Microbiology 147:851–860 [DOI] [PubMed] [Google Scholar]
- 18.Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kuhlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. 2004. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339:379–394. 10.1016/j.jmb.2004.03.068 [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, Lan R, Wang Y, Wang J, Luo X, Zhang S, Li P, Ye C, Jing H, Xu J. 2011. Genesis of a novel Shigella flexneri serotype by sequential infection of serotype-converting bacteriophages SfX and SfI. BMC Microbiol. 11:269–274. 10.1186/1471-2180-11-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, Jin D, Shashkov AS, Xia SAV, Perepelov Chen Q, Wang Y, Wang HJJX. 2012. A novel plasmid-encoded serotype conversion mechanism through addition of phosphoethanolamine to the O-antigen of Shigella flexneri. PLoS One 7:e46095. 10.1371/journal.pone.0046095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Lan R, Wang J, Xia S, Wang Y, Jin D, Yu B, Knirel YA, Xu J. 2013. Identification and characterization of a novel Shigella flexneri serotype Yv in China. PLoS One 8:e70238. 10.1371/journal.pone.0070238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perepelov AV, L'vov VL, Liu B, Senchenkova SN, Shekht ME, Shashkov AS, Feng L, Aparin PG, Wang L, Knirel YA. 2009. A new ethanolamine phosphate-containing variant of the O-antigen of Shigella flexneri type 4a. Carbohydr. Res. 344:1588–1591. 10.1016/j.carres.2009.03.022 [DOI] [PubMed] [Google Scholar]
- 23.Knirel YA, Lan R, Senchenkova SN, Wang J, Shashkov AS, Wang Y, Perepelov AV, Xiong Y, Xu J, Sun Q. 2013. O-antigen structure of Shigella flexneri serotype Yv and effect of the lpt-O gene variation on phosphoethanolamine modification of S. flexneri O-antigens. Glycobiology 23:475–485. 10.1093/glycob/cws222 [DOI] [PubMed] [Google Scholar]
- 24.Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, Shashkov AS, Wang Y, Wang Y, Luo X, Xu J. 2014. Dissemination and serotype modification potential of pSFxv_2, an O-antigen PEtN modification plasmid in Shigella flexneri. Glycobiology 24:305–313. 10.1093/glycob/cwt115 [DOI] [PubMed] [Google Scholar]
- 25.Perepelov AV, Shevelev SD, Liu B, Senchenkova SN, Shashkov AS, Feng L, Knirel YA, Wang L. 2010. Structures of the O-antigens of Escherichia coli O13, O129, and O135 related to the O-antigens of Shigella flexneri. Carbohydr. Res. 345:1594–1599. 10.1016/j.carres.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 26.Perepelov AV, L'Vov VL, Liu B, Senchenkova SN, Shekht ME, Shashkov AS, Feng L, Aparin PG, Wang L, Knirel YA. 2009. A similarity in the O-acetylation pattern of the O-antigens of Shigella flexneri types 1a, 1b, and 2a. Carbohydr. Res. 344:687–692. 10.1016/j.carres.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 27.Kubler-Kielb J, Vinogradov E, Chu C, Schneerson R. 2007. O-acetylation in the O-specific polysaccharide isolated from Shigella flexneri serotype 2a. Carbohydr. Res. 342:643–647. 10.1016/j.carres.2006.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Knirel YA, Lan R, Senchenkova SN, Luo X, Perepelov AV, Wang Y, Shashkov AS, Xu J, Sun Q. 2014. Identification of an O-acyltransferase gene (oacB) that mediates 3- and 4-O-acetylation of rhamnose III in Shigella flexneri O antigens. J. Bacteriol. 196:1525–1531. 10.1128/JB.01393-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Lan R, Knirel YA, Luo X, Senchenkova SN, Shashkov AS, Xu J, Sun Q. 2014. Serological identification and prevalence of a novel O-antigen epitope linked to 3- and 4-O-acetylated rhamnose III of lipopolysaccharide in Shigella flexneri. J. Clin. Microbiol. 52:2033–2038. 10.1128/JCM.00197-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. 10.1016/0378-1119(87)90095-3 [DOI] [PubMed] [Google Scholar]
- 31.Pradel N, Ye C, Livrelli V, Xu J, Joly B, Wu LF. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908–4916. 10.1128/IAI.71.9.4908-4916.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shashkov AS, Senchenkova SN, Sun Q, Lan R, Wang J, Perepelov AV, Knirel YA, Xu J. 2013. Structure of the O-antigen of a novel Shigella flexneri serotype, 1d (I: 7,8). Carbohydr. Res. 373:93–96. 10.1016/j.carres.2013.03.015 [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 34.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freter R. 1957. Agglutinating efficiency and combining capacity of Shigella and Vibrio antisera from rabbits at different stages of immunization. J. Exp. Med. 105:623–634. 10.1084/jem.105.6.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morona R, Brown MH, Yeadon J, Heuzenroeder MW, Manning PA. 1991. Effect of lipopolysaccharide core synthesis mutations on the production of Vibrio cholerae O-antigen in Escherichia coli K-12. FEMS Microbiol. Lett. 66:279–285 [DOI] [PubMed] [Google Scholar]
- 37.Daniels C, Morona R. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181–194. 10.1046/j.1365-2958.1999.01591.x [DOI] [PubMed] [Google Scholar]
- 38.Westphal O, Jann K. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83–90 [Google Scholar]
- 39.George DT, Stephenson DP, Tran E, Morona R, Verma NK. 2013. Complete genome sequence of SfII, a serotype-converting bacteriophage of the highly prevalent Shigella flexneri serotype 2a. Genome Announc. 1:e00626-13. 10.1128/genomeA.00626-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staaf M, Widmalm G, Weintraub A, Nataro JP. 1995. Structural elucidation of the O-antigenic polysaccharide from Escherichia coli O44:H18. Eur. J. Biochem. 233:473–477. 10.1111/j.1432-1033.1995.473_2.x [DOI] [PubMed] [Google Scholar]
- 41.von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, Canh do, Chaicumpa GW, Agtini MD, Hossain A, Bhutta ZA, Mason C, Sethabutr O, Talukder K, Nair GB, Deen JL, Kotloff K, Clemens J. 2006. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353. 10.1371/journal.pmed.0030353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye C, Lan R, Xia S, Zhang J, Sun Q, Zhang S, Jing H, Wang L, Li Z, Zhou Z, Zhao A, Cui Z, Cao J, Jin D, Huang L, Wang Y, Luo X, Bai X, Wang P, Xu Q, Xu J. 2010. Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. J. Clin. Microbiol. 48:419–426. 10.1128/JCM.00614-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.