Abstract
Thermoacidophilic archaea, such as Metallosphaera sedula, are lithoautotrophs that occupy metal-rich environments. In previous studies, an M. sedula mutant lacking the primary copper efflux transporter, CopA, became copper sensitive. In contrast, the basis for supranormal copper resistance remained unclear in the spontaneous M. sedula mutant, CuR1. Here, transcriptomic analysis of copper-shocked cultures indicated that CuR1 had a unique regulatory response to metal challenge corresponding to the upregulation of 55 genes. Genome resequencing identified 17 confirmed mutations unique to CuR1 that were likely to change gene function. Of these, 12 mapped to genes with annotated function associated with transcription, metabolism, or transport. These mutations included 7 nonsynonymous substitutions, 4 insertions, and 1 deletion. One of the insertion mutations mapped to pseudogene Msed_1517 and extended its reading frame an additional 209 amino acids. The extended mutant allele was identified as a homolog of Pho4, a family of phosphate symporters that includes the bacterial PitA proteins. Orthologs of this allele were apparent in related extremely thermoacidophilic species, suggesting M. sedula naturally lacked this gene. Phosphate transport studies combined with physiologic analysis demonstrated M. sedula PitA was a low-affinity, high-velocity secondary transporter implicated in copper resistance and arsenate sensitivity. Genetic analysis demonstrated that spontaneous arsenate-resistant mutants derived from CuR1 all underwent mutation in pitA and nonselectively became copper sensitive. Taken together, these results point to archaeal PitA as a key requirement for the increased metal resistance of strain CuR1 and its accelerated capacity for copper bioleaching.
INTRODUCTION
Thermoacidophilic archaea include taxa that are lithoautotrophs with high levels of metal resistance, such as Sulfolobus, Metallosphaera, and Acidianus (1, 2). They occur in environments that are sulfur rich and pyritic, including mining-related heaps of extracted ores. These organisms are of particular importance in the extraction of base and precious metals through the bioleaching process (3). Metal bioleaching has been proposed to use both direct and indirect mechanisms that convert metals to soluble forms (4, 5). Metal release may occur via biooxidation of the sulfur and iron component of minerals, thereby releasing complexed metals, or by direct metal oxidation involving cell contact with metal surfaces (6). Advances in understanding this type of lithoautotrophic metabolism are coming from genome sequencing of biomining organisms (7–10), transcriptomic analyses (10–12), and metagenomic investigations (13).
Copper resistance mechanisms among the archaea are understood primarily from genome sequencing, where the distribution of CopA (P-type ATPase) copper efflux transporters is broad (1, 3). Biochemical studies have been conducted on CopA from the sulfate reducer Archaeoglobus fulgidus (14, 15) and CopB from Sulfolobus solfataricus (16, 17). Multiple mechanisms of copper resistance also have been examined using genetic and molecular approaches in S. solfataricus, including copA and copB (P-type ATPase efflux pumps), copR (transcriptional regulator), and copT (a putative metallochaperone) (16, 18–20). An inorganic polyphosphate symport system with importance in copper resistance also has been identified in Sulfolobus metallicus (21). Intracellular polyphosphate hydrolysis catalyzed by exopolyphosphatase (Ppx) could provide a counter ion for metal complexation whose subsequent symport through a phosphate efflux system would reduce metal burden. Interestingly, polyphosphate is produced by both chemoheterotrophic and lithoautotrophic thermoacidophilic archaea, but only the latter organisms accumulate significant levels, perhaps in concert with increased levels of metal resistance (21, 22).
Metallosphaera sedula is a lithoautotrophic archaeon that grows optimally at 75°C over a pH range of 2.0 to 4.5 (23). It differs greatly from its relatives, S. solfataricus and S. acidocaldarius, because these organisms are strict chemoheterotrophs (1, 24). Autotrophy in M. sedula uses a modified 3-hydroxypropionate cycle for carbon fixation (25, 26). M. sedula also is more resistant to metals than chemoheterophic Sulfolobus species, perhaps as a consequence of its metal-rich habitat (19, 23, 27). The first genetic system for M. sedula was developed recently to investigate parameters that limit bioleaching of chalcopyrite (CuFeS2) (27). Cross-species complementation using a S. solfataricus copR mutant (19) demonstrated the M. sedula copRTA operon was active and that gene expression signals are conserved between these genera. Inactivation of the M. sedula copper efflux protein, copA, using targeted recombination based on the pyrE genetic marker compromised metal resistance and eliminated copper bioleaching from chalcopyrite. In contrast, a spontaneous M. sedula strain, called CuR1, that had supranormal metal resistance leached copper from chalcopyrite at an accelerated rate. Taken together, these data showed that metal resistance was integral to copper bioleaching. While CuR1 exhibited normal growth rates and cell yields under chemoheterotrophic conditions, transcriptomic analysis identified changes in the expression of 22 proteins by 3-fold or more and 13 proteins by 5-fold or more 15 min after copper shock than the wild type grown under the same conditions. These included putative transporter-encoding AAA-ATPase motifs. In this study, transcriptomics, genome resequencing, solute transport, and classical genetics were used to identify regulatory and genetic differences resulting in extreme copper resistance of this cell line, including a specific role for phosphate transport.
MATERIALS AND METHODS
Archaeal strains, cultivation, and microscopy.
Cell lines used in this study included Metallosphaera sedula (DSM 5348T) (wild type) and the CuR1 copper-resistant derivative (27). They were grown in a basal salts medium (BSM) (28) adjusted to pH 2.0 using sulfuric acid. The cultures were incubated at 75°C in either glass screw-cap flasks with aeration in orbital baths or in glass screw-cap test tubes, which were placed in rotary drum agitators that were mounted in incubators that had external DC motors. Chemoheterotrophic growth used tryptone at 0.05% (wt/vol). Cultures were frozen at −80°C as described for other thermoacidophiles (29). Prior to inoculation into liquid medium, cultures were streaked on a solid complex medium prepared using BSM mixed with 0.6% (wt/vol) gelrite (Kelco) and supplemented with 0.05% (wt/vol) tryptone. Planktonic growth was monitored by light adsorption at a wavelength of 540 nm. Cultures were supplemented with different concentrations of potassium arsenate (Sigma) to determine its effect on growth in both strains. Arsenate-adapted derivatives of CuR1 were isolated by growing the cells in 15 mM potassium arsenate for two growth cycles and then subculturing into 20 mM arsenate followed by 25 mM arsenate. To isolate arsenate-resistant mutants of CuR1, CuR1 cultures that grew in 25 mM arsenate were plated on solid complex medium (without arsenate), and colonies were screened for the ability to grow in the presence of 25 mM arsenate upon inoculation back into liquid medium. The Msed_1517 allele of five of these arsenate-resistant isolates were determined.
Transcriptomics methods.
For transcriptomic experiments, M. sedula (DSM 5348T) (wild type) and the CuR1 copper-resistant derivative were cultured aerobically in DSM88 medium adjusted to pH 2.0 using sulfuric acid, supplemented with 0.05% (wt/vol) tryptone and 25 mg/liter uracil, and incubated at 70°C in an oil bath with a shaking rate of 100 rpm. The first global transcriptional response experiment was designed to compare wild-type and CuR1 strains under normal growth conditions (i.e., no copper challenge). The second was aimed at comparing the response of wild-type and CuR1 strains under subinhibitory copper challenge (2.0 mM copper sulfate). For the normal condition comparison, cells were grown to a cell density of approximately 1.0 × 108 cells/ml (optical density at 540 nm [OD540], 0.1) and harvested, while for the copper challenge comparison, cells were grown to a similar density shocked by addition of 2.0 mM copper and harvested 15 and 60 min postshock. For both experiments, cells were harvested by quickly chilling, centrifuging at 6,080 × g for 10 min, and washing the cell pellet with Tris-EDTA buffer to remove residual media. Epifluorescence microscopy, using acridine orange stain, was used for determination of cell densities.
RNA was extracted by phase separation with TRIzol reagent (Invitrogen), purified using an RNeasy minikit (Qiagen), reverse transcribed with Superscript III (Invitrogen), repurified, labeled with either Cy3 or Cy5 dye (GE Healthcare), and hybridized onto the M. sedula array, constructed as previously described (7, 12, 30). RNA from biological replicates was pooled prior to the reverse transcription to cDNA. Slides were scanned using a Genetix 4000B scanner, and average intensities for all spots were normalized using an analysis of variance (ANOVA) mixed-effects model and analyzed using JMP Genomics (SAS Institute, Cary, NC). Bonferroni's correction was utilized to determine the statistical significance of differential transcription, and it was 5.00 for the normal condition and 5.53 for the copper challenge.
Genome sequencing and bioinformatics.
High-molecular-weight genomic DNA was prepared from clonal cultures of M. sedula strains as described previously (7, 29), and its integrity and purity were confirmed by agarose gel electrophoresis and spectroscopy. The genomic DNA samples were dissolved in 10 mM Tris-Cl, pH 8.5, and sonicated using a Misonix ultrasonic liquid processor (model 4000) at a setting of 7 for 30 s with 1-min intervals at 20% power output. The sheared DNA ranged in size from 100 bp to 800 bp, with a peak area of 250 to 500 bp, and was used to prepare DNA libraries. DNA was sequenced using an Illumina GAIIx to obtain 75-bp single-end reads using Illumina FC-102-1001 sequence kits, a TruSeq SR cluster generation kit, v5-SCS (v2.8 protocol), and two 36-cycle TruSeq sequencing kits, v5 (v8 protocol). M. sedula wild-type strain sequencing yielded 32 million 75-bp reads (2.3 Gb), while the mutant strain, CuR1, had 25 million 75-bp reads (1.8 Gb). The genomes of both strains then were assembled by de novo assembly. Raw sequence data files were compiled into single FASTq files for each M. sedula strain. Initial contigs were assembled from these files using VELVET (version 1.2.07), a sequence assembler for very short reads (31), while k-mer length settings were manually set to 49. The outputs from Velvet were 70 contigs ranging from <100 to nearly 250,000 bp for each genome. Full genomes were assembled from the contigs using the CODON CODE ALIGNER genome assembly program (CodonCode). Mutations that were located within open reading frames and identified through sequence comparisons were analyzed in more detail by cross-referencing the genome coordinates and type of mutation at each position to these same positions in the NCBI gene database to determine their codon positions and the effects they would have on protein sequence. The coordinates of each mutation also were cross referenced to locations of the known domains in each protein to verify whether or not any mutations occurred within important functional domains. Nonsynonymous changes that were found then were verified by PCR and sequencing, followed by comparison to the genome resequencing results of the same region.
In addition, a guided assembly was conducted using both sets of genome sequence reads and were aligned uniquely via bowtie2 (version 2.0.0-beta6) to the reference Metallospaera sedula DSM 5348 genome (RefSeq accession no. NC_009440.1). Matching reads were extracted via SAMtools (version 0.1.18) and converted into regions via bedtools (version 2.16.1), and FASTA sequences of these regions were extracted from NC_009440.1 to serve as the reference regions for the genome assembly. Velvet (version 1.2.07) (31) was used again for the genome assemblies with a k-mer value of 57, the extracted reference in the case of the guided assembly, and automatic determination of expected coverage and cutoffs, as these were very similar to manual cutoffs determined by visual inspection (wild type, expected coverage of 291 and cutoff of 145; CuR1, expected coverage of 219 and cutoff of 109). The resulting contigs for each assembly were reordered against NC_009440.1 via the progressive Mauve tool (version 2.0), and a draft genome compiled from the contigs was interspersed with lowercase N nucleotides to the estimated size of any gaps in the genome compared to the reference (in-house Perl script).
Phosphate transport assays.
Cells were grown to mid-log phase in BSM containing 0.05% tryptone and then washed twice and resuspended in uptake media (BSM lacking phosphate). After a 5-min incubation at 75°C, 10 μCi of 32PO43− (specific activity, 900 to 1,100 mCi/mmol; PerkinElmer) was added to the suspension, and the suspension was sampled in 0.5-ml amounts every 15 s over a period of 1 min. The samples were applied to 0.45-μm filters (Whatman Nuclepore track-etch membrane) supported on a filtration manifold. The filters containing the labeled cells then were washed with 3 ml of 100 mM LiCl, dried, and placed in scintillation vials with 5 ml of scintillation fluid (EcoLite). Radioactivity was measured using a Packard liquid scintillation counter. Gramicidin (32) was added to the cells to a final concentration of 12.5 μM immediately prior to the 5-min preincubation period and before 32PO43− addition. Phosphate transport rates were determined as described previously (33–35) and were determined over a range of 0 to 200 μM external phosphate. Phosphate transport rates also were determined over a range of 0 to 200 μM potassium arsenate at a constant phosphate concentration of 200 μM. Km values for phosphate and Ki values for arsenate for both strains were extrapolated from a Michaelis-Menten plot. All rates were verified in replicate.
Nucleotide sequence and transcriptomic data accession numbers.
The M. sedula strain CuR1 genome sequence is available at GenBank under the accession number CP008822. The transcriptomic data for wild-type M. sedula and CuR1 are available at the Gene Expression Omnibus under the following accession numbers: GSM1431885, GSM1431886, GSM1431887, GSM1431888, and GSM1431889.
RESULTS
Transcriptomics of copper shock.
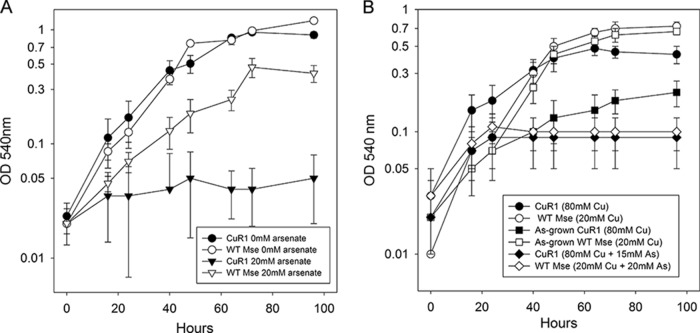
M. sedula strain CuR1 has increased resistance to copper relative to that of the wild-type strain (27). When an exponentially growing culture of this strain was challenged by addition of 75 mM copper [cupric ion, or Cu(II)], there was no apparent change in growth rate. In contrast, when a wild-type culture was challenged at the same metal dose, there was a prolonged lag phase eventually followed by the resumption of growth (Fig. 1A). This demonstrated that increased copper resistance in strain CuR1 does not depend on a period of adaptation but instead occurs in a constitutive manner. The immediate demonstration of resistance suggested genes involved in this trait will be evident through transcriptomic analysis without prior metal challenge. Transcriptomic analysis was conducted initially using RNA from cultures of exponentially growing cells in media lacking high levels of copper. Only two genes were upregulated in CuR1 relative to the wild type: Msed_1525, a putative peroxiredoxin (cd3016 PFAM0578), and Msed_0842, a hypothetical protein. Both were expressed at levels 2-fold above those in the wild-type strain. As orthologs of both genes are in the genomes of the copper-sensitive organisms S. solfataricus and S. acidocaldarius, they were not pursued further. Instead, other mechanisms were considered, such as a rapid regulatory response to metal challenge that resulted in high metal resistance. Transcriptomic analysis was conducted by challenging both strains with 2.0 mM copper, a relatively low dose of copper that did not inhibit the growth of either strain (Fig. 1B). The regulatory response to copper shock, measured after 15 min and 60 min, revealed distinct differences between the wild type and CuR1 M. sedula strains (Fig. 2). Two patterns were evident: a more rapid response to metal challenge in strain CuR1 and hyperinduction of several genes. The most responsive gene in the comparison between the wild-type and CuR1 strains 15 min after metal addition was Msed_1076, a hypothetical protein that showed 47.3-fold upregulation in CuR1. This protein contains a lycopene cyclase region/domain that might have a role in carotenoid biosynthesis. After both 15 min and 60 min, the primary copper efflux protein, Msed_0490, was 3-fold upregulated in CuR1. The expression of additional transporters that could promote metal efflux also was increased in CuR1, including Msed_0805, Msed_0960, and Msed_1772. Their increased expression ranged from 5- to 9-fold. In addition, two differentially expressed transcription factors were identified. Msed_1818 was upregulated 4.6-fold in CuR1 15 min after copper challenge. This gene is annotated as a bacterium-opsin activator with an HTH domain and may be a metal response transcriptional regulator. Similarly, Msed_2265 was upregulated 2-fold in CuR1 60 min after metal challenge (not shown). This gene also responds 15 min after metal challenge but with 6.3-fold upregulation (Fig. 2). Msed_2265 is annotated as a zinc finger TFIIB-type domain-containing protein and is a transcription initiation factor associated with RNA polymerase II. The expression of two other functional classes of proteins also were noted. These consisted of hydrolases (GTPases and phosphatases), including Msed_0499, Msed_0050, Msed_2061, and Msed_2226, whose increased expression ranged from 4- to more than 10-fold. Finally, a second group of proteins with probable antioxidant activity (in addition to Msed_1076), along with class II toxin and antitoxin genes, were identified. These included Msed_0014, Msed_0015, Msed_0046, Msed_0047, and Msed_0048. Increased expression of toxins could position the cell for rapid readjustment in transcript abundance as part of an overall response to metal challenge.
FIG 1.
Response of M. sedula and CuR1 to copper challenge. (A) Effect of 75 mM copper [Cu(II)]. WT Mse, wild-type M. sedula. (B) Effect of 2.0 mM copper shock.
FIG 2.
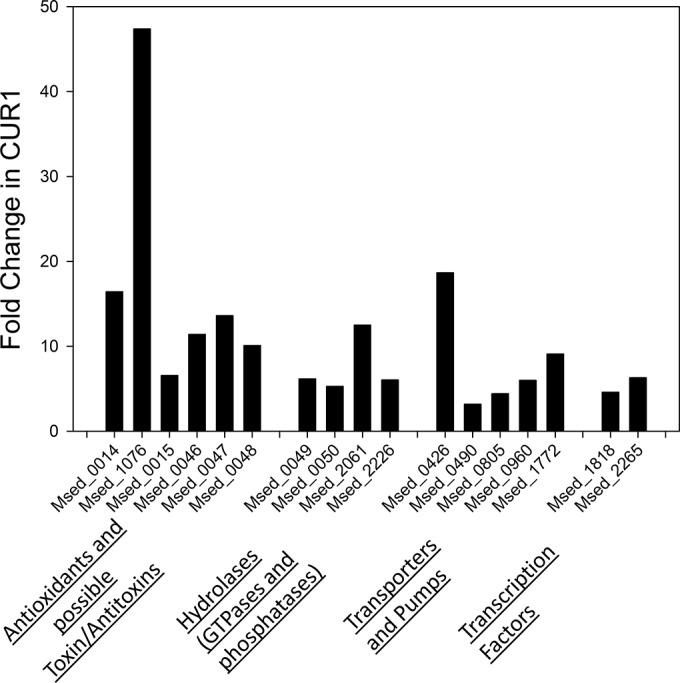
Transcriptomic profile of CuR1 in response to copper shock. Fold changes in CuR1 are relative to the level for the wild type and represent observed differences at 15 min postshock. In total, 40 genes showed increased expression (>2-fold), while only 1 showed decreased expression (>2-fold) relative to the wild type. Classes of affected genes are shown below the figure.
Genome resequencing.
Differences in gene expression patterns between the wild-type and CuR1 strains could be caused by mutations arising during the isolation of CuR1. Therefore, genome resequencing was conducted to map possible sequence differences between these strains. At a genome size of 2.19 Mb (7), this resulted in depths of sequence coverage for the two strains of 1,050-fold and 821-fold, respectively (Table 1). Since the goal was to identify genetic changes causing the metal-resistant CuR1 phenotype, the only mutations that were examined were those that were distinct between the CuR1 strain, the resequenced wild-type genome, and the previously determined M. sedula genome deposited at NCBI (NC_009440.1, GI:146302785). Of these, 20 were pursued further because they were most likely to alter gene function and were not merely synonymous changes in open reading frames or in intergenic regions. DNA spanning these 20 sequence differences then was PCR amplified from the CuR1 genome and resequenced to verify their occurrence. Seventeen of these putative mutations were confirmed. While 5 of these mutations mapped to unannotated open reading frames (ORFs), 12 mapped to 11 genes with annotated function. These included 7 nonsynonymous substitutions, 4 insertions, and 1 deletion. A tabulated summary is presented of the confirmed mutations along with gene annotation, gene/protein length, numerical position of the mutational change, and its proximity to identified domains and/or motifs (Table 2).
TABLE 1.
Summary of sequence coverage and mutations
| M. sedula strain | Depth of coverage by Illumina reads (fold) | No. of substitutions in ORFs | No. of substitutions in intergenic regions |
|---|---|---|---|
| Wild type | 1,050 | 24 | 3 |
| CuR1 | 821 | 17a | 3 |
No overlap with mutations detected in wild-type M. sedula.
TABLE 2.
Confirmed mutations in CuR1
| Genome coordinate | Substitution and/or description | Msed ORF | Protein | Mutation location | Domain affected |
|---|---|---|---|---|---|
| 155,137 | C→T, nonsynonymous (Glu→Lys) | 0191 | Ornithine cyclodeanimase | nt 691/939; aa 231/312 | NADB Rossman superfamily |
| 448,008 | C→A, nonsynonymous (Gly→Val) | 0490 | Heavy-metal translocating ATPase | nt 5/2235; aa 2/744 | Leader sequence |
| 500,688 | C→T, nonsynonymous (Glu→Lys) | 0545 | Hypothetical | nt 187/207; aa 63/68 | |
| 898,334 | C→T, nonsynonymous (Pro→Ser) | 0972 | Major facilitator transporter | nt 1009/1164; aa 337/387 | MFS superfamily (in region with putative substrate translocation pores) |
| 1,187,052 | 1-nt insertion of G; frameshift starting at aa 160 and premature stop after aa 199 | 1213 | Pyruvate dehydrogenase | nt 478/1035; aa 160/344 | TPP enzyme superfamily |
| 1,255,967 | C→T, nonsynonymous (Ser→Leu) | 1279 | Transcriptional regulator, MarR family | nt 344/453; aa 115/150 | HTH superfamily |
| 1,318,092 | 1-nt deletion; frameshift starting at aa73 | 1343 | Hypothetical (close BLAST hit to nucleotide binding domain-containing protein) | nt 217/243; aa 73/81 | Outside domains (close to C-terminal end of protein) |
| 1,314,441 | T→C, nonsynonymous (Leu→Pro) | 1338 | Hypothetical (close hit to bacterial regulatory protein, ArsR family) | nt 245/846; aa 82/281 | Outside domains |
| 1,377,170 | C→T, nonsynonymous (Tyr→Asn) | 1408 | Hypothetical | nt 175/309; aa 59/103 | |
| 1,377,775 | T→G, nonsynonymous (Thr→Pro) | 1409 | Hypothetical | nt 46/474, 16/157 | |
| 1,397,090 | T→G | 1427 | Pseudogene | nt 597/1217 | |
| 1,479,994 | T→C, nonsynonymous (Leu→Ser) | 1516 | Hypothetical (low-score BLAST hit to sodium-dependent phosphate transporter) | nt 806/975; aa 269/374 | No conserved domains in protein |
| 1,481,054 | 1-nt insertion of G; extends ORF by 627 nt | 1517 | Pseudogene | nt 170/854 | Extends ORF to include PHO4 superfamily |
| 1,783,624-1,783,625 | 2-nt insertion | 1838 | Pseudogene (upregulated in transcriptomic data but no coding sequence) | nt 631/2282 | Pseudogene |
| 1,783,847 | 1-nt insertion | 1838 | Pseudogene | nt 855/2282 | Pseudogene |
| 1,784,992 | T→C | 1838 | Pseudogene | nt 1838/2282 | Pseudogene |
| 1,827,819 | T→C, nonsynonymous (Leu→Ser) | 1884 | Glutamate synthase | nt 1512/2139; aa 504/712 | Outside domains |
Mutations in the CuR1 genome were identified in two transcription factors, Msed_1279 and Msed_1338, both annotated as multiple antibiotic resistance regulator (MarR) family members. These changes could be responsible for the differences in transcriptional response to copper shock. The MarR family regulates the expression of proteins conferring resistance to oxidative stress agents, multiple antibiotics, organic solvents, and pathogenic factors (36, 37). While the mutation in Msed_1338 resulted in the loss of a charged residue (Glu133 to Gly133), it did not map to the putative MarR family domain. In contrast, mutation of Msed_1279, another MarR family member belonging to the OhrR subfamily, was mutated from Ser115 to Leu115 within alpha helix 5 of the MarR domain. This change could impact DNA binding and alter transcription factor activity.
Two distinct genes were mutated in CuR1 encoding proteins with important metabolic function. Msed_1213 encodes the alpha subunit of pyruvate dehydrogenase that catalyzes conversion of pyruvate into acetyl coenzyme A (CoA). Msed_1213 underwent a frameshift mutation resulting from insertion of a G and corresponding premature chain termination such that the protein was reduced from 344 to 199 amino acids (aa). This event ablated the conserved domain. However, pyruvate-ferredoxin oxidoreductase may provide an alternative route for acetyl-CoA formation, as demonstrated in S. solfataricus (36, 37). Orthologs of this enzyme are found in M. sedula as Msed_0305, Msed_0306, Msed_0307, Msed_0308, and Msed_0309. Msed_0191 encodes ornithine cyclodeaminase, which is required for proline biosynthesis. In addition, Msed_0191 had a nonsynonymous mutation replacing Glu230 with Lys (acidic to basic) within the region coding for the NADB-Rossman superfamily domain that overlaps both the ornithine cyclodeaminase and amino acid deaminase domains. This event may change proline abundance and resistance to metals (38).
Several genes were mutated in CuR1-encoding proteins involved in phosphorylation. Msed_1343 contains the P-loop_NTPase superfamily domain, and the adjacent Msed_1342 has homology to chromosome partitioning proteins, suggesting these genes are involved in cell division processes. There is an 8-bp overlap between the Msed_1343 and Msed_1344 genes. The 1-nt deletion within Msed_1343 extends the reading frame an additional 195 aa. Msed_0545 is a small gene (68 amino acids) whose closest BLAST hit to a nonhypothetical protein was to an ROK (repressor, ORF, kinase) family protein. ROK is a RhoA-binding kinase that plays a role in actin and other facets of cytoskeletal reorganization (39). Expression studies have shown that upregulating ROK promotes formation of stress fibers and adhesion complexes, while an N-terminally truncated ROK instead induces disassembly of these.
The genes of the last functional group that were mutated in CuR1 were involved in transport. Msed_0972 is annotated as a major facilitator transporter, but its substrate is not known. The mutation in this gene changed Ala28 to Val within the major facilitator superfamily (MFS) domain. The second mutated transporter was Msed_0490, which encodes the CopA efflux protein required for copper resistance in M. sedula (27). A nonsynonymous mutation occurred within the leader sequence, changing Gly18 to Val. Since this region is likely to be removed upon membrane insertion, its impact on efflux activity should be nominal. The third transporter that underwent a mutation, Msed_1516, had a change of Leu218 to Ser. Msed_1516 shares homology with the Pho80 family of phosphate transporters.
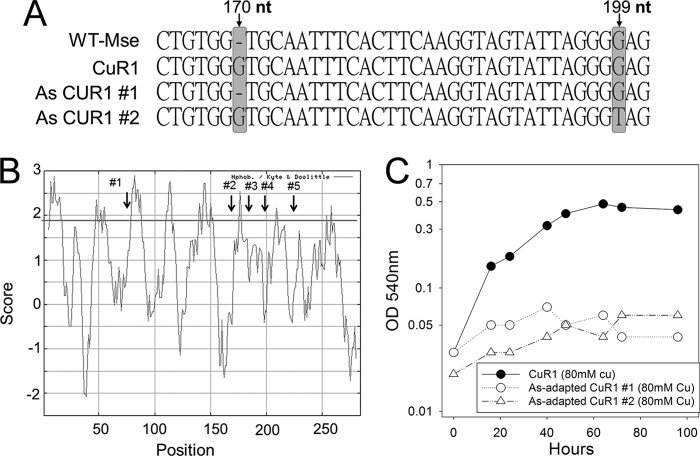
The third example of a transporter that was altered in CuR1 involves Msed_1517. In this case, however, the mutation appeared to result in a gain of function. In the genome sequence of M. sedula, Msed_1517 is annotated as a pseudogene, and this was verified by genome resequencing of this strain. The pseudogene could produce a 75-aa protein. In strain CuR1, a 1-nucleotide (nt) insertion of a G at position 170 reverted a natural frameshift, thereby extending the coding sequence an additional 640 nucleotides. The deduced amino acid sequence encoded by the extended allele has high identity to the Pho4 superfamily of phosphate transporters along with strong identity to the PitA domain; over a coding length of 305, residues of this sequence matched the PitA (COG0306) phosphate/sulfate permeases (e value of 9.22e−29). The restored Msed_1517 coding sequence identified in strain CuR1 also had high identity to homologs conserved in other Metallosphaera species. These alignments indicated the CuR1 Msed_1517 allele remained truncated at the C-terminal end, as the reading frames of homologous sequences in related species were 36 amino acids longer (Fig. 3).
FIG 3.
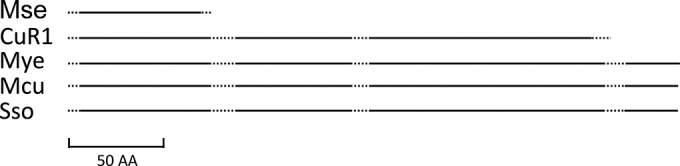
Amino acid alignment of Msed_1517 variants from thermoacidophiles. The line length corresponds to the open reading frame. The solid portions indicate regions of conservation, and the dashed portions indicate regions of divergence. Mse, Metallosphaera sedula DSM5348T; CuR1, spontaneous copper resistant derivative of M. sedula; Mye, Metallosphaera yellowstonensis MK1; Mcu, Metallosphaera cuprina Ar-4; Sso, Sulfolobus solfataricus 98/2.
Characterization of phosphate transport.
To determine whether Msed_1517 was a phosphate transport protein, rates of radiolabeled phosphate uptake were determined for CuR1 versus wild-type M. sedula under physiologic conditions (pH 2.5, 75°C). Km values for phosphate uptake were 2 μM for wild-type M. sedula and 36 μM for CuR1. Vmax values (nmol/min) for the wild-type and CuR1 strains at the phosphate concentrations indicated in parentheses were 2.4 (0 μM), 11.8 (2 μM), 14.25 (20 μM), and 22.55 (200 μM). These values are similar to previously reported values for PitA in E. coli (33) and describe a low-affinity, high-velocity phosphate transport protein. A second hallmark of the secondary transporter PitA is its reliance on the proton motive force for energy (34). Gramicidin D treatment was used as described previously (32) to measure the impact on phosphate uptake through dissipation of the proton motive force. Treatment for 5 min at 12.5 μM concentration prior to addition of labeled phosphate reduced phosphate uptake to 16% of that of untreated cells for CuR1 and to 61% of that of untreated cells for the wild type, indicating a greater reliance of the PitA system on proton motive force than other unidentified phosphate transporters in this organism.
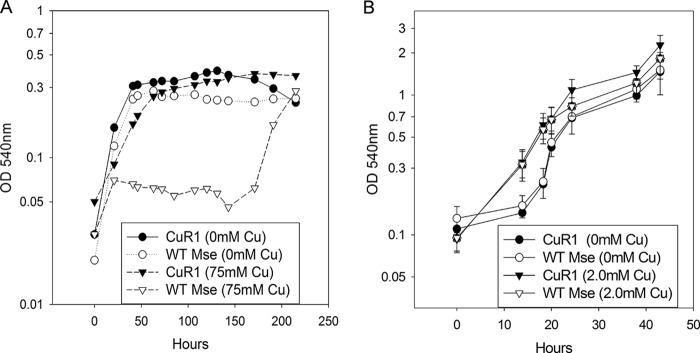
Studies of E. coli have shown that PitA transports arsenate in addition to phosphate and that PitA mutants are arsenate resistant (35). Rates of radiolabeled phosphate uptake also were measured under different arsenate concentrations to determine if there was competition for phosphate uptake by arsenate. The Ki values for arsenate were 16 μM for the wild type and 32.4 μM for CuR1. As expected, the Ki value is nearly 10-fold higher than the Km value for the wild type, indicating that it has much higher affinity for phosphate than arsenate, while in CuR1 the affinity for phosphate and arsenate is very similar. CuR1 showed increased arsenate sensitivity relative to wild-type M. sedula, consistent with the allelic state of pitA (functional in CuR1, nonfunctional in the wild type) (Fig. 4A). CuR1 showed a decreased growth rate and lower cell yields at 15 mM arsenate, and growth was inhibited by 20 mM arsenate. In contrast, wild-type M. sedula showed only slight growth inhibition at 20 mM arsenate. This observation suggested a link between copper, arsenate, and Msed_1517 and the possibility that arsenate, through its competition with phosphate, antagonizes copper resistance. This was tested by examining the effect of arsenate exposure on copper resistance (Fig. 4B). Arsenate-exposed CuR1 showed increased copper sensitivity, but arsenate-grown wild-type cells showed no difference in metal resistance between arsenate-exposed and unexposed cells. In contrast, growth in medium containing both copper and arsenate was inhibitory for both strains. Based on these observations, it was reasonable to propose that spontaneous arsenate-resistant derivatives of CuR1 arise by mutation of Msed_1517. Seven spontaneous arsenate-resistant isolates were recovered, and DNA sequence analysis indicated each had undergone mutation in Msed_1517. Mutant CuR1 As#1 underwent a single-nucleotide deletion recreating the wild-type Msed_1517 truncated allele, while mutant CuR1 As#2 underwent a G→T substitution at nt 199 (Fig. 5A). In both cases, these mutations produced highly truncated open reading frames. Three additional alleles were identified with nonsynonymous mutations that occurred much later in the Msed_1517 coding sequence. Two of them comprised changes at two positions. These alleles consisted of the following changes at the indicated nucleotide positions: G→A, 77; T→G, 173; T→A, 180; T→A, 193; and G→C, 211 (Fig. 5B). Interestingly, all five of these mutations mapped to predicted membrane-spanning domains, as indicated by Kyte-Doolittle analysis (ExPasy) (Fig. 5B). The arsenate-resistant CuR1 derivatives also showed decreased copper resistance compared to the parental CuR1 strain (Fig. 5C) and a reduced rate of phosphate transport, resulting in 28% of the parental CuR1 strain transport rate. Taken together, these data identify Msed_1517 as a PitA homolog and a critical component of the metal resistance mechanisms found in thermoacidophilic archaea. A model for copper resistance in M. sedula is presented (Fig. 6) that integrates these results with previous findings.
FIG 4.
Response of M. sedula and CuR1 growth to arsenate challenge and its effect on copper resistance. (A) Effect of arsenate. (B) Effect of arsenate on growth at maximum tolerated levels of cupric ion.
FIG 5.
Location and copper resistance of arsenate-resistant derivatives of CuR1. (A) DNA sequences of Msed_1517 in wild-type M. sedula, strain CuR1, and two spontaneous arsenate-resistant mutants resulting in early sequence truncation. Bars indicate the location of sequence alterations. (B) Association of spontaneous arsenate-resistant mutations and predicted Msed_1517 membrane-spanning regions. Membrane-spanning regions were determined using Kyte-Doolittle hydropathy analysis. (C) Growth in copper medium (80 mM).
FIG 6.
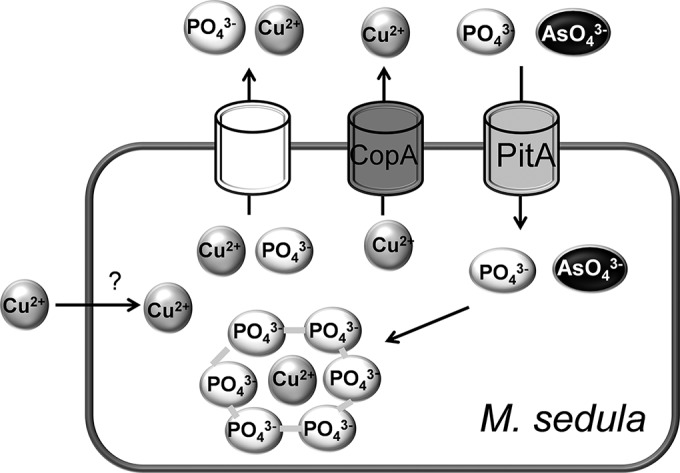
Copper resistance model in M. sedula. CopA catalyzes copper efflux. Archaeal PitA (Msed_1517) catalyzes the uptake of both phosphate and arsenate. An unidentified transporter may catalyze the efflux of copper phosphate complexes. The location of intracellular copper is unknown but may be phosphate associated.
DISCUSSION
Of the 17 mutations distinguishing M. sedula strain CuR1 from the wild type, the gain-of-function mutation in pseudogene Msed_1517 was the most informative. This mutation restored nearly the entire reading length of a gene encoding the PitA/Pho4 domain that is conserved across the thermoacidophilic archaea. Therefore, wild-type M. sedula (DSM 5348T) would be a pitA mutant. Based on comparisons between cell lines, archaeal PitA appears to promote copper resistance and may do so by increasing phosphate-metal efflux. This type of resistance mechanism has been proposed as the basis for high-level metal resistance in M. sedula and related lithoautotrophic archaea (3). The ability to increase phosphate ion efflux in complex with copper ions would decrease intracellular metal concentrations. Phosphate ions would derive from polyphosphate granules shown previously to accumulate to high levels only in the highly metal-resistant group of thermoacidophilic archaea (21). In bacteria such as E. coli, inorganic phosphate transport involves two transport systems, including a high-affinity, low-velocity Pst (phosphate-specific transport) system and a low-affinity, high-velocity Pit (Pi transport) system (35). These systems have been distinguished by their contribution to arsenate resistance; cells having only Pst (mutated for Pit) will grow in the presence of arsenate, while those having only Pit (mutated in Pst) cannot grow in the presence of an arsenate-to-phosphate ratio of 10 in the medium (35). If Msed_1517 constitutes the archaeal Pit transport system, then restoration would support increased phosphate efflux and metal symport through the Pit-like system. This type of mechanism has been suggested to operate in E. coli (33, 40). In previous studies on metal resistance in thermoacidophilic archaea, the presence of another transporter typified by the Pho84 domain was noted (21). Pho84, like Pit, belongs to the family of phosphate-proton symporters and the major facilitator superfamily. Perhaps acting together, Msed_1517 and Pho84 comprise dual phosphate transporters that are inherent to these organisms and are critical for high-level metal resistance.
Genome resequencing of CuR1 also provided the first measure of mutation rate in lithoautotrophic archaea. After 24 generations over 6 passages, CuR1 accumulated a total of 20 mutations, of which 17 could be experimentally verified. This resulted in a mutation rate of 3.44 × 10−7 mutations per cell division and was comparable to mutation rates seen in heterotrophic thermoacidophilic archaea (41). This frequency will be of use to assess the mutation rates experienced by such organisms in natural settings.
Transcriptomic analysis of CuR1 also demonstrated that increased metal resistance reflected a more rapid and stronger response to metal challenge. The key genes involved in this response included two transcription factors, of which one, TFB, constitutes a generalized transcription factor. Changes in the abundance of both of these transcription factors are likely to change gene expression. In addition, several transporters exhibited increased expression and could assist in the export of metal ions to achieve metal homeostasis. Finally, the induction of toxin-antitoxin proteins could provide an additional mechanism to adjust the abundance of transcripts critical to meet the needs of a copper-challenged cell, much as has been described recently for heat shock in the related S. solfataricus (42).
The type species of Metallosphaera sedula, DSM 5348T, unexpectedly was found to be naturally mutated for Msed_1517. The restoration of the function of this gene is a key component of the elevated copper resistance of the spontaneous copper-resistant derivative strain, CuR1. However, the gain-of-function mutation did not completely restore the full length of Msed_1517 (pitA) relative to the alleles in other genome sequences of thermoacidophilic archaeal lineages. Like bacterial PitA, archaeal PitA was found to be a low-affinity, high-velocity secondary transporter dependent on the proton motive force. While a role for PitA has been described as a mediator of arsenate sensitivity and loss-of-function mutations result in arsenate resistance (43), archaeal PitA also is required for high-level copper resistance, and its inactivation resulted in a 3-fold reduction of adaptive levels of this trait (200 mM versus 76 mM) (27). To explain the combined roles of PitA as a transporter (of phosphate and arsenate) and a mediator of high-level copper resistance, it is necessary to suggest that intracellular phosphate is limiting in the absence of PitA. It has been suggested in extreme thermoacidophiles that intracellular polyphosphate provides a source of soluble phosphate that can be effluxed bound to copper ions (21, 44). Here, PitA could be shown to mediate both phosphate and arsenate uptake; therefore, it seems unlikely that PitA also mediates phosphate-copper complex efflux but rather that some unidentified transport protein is responsible for this process.
ACKNOWLEDGMENTS
This research was supported by funding from the NIH (1R01GM090209), DTRA (HDTRA1-09-0030), and AFOSR (FA9550-13-1-0236) to R.K. and P.B. and the UNL Cell Development Facility to P.B.
Footnotes
Published ahead of print 4 August 2014
REFERENCES
- 1.Maezato Y, Blum P. 2012. Survival of the fittest: overcoming oxidative stress at the extremes of acid, heat and metal. Life 2:229–242. 10.3390/life2030229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huber H, Prangishvili D. 2006. The Sulfolobales, p 23–50 In Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E. (ed), The prokaryotes, vol 3, 3rd ed. Springer, New York, NY [Google Scholar]
- 3.Orell A, Navarro CA, Arancibia R, Mobarec JC, Jerez CA. 2010. Life in blue: copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 28:839–848. 10.1016/j.biotechadv.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 4.Rawlings DE. 2005. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 4:13. 10.1186/1475-2859-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schippers A, Jozsa P-G, Sand W. 1996. Sulfur chemistry in bacterial leaching of pyrite. Appl. Environ. Microbiol. 62:3424–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schippers A, Sand W. 1999. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 65:319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auernik KS, Maezato Y, Blum PH, Kelly RM. 2008. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74:682–692. 10.1128/AEM.02019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu LJ, You XY, Zheng H, Wang S, Jiang CY, Liu SJ. 2011. Complete genome sequence of Metallosphaera cuprina, a metal sulfide-oxidizing archaeon from a hot spring. J. Bacteriol. 193:3387–3388. 10.1128/JB.05038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdes J, Ossandon F, Quatrini R, Dopson M, Holmes DS. 2011. Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J. Bacteriol. 193:7003–7004. 10.1128/JB.06281-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auernik KS, Kelly RM. 2010. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic, and mixotrophic growth. Appl. Environ. Microbiol. 76:931–935. 10.1128/AEM.01336-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kappler U, Sly LI, McEwan AG. 2005. Respiratory gene clusters of Metallosphaera sedula–differential expression and transcriptional organization. Microbiology 151:35–43. 10.1099/mic.0.27515-0 [DOI] [PubMed] [Google Scholar]
- 12.Auernik KS, Kelly RM. 2010. Impact of molecular hydrogen on chalcopyrite bioleaching by the extremely thermoacidophilic archaeon Metallosphaera sedula. Appl. Environ. Microbiol. 76:2668–2672. 10.1128/AEM.02016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valenzuela L, Chi A, Beard S, Orell A, Guiliani N, Shabanowitz J, Hunt DF, Jerez CA. 2006. Genomics, metagenomics and proteomics in biomining microorganisms. Biotechnol. Adv. 24:197–211. 10.1016/j.biotechadv.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 14.Agarwal S, Hong D, Desai N, Sazinsky M, Arguello J, Rosenzweig A. 2010. Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA. Proteins 78:2450–2458. 10.1002/prot.22753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Guerrero M, Arguello JM. 2008. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U. S. A. 105:5992–5997. 10.1073/pnas.0711446105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vollmecke C, Drees SL, Reimann J, Albers SV, Lubben M. 2012. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 158:1622–1633. 10.1099/mic.0.055905-0 [DOI] [PubMed] [Google Scholar]
- 17.Deigweiher K, Drell TL, Prutsch A, Scheidig AJ, Lubben M. 2004. Expression, isolation, and crystallization of the catalytic domain of CopB, a putative copper transporting ATPase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Bioenerg. Biomembr. 36:151–159. 10.1023/B:JOBB.0000019607.05233.4c [DOI] [PubMed] [Google Scholar]
- 18.Ettema TJ, Brinkman AB, Lamers PP, Kornet NG, de Vos WM, van der Oost J. 2006. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology 152:1969–1979. 10.1099/mic.0.28724-0 [DOI] [PubMed] [Google Scholar]
- 19.Villafane A, Voskoboynik Y, Ruhl I, Sannino D, Maezato Y, Blum P, Bini E. 2011. CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology 157:2808–2817. 10.1099/mic.0.051862-0 [DOI] [PubMed] [Google Scholar]
- 20.Ettema TJ, Huynen MA, de Vos WM, van der Oost J. 2003. TRASH: a novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends Biochem. Sci. 28:170–173. 10.1016/S0968-0004(03)00037-9 [DOI] [PubMed] [Google Scholar]
- 21.Remonsellez F, Orell A, Jerez CA. 2006. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology 152:59–66. 10.1099/mic.0.28241-0 [DOI] [PubMed] [Google Scholar]
- 22.Cardona ST, Chavez FP, Jerez CA. 2002. The exopolyphosphatase gene from Sulfolobus solfataricus: characterization of the first gene found to be involved in polyphosphate metabolism in archaea. Appl. Environ. Microbiol. 68:4812–4819. 10.1128/AEM.68.10.4812-4819.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber G, Spinnler C, Gambacorta A, Stetter K. 1989. Metallosphaera sedula gen. and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst. Appl. Microbiol. 12:38–47. 10.1016/S0723-2020(89)80038-4 [DOI] [Google Scholar]
- 24.Grogan DW. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786. 10.1126/science.1149976 [DOI] [PubMed] [Google Scholar]
- 26.Alber BE, Kung JW, Fuchs G. 2008. 3-Hydroxypropionyl-coenzyme A synthetase from Metallosphaera sedula, an enzyme involved in autotrophic CO2 fixation. J. Bacteriol. 190:1383–1389. 10.1128/JB.01593-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maezato Y, Johnson T, McCarthy S, Dana K, Blum P. 2012. Metal resistance and lithoautotrophy in the extreme thermoacidophile Metallosphaera sedula. J. Bacteriol. 194:6856–6863. 10.1128/JB.01413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen MB. 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Mikrobiol. 32:270–277. 10.1007/BF00409348 [DOI] [PubMed] [Google Scholar]
- 29.Maezato Y, Dana K, Blum P. 2011. Engineering thermoacidophilic archaea using linear DNA recombination. Methods Mol. Biol. 765:435–445. 10.1007/978-1-61779-197-0_26 [DOI] [PubMed] [Google Scholar]
- 30.Auernik KS, Kelly RM. 2008. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74:7723–7732. 10.1128/AEM.01545-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moll R, Schäfer G. 1988. Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 232:359–363. 10.1016/0014-5793(88)80769-5 [DOI] [Google Scholar]
- 33.Van Dien SJ, Keyhani S, Yang C, Keasling JD. 1997. Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl. Environ. Microbiol. 63:1689–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konings WN, Rosenberg H. 1978. Phosphate transport in membrane vesicles from Escherichia coli. Biochim. Biophys. Acta 508:370–378. 10.1016/0005-2736(78)90339-5 [DOI] [PubMed] [Google Scholar]
- 35.Willsky GR, Malamy MH. 1980. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144:356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiorentino G, Ronca R, Cannio R, Rossi M, Bartolucci S. 2007. MarR-like transcriptional regulator involved in detoxification of aromatic compounds in Sulfolobus solfataricus. J. Bacteriol. 189:7351–7360. 10.1128/JB.00885-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siripornadulsil S, Traina S, Verma DP, Sayre RT. 2002. Molecular mechanisms of proline-mediated tolerance to toxic heavy metals in transgenic microalgae. Plant Cell 14:2837–2847. 10.1105/tpc.004853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kher SS, Worthylake RA. 2011. Regulation of ROCKII membrane localization through its C-terminus. Exp. Cell Res. 317:2845–2852. 10.1016/j.yexcr.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keasling JD. 1997. Regulation of intracellular toxic metals and other cations by hydrolysis of polyphosphate. Ann. N. Y. Acad. Sci. 829:242–249 [DOI] [PubMed] [Google Scholar]
- 41.Jacobs KL, Grogan DW. 1997. Rates of spontaneous mutation in an archaeon from geothermal environments. J. Bacteriol. 179:3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maezato Y, Daugherty A, Dana K, Soo E, Cooper C, Tachdjian S, Kelly RM, Blum P. 2011. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA 17:1381–1392. 10.1261/rna.2679911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard SJ, Hashim R, Wu G, Binet MR, Hughes MN, Poole RK. 2000. Evidence for the transport of zinc(II) ions via the pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol. Lett. 184:231–235. 10.1111/j.1574-6968.2000.tb09019.x [DOI] [PubMed] [Google Scholar]
- 44.Alvarez S, Jerez CA. 2004. Copper ions stimulate polyphosphate degradation and phosphate efflux in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 70:5177–5182. 10.1128/AEM.70.9.5177-5182.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]