Abstract
Different models have been proposed explaining how eukaryotic gene transcription is terminated. Recently, Nsi1, a factor involved in silencing of ribosomal DNA (rDNA), was shown to be required for efficient termination of rDNA transcription by RNA polymerase I (Pol I) in the yeast Saccharomyces cerevisiae. Nsi1 contains Myb-like DNA binding domains and associates in vivo near the 3′ end of rRNA genes to rDNA, but information about which and how DNA sequences might influence Nsi1-dependent termination is lacking. Here, we show that binding of Nsi1 to a stretch of 11 nucleotides in the correct orientation was sufficient to pause elongating Pol I shortly upstream of the Nsi1 binding site and to release the transcripts in vitro. The same minimal DNA element triggered Nsi1-dependent termination of pre-rRNA synthesis using an in vivo reporter assay. Termination efficiency in the in vivo system could be enhanced by inclusion of specific DNA sequences downstream of the Nsi1 binding site. These data and the finding that Nsi1 blocks efficiently only Pol I-dependent RNA synthesis in an in vitro transcription system improve our understanding of a unique mechanism of transcription termination.
INTRODUCTION
Cellular multisubunit RNA polymerases of all three domains of life share common structural and functional features. After initiation of transcription at their cognate gene promoters, they extend the transcripts until they reach specific DNA cis elements, at which transcription is terminated. Termination occurs when the contacts of the RNA-DNA hybrid within the elongating RNA polymerase are destabilized. Termination finally results in the stopping of RNA synthesis, release of the transcript, and dissociation of the RNA polymerase from the DNA template (reviewed in reference 1). A first step in the termination pathway can be the pausing of the elongating polymerase, which is then followed by the disruption of the elongation complex.
Dissociation of the ternary elongation complex formed by RNA polymerase, RNA, and DNA can be caused either by auxiliary protein factors or solely by interactions of DNA and RNA with the transcribing RNA polymerase. Both principles, factor-dependent and intrinsic termination, have been described for bacterial transcription termination (reviewed in reference 1). In the case of eukaryotic RNA polymerase II (Pol II), either 3′-end processing factors of the mRNA or the yeast RNA-DNA helicase Sen1 (human senataxin) was suggested to disrupt the ternary complex (reviewed in references 2 and 3). This might resemble Rho-dependent termination in bacteria in which the ATP-dependent RNA-DNA helicase Rho destabilizes the elongation complex (4, 5; for a review, see reference 1). RNA polymerase III (Pol III) termination resembles the other bacteria-like termination mechanism, the intrinsic termination, in which the RNA-DNA hybrid is destabilized by a stem-loop in the nascent RNA (6, 7) (8).
In contrast to these mechanisms of transcription termination, Pol I requires a termination factor which binds to a specific DNA sequence to pause elongation 15 bp upstream of its binding site and to release the transcripts (reviewed in reference 9). In mouse, binding of the transcription termination factor TTF1 to a conserved 18-bp DNA element was shown to be sufficient to stop Pol I transcription in vitro (10–12) and to release the transcripts with the help of Pol I transcript release factor (PTRF) (13, 14). The same DNA cis element was able to terminate transcription in vivo (12). Adjacent DNA elements could modulate termination efficiency. In the yeast Saccharomyces cerevisiae association of the TTF1 and Myb homologue Reb1 protein to its cognate 11-bp binding site pauses the elongating Pol I in vitro. The presence of a 10- to 15-bp T-rich stretch (T-rich 1) (see Fig. 1A) 12 to 20 bp upstream of the Reb1 binding site supported pausing of transcription and release of the transcript, suggesting that Reb1 binding in addition to the T-rich stretch is sufficient to terminate transcription in vitro (15, 16). In vivo analyses of yeast Pol I termination further pointed to the importance of the Reb1 binding site and the upstream T-rich 1 stretch for termination (17). Pol I readthrough at the minimal terminator element resulted in transcription termination at a downstream T-rich element (T-rich 2) (17–19) or at the replication fork barrier (RFB) (20). The latter was dependent on the presence of the “Fork blocking less” protein Fob1 (21, 22) (for structural organization of the Pol I terminator cis elements see Fig. 1A).
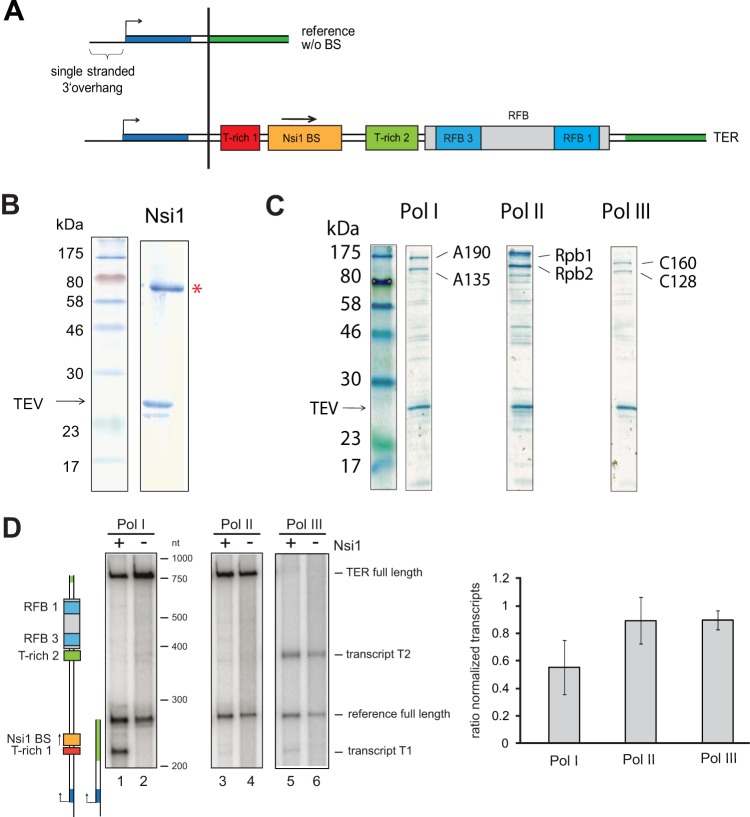
FIG 1.
Nsi1 pauses efficiently Pol I transcription at the Nsi1 binding site in tailed-template transcription assays. (A) Schematic representation of the DNA matrices for tailed-template assays. Transcription is initiated on a 24-nt single-stranded 3′ extension of the template strain. The transcription start site is indicated with a black arrow. Yeast 35S rDNA terminator region cis elements are depicted with red (T-rich 1), orange (Nsi1 binding site), and light green (T-rich 2) boxes. Two cyan boxes embedded in a gray rectangle represent the RFB 1 and RFB 3 regions within the replication fork barrier. The DNA stretches shown in dark blue and dark green of the reference template and of template TER denote sequence identity of the respective template sections. TER refers to the region from bp +70 to bp +414 with respect to the 3′ end of the 25S rDNA and with the Nsi1 binding site stretching from +109 to +119. Note that the DNA sequence coding for the Rnt1 cleavage site is not included. The reference template (254 nt) contains the same 3′ extension to start transcription but no DNA sequence from the terminator region. (B) Coomassie-stained gel of recombinant Nsi1 purified from Sf21 cells via FLAG immunoprecipitation (see Materials and Methods). Ten percent of the eluate derived from lysates of 50 × 106 infected cells was separated on an SDS gel. A red asterisk marks the band representing Nsi1, and an arrow indicates TEV protease. (C) Coomassie-stained gel of affinity-purified yeast RNA Pol I, II, and III. Pol I, II, and III were purified from yeast strain y2423, y2424, and y2425 in which subunits A135, Rpb2, and C128 are expressed with C-terminal protein A tags, respectively (see Materials and Methods). Ten percent of affinity-purified Pol I/II/III from a cell lysate representing an 800-ml culture volume at an OD600 of 1.5 was loaded on an SDS gel. Bands representing the two large subunits of each polymerase and TEV protease are indicated (see also Fig. S2 in the supplemental material for comparative mass spectrometric analysis). (D) In vitro transcription reactions were carried out in the presence of a template carrying the complete 35S rDNA terminator region (TER) and the reference template which did not contain putative termination elements. Experiments were conducted with one-step purified Pol I, II, and III. Final template concentration was 10 nM each. Nsi1 was preincubated with the template (+) and the nucleoside triphosphates for 15 min at room temperature before the reaction was started by addition of 7.5 nM RNA Pol I. Nsi1 concentration was 88 nM. Control reactions (−) were supplemented with Nsi1 storage buffer and processed equally. Transcription was stopped after 30 min by addition of proteinase K. The RNAs were extracted and separated on a denaturing polyacrylamide urea gel, and the radiolabeled transcripts were visualized side-by-side with an RNA marker which was in vitro transcribed with T7 RNA polymerase. Cartoons visualize the length and identity of the in vitro-transcribed RNAs. For quantification, signal intensities of TER full-length transcripts (757 nt) were normalized to reference full-length transcript (254 nt) signal intensities in the respective lanes. Then, the ratio of the normalized TER full-length transcript signal intensities in presence (+) versus absence (−) of Nsi1 was calculated. Experiments were conducted at least in duplicate, and mean values were plotted. Error bars represent ±1 standard deviation of the sample. (see Fig. S1 in the supplemental material for template specific transcription reactions).
More recently, proteins involved in 3′-end processing of pre-rRNA were suggested to be involved in Pol I termination, and a more complex in vivo termination model arose (20, 23–25). This model implicated cotranscriptional cleavage of a stem-loop in pre-rRNA downstream of the rRNA coding region by the endonuclease Rnt1. The newly formed 5′ RNA end serves as an entry point for the exonuclease Rat1. Subsequently, Rat1 progressively degrades the Pol I-associated transcript from the 5′ end with the help of the 5′ polynucleotide kinase Grc3 and the helicase Sen1. After Rat1 reaches Pol I in a torpedo-like fashion, the ternary transcription elongation complex is thought to be dissociated. In accordance with the described in vitro model, pausing of the elongating Pol I at the Reb1 binding site was also considered an important prerequisite to trigger termination in the in vivo torpedo model. However, no significant association of Reb1 with its bona fide binding site at the terminator could be observed in vivo. In contrast, the Reb1 homologue Nsi1 was found to be associated with the Reb1 binding site at the yeast terminator (26, 27). Genome-wide DNA association studies indicated that Nsi1 recognizes the same DNA sequence motif as Reb1 (28). Accordingly, we refer in the following to the previously described putative Reb1-binding sequence at the terminator as the Nsi1 binding site. Similar to Reb1 and to the mammalian termination factor TTF1, Nsi1 contains three Myb-like DNA binding domains. It was shown that Nsi1 binding to the 322-bp terminator element TER, which contains the 3′ end of the rRNA gene including the T-rich 1 element, the Nsi1 binding site, the T-rich 2 element, and the replication fork barrier RFB (see also Fig. 1A), is required for efficient transcription termination (26). Whether other cis elements of the terminator region contribute to Nsi1-dependent transcription termination and the specific role of Nsi1 in the termination process remain to be elucidated. In particular, the following questions were addressed in this work: (i) Which cis elements are involved in Nsi1-dependent Pol I transcription termination in vitro and in vivo? (ii) Does Nsi1 interact with all nuclear RNA polymerases in in vitro transcription reactions? (iii) Is Nsi1 binding sufficient for pausing and/or release of transcripts?
In vitro experiments using highly purified protein factors were used to determine the minimal terminator elements required for pausing and release of transcripts. The role of these terminator elements in transcription termination in vivo were then analyzed using a recently published reporter assay (26). In summary, these experiments suggested that the presence of the Nsi1 binding site in the correct orientation and Nsi1 binding are sufficient to terminate Pol I transcription upstream of its binding site in vitro. In agreement with this, Nsi1 appears to be strictly required for efficient Pol I termination in vivo.
MATERIALS AND METHODS
Yeast strains, plasmids, oligonucleotides, and construction of transcription templates.
Oligonucleotides, plasmids, and yeast strains used in this work are listed in Tables S1, S2, and S3 in the supplemental material. Molecular biological methods and transformation of yeast cells were performed according to standard protocols (29–31). The generation of transcription templates is described in the supplemental material.
Purification of Nsi1.
Nsi1 was expressed as an N-terminal FLAG-tagged fusion protein in Sf21 insect cells. A recognition site for tobacco etch virus (TEV) protease was located between the FLAG tag and Nsi1. A total of 50 × 106 infected Sf21 cells were resuspended in 40 ml of TAP 300 buffer (see Table S4 in the supplemental material for components), and a crude cell extract was prepared by sonication for 5 min with a pattern of a 30-s pulse and a 30-s pause using the macrotip (output 5; Branson Sonifier). Cell debris was removed by centrifugation (4,000 × g for 15 min at 4°C), and the supernatant was added to 50 μl (100-μl slurry) of anti-FLAG M2 beads (Sigma-Aldrich) equilibrated three times with 5 ml of TAP 300 buffer. The suspension was incubated on a rotating wheel at 4°C for 2 h. After centrifugation (130 × g for 5 min at 4°C) the supernatant was removed, and the beads were washed three times with 10 ml of TAP 100 buffer (see Table S4 in the supplemental material) (without protease inhibitors [PIs]) each. For elution, the beads were resuspended in 100 μl of TAP 100 buffer (without PIs), transferred to a microreaction tube, and incubated with 8 μl of TEV protease (2.6 mg/ml) for 2 h at 16°C with gentle shaking. The suspension was centrifuged at 16, 000 × g for 5 min at 4°C, and the supernatant was loaded on a MobiCol column (MoBiTec). For removal of the remaining beads, the column was spun again (16,000 × g for 5 min at 4°C). Each sample taken during the purification process was analyzed via SDS-PAGE to monitor the purification success, and the protein concentration in the elution fraction was determined.
Purification of yeast RNA polymerases.
Wild-type (WT) RNA polymerases I, II, and III (Pol I, II, and III) were purified from yeast strains y2423 (Pol I), y2424 (Pol II), and y2425 Pol III (see Table S3 in the supplemental material) via a protein A (ProtA) affinity tag. In each strain, the second largest subunit of one polymerase is expressed as a C-terminal fusion protein with a protein A tag. Between the C terminus of the subunit and the protein A part, a recognition site for TEV protease is located. A 20-ml YPD (yeast extract, peptone, dextrose) culture was grown to stationary phase at 30°C. From this culture, 2 liters of YPD was inoculated to an optical density at 600 nm (OD600) such that it would result in an OD600 of 1.5 after overnight cultivation at 30°C. At an OD600 of 1.5, the cells were harvested (4,000 × g for 6 min at room temperature [RT]) and washed with ice-cold water. Cells were split in aliquots representing a 400-ml culture volume and again spun down (4,000 × g for 6 min at 4°C). Cell aliquots were frozen in liquid nitrogen and stored at −20°C.
Polymerase purification was performed with one to four cell aliquots in a 4°C room. Cells were thawed, washed in 5 ml of P1 buffer (see Table S4 in the supplemental material) supplemented with protease inhibitors (PIs) and spun down again (4,000 × g for 6 min at 4°C). The pellet was weighed and resuspended in 1.5 ml of P1 buffer plus PIs per gram; 0.7 ml of this solution was added to 2-ml reaction tubes containing 1.4 g of glass beads (diameter, 0.75 to 1 mm; Roth). Cells were lysed on an IKA Vibrax VXR basic shaker at 2,200 rpm at 4°C for 15 min, followed by a 5-min incubation on ice. This procedure was repeated four times. The cell extract was cleared from glass beads by perforation of the cup at the bottom and cap and a centrifugation step (150 × g for 1 min at 4°C). Cell debris was removed by centrifugation at 16,000 × g and 4°C for 30 min. The protein content of the supernatant was determined using a Bradford assay. The lysate was supplemented with NP-40 to a concentration of 0.5%, and PIs were added. Equal protein amounts (usually 1 ml of cell extract, 30 to 40 mg) were incubated with 200 μl of IgG (rabbit serum, I5006-100MG; Sigma)-coupled magnetic bead slurry (1-mm BcMag, FC-102; Bioclone) for 1 h on a rotating wheel. The slurry had previously been equilibrated three times with 500 μl of P2 KCl buffer (see Table S4 in the supplemental material). The beads were washed three times with 700 μl of the respective P2 KCl buffer plus PIs and then washed three times with 700 μl of P2 potassium acetate (KOAc) buffer. For elution, the beads were resuspended in 25 μl of P2 KOAc buffer supplemented with 3 μl of TEV protease (2.6 mg/ml). Cleavage was performed for 2 h at 16°C in a thermomixer at 1,000 rpm. The supernatant was collected, the beads were washed with 25 μl of P2 KOAc buffer, and the two fractions were combined. The elution fraction containing either Pol I, II, or III was split into 10-μl aliquots, frozen in liquid nitrogen, and stored at −80°C. Each sample taken during the purification process was analyzed by SDS-PAGE and Coomassie staining to monitor the purity of the elution fraction. Protein concentration was determined by comparing Coomassie staining of RNA polymerase subunits to the Coomassie staining of a bovine serum albumin (BSA) titration series.
The initiation-active Pol I-Rrn3 complex was purified from strain BSY420 Rrn3-ProtA (strain number y2183) which overexpresses Rrn3-TEV-ProtA-His7 under the control of an GAL1/10 promoter on plasmid YCplac111-GAL-Rrn3-ProtA (plasmid number 729). The strain was grown in YPR (yeast extract, peptone, raffinose) medium for 24 h, and Rrn3 overexpression was induced with 2% galactose for 3 h. The cells were lysed and proceeded to yield the PA600 fraction, which contains all factors required for Pol I transcription initiation in vitro, as described by H. Tschochner (32). Imidazole and NP-40 were added to the PA600 fraction to a concentration of 5 mM and 0.1%, respectively. For affinity purification, Talon affinity resin (Clontech) was equilibrated in buffer 1 (buffer P600 consisting of 20 mM HEPES, pH 7.8, 20% glycerol, 600 mM potassium acetate, and 1 mM MgCl2 supplemented with 5 mM imidazole, 0.1% NP-40, 5 mM β-mercaptoethanol). One hundred microliters of affinity resin was used per ml of the PA600 fraction (2.5 mg/ml). After 2 h of incubation the suspension was poured into a disposable column. Flowthrough was collected, and the column was washed with 30 ml of buffer B (20 mM HEPES, pH 7.8, 20% glycerol, 1.5 M KOAc, 5 mM MgCl2, 0,1% NP-40, 5 mM β-mercaptoethanol, 2 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and 30 ml of buffer C (buffer B but with 600 mM KOAc). Proteins were eluted with 100 mM imidazole in buffer C. Pol I subunits including subunit A12.2 were identified by mass spectrometry.
Generation and purification of recombinant core factor in baculovirus-infected insect cells through FLAG-tagged Rrn7.
cDNAs coding for the core factor subunits Rrn6, Rrn7, and Rrn11 were amplified by PCR. Rrn6 with six copies of a His tag and three copies of a hemagglutinin tag (Rrn6-6His-3×HA) and Rrn7-FLAG were both inserted in plasmid pSPL; Rrn11 was cloned in plasmid pFL (33, 34) by standard cloning techniques (29). Sf21 insect cultures were maintained and infected with recombinant baculoviruses as described previously (35). Oligonucleotides and plasmids are listed in Tables S1 and S2, respectively, in the supplemental material. Purification of core factor from Sf21 cells by FLAG-tagged Rrn7 was performed according to Hierlmeier et al. (35) with some modifications. A total of 50 × 106 cells were lysed in 30-ml of ice-cold TAP 100 buffer [50 mM HEPES-KOH, pH 7.8, 100 mM KCl, 5 mM Mg(OAc)2, 2 mM benzamidine, 1 mM PMSF, 0.1% Tween 20] using a Branson Sonifier 250 (output 5, duty cycle 40%, 30-s pulse, and 30-s cooling; six repeats), and cell debris was removed by centrifugation (at 4°C for 20 min at 3,300 × g).The cleared cell lysate was incubated with 200 μl of previously equilibrated anti-FLAG M2 affinity agarose (Sigma) for 2 h at 4°C while being inverted on a rotating wheel. The agarose was washed three times with 10 ml of TAP 100 buffer and TAP 200 (TAP100 but with 200 mM KCl) buffer. For elution of bound proteins, 50 to 100 μl of TAP 200 buffer was added to the beads together with 0.2 mg/ml FLAG peptide. After incubation for 3 h at 4°C and inversion, the whole suspension was loaded on a disposable spin column (Mobicol), and the eluted proteins were separated from the agarose by centrifugation for 1 min at 2,000 rpm and 4°C.
In vitro transcription.
In vitro transcription using tailed templates was performed according to H. Tschochner (32) with some modifications. Templates (10 nM) were preincubated in reaction buffer (see Table S4 in the supplemental material) with 0.5 mM each ATP, UTP, and GTP and 24 μM CTP for 15 min at room temperature. Then 1.25 μM [α-32P]CTP was added for RNA detection. If applicable, preincubation was done in the presence of 88 nM Nsi1. Controls contained an equal amount of Nsi1 elution buffer. Reactions were started by addition of the RNA polymerase and allowed to proceed for 30 min at 30°C. Reactions were stopped by addition of proteinase K, and the RNAs were extracted by ethanol precipitation. Transcripts were separated on a denaturing polyacrylamide gel and directly visualized. For quantification, signals were normalized to the signal intensity of the reference transcript in each lane using Multi Gauge (Fuji). Then, the ratio of the full-length transcripts in the presence versus absence of Nsi1 was calculated, and the result is indicated as pausing percentage. Promoter-dependent transcription was performed as previously described (32).
Modification of the ribosomal DNA (rDNA) locus in S. cerevisiae.
Insertion of DNA elements into the internal transcribed spacer 1 (ITS1) locus of yeast strains was performed as previously described (26). Details about plasmid construction are described in the supplemental material.
Analysis of generation times.
Strains containing the empty YCplac33 plasmid or plasmid pNsi1::Nsi1 were precultivated in selection medium (lacking Ura [−Ura]) containing glucose. The generation time was determined by measuring the increase in cell density upon cultivation in 96-well plates at 30°C in selection medium (−Ura) containing glucose using a Tecan (Infinite 500) reader. Strains containing empty plasmid Ycplac181 or pGAL::Nsi1 were precultivated in selection medium (−Leu) containing raffinose, and galactose was added to a final concentration of 2% when galactose induction was required.
RESULTS
Nsi1 pauses Pol I-dependent transcription in an in vitro transcription assay.
Previous in vivo experiments revealed that Nsi1 bound to the 3′ end of rRNA genes is required for efficient Pol I termination in yeast. In a first attempt to address more directly the role of Nsi1 in transcription termination, we made use of a tailed-template in vitro transcription assay (36–39) in which RNA polymerases can initiate transcription in the absence of specific promoter sequences or transcription initiation factors. Thereby, the effect of Nsi1 on transcriptional pausing could be assayed under highly defined conditions in which purified RNA polymerase and Nsi1 were the only protein fractions added. This allowed a direct comparison of transcription elongation of yeast RNA polymerases I, II, and III in the absence and presence of Nsi1.
It was important to distinguish between the general impact of Nsi1 addition on the in vitro transcription reaction and the effect of Nsi1 mediated by the DNA cis element. Therefore, each reaction mixture contained a tailed template including the putative Nsi1 binding site and, in addition to that, a control template (254 bp), which did not contain rDNA terminator elements (Fig. 1A; see also Fig. S1 in the supplemental material). Highly purified recombinant Nsi1 was obtained from baculovirus-infected insect cells (Fig. 1B). Furthermore, tagged RNA polymerase I, II, or III was affinity purified from yeast cell extracts using a one-step affinity purification protocol (see Materials and Methods). Affinity-purified RNA polymerases were highly enriched (Fig. 1C) without significant cross-contamination with other RNA polymerases or with elongation factors as monitored by comparative mass spectrometry (see Fig. S2 in the supplemental material). Salt conditions of the respective transcription reaction mixtures were adjusted such that all polymerases synthesized RNA in an identical buffer with approximately the same efficiency (see Fig. S3).
Purified Nsi1 blocked Pol I elongation just upstream of the Nsi1-binding site and gave rise to a transcript of an approximate length of 210 nucleotides (nt) (Fig. 1D). The amount of readthrough transcript was reduced to about 50% (Fig. 1D, compare lanes 1 and 2; note graph at right). In contrast, Pol II transcription was not significantly affected in the presence of Nsi1 (Fig. 1D). Pol III transcription was also slightly affected by Nsi1 since a shorter transcript became visible in the presence of Nsi1 (Fig. 1D, lanes 5 and 6). Most Pol III-dependent transcripts were, however, stopped at the T-rich 2 element in the template DNA, which contains a stretch of 16 consecutive thymidine nucleotides. This is consistent with previous observations that transcribing the Pol III complex can be paused and eventually disrupted when Pol III encounters a poly(T) stretch of more than 7 T residues (40, 41). These findings suggest that Nsi1 can specifically stop elongating Pol I in proximity of its binding site in vitro.
Identification of DNA elements required to pause elongating Pol I in the presence of Nsi1.
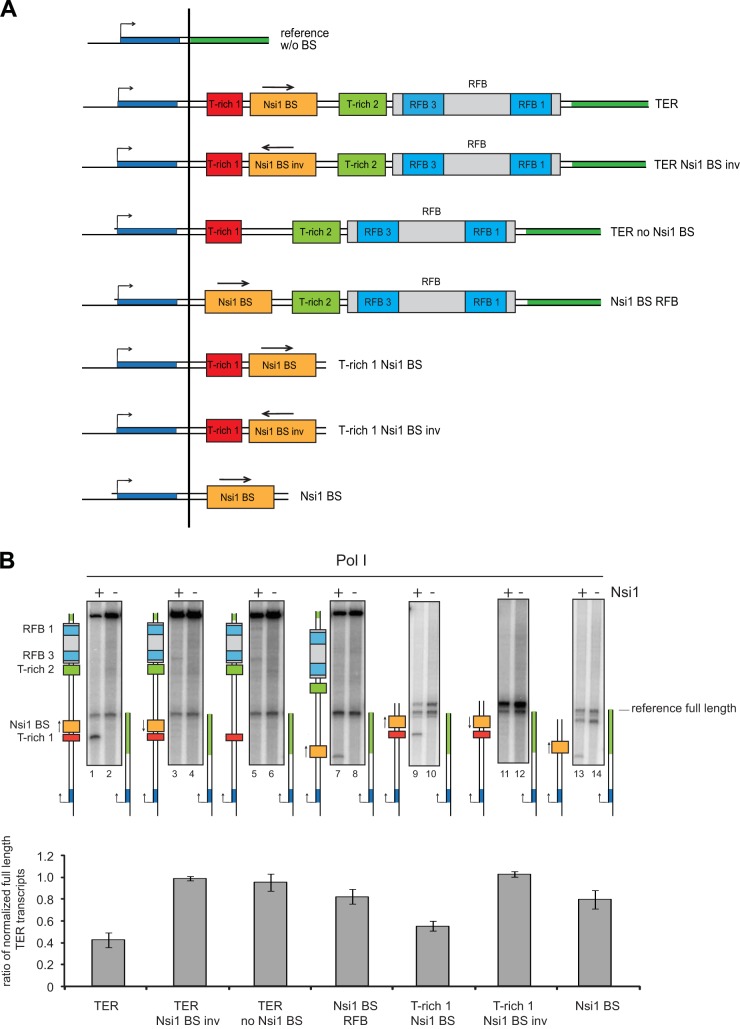
To analyze which DNA cis element of the terminator region contributes to pause Nsi1-dependent Pol I transcription, a set of tailed templates was constructed (Fig. 2A). DNA templates contained different combinations of DNA elements previously implicated in yeast Pol I termination. Thus, templates included the Nsi1 binding site in both orientations in combination with the T-rich 1 element and the further downstream elements, the T-rich 2 element and the replication fork barrier (RFB) (Fig. 2A). Figure 2B shows that almost no shorter transcripts could be detected if the Nsi1 binding site was inverted or omitted (Fig. 2B, compare lane 1 with lanes 3 and 5 and corresponding quantitation). Paused transcripts could also be detected in the absence of the upstream T-rich 1 element from the DNA template (compare lanes 1 and 7). However, pausing was not as efficient when the T-rich 1 element was lacking (46% versus 80% full-length transcripts, respectively). On the other hand, removal of the RFB including the T-rich 2 element did not significantly weaken transcriptional pausing (Fig. 2B, lanes 9 to 14). This indicates that the DNA elements downstream of the Nsi1 binding site have almost no impact on Nsi1-dependent Pol I pausing in vitro.
FIG 2.
Association of Nsi1 to the Nsi1 binding site (BS) is sufficient to pause Pol I elongation. (A) The schematic representation of the tailed-template constructs is explained in the legend to Fig. 1. An arrow indicates whether the respective cis element is in physiological or inverted (inv) orientation regarding the direction of the elongating polymerase. Templates missing the T-rich 1 element start at position +96 with respect to the 3′ end of the 25S rDNA. (B) Transcription reactions were carried out as described in the legend of Fig. 1 in the presence of a template carrying the complete 35S rDNA terminator region (TER) or derivatives thereof (Fig. 1A) and the reference template. Nsi1 and Pol I concentrations were 88 nM and 7.5 nM, respectively. The length of the readthrough transcript depended on the length of the inserted cis elements and varied from 757 nt (template containing the entire TER) to 269 nt when only the Nsi1 binding site was present. The presence (+) or absence (−) of Nsi1 is indicated. Calculations and error analyses were conducted as outlined in the legend to Fig. 1.
The experiments therefore indicated that the crucial DNA element for Nsi1-dependent transcriptional pausing is the Nsi1 binding site and that pausing is further enhanced by the presence of the upstream T-rich 1 stretch. These results largely resemble those of previous experiments studying Reb1-dependent termination of Pol I transcription in vitro (15).
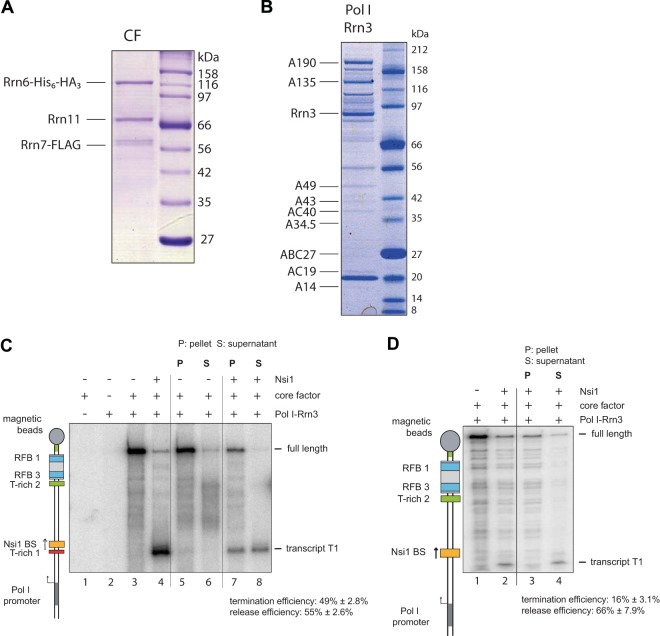
DNA-bound Nsi1 is sufficient to stop Pol I transcription and to release paused transcripts in a promoter-dependent in vitro transcription assay.
Tailed-template transcription could occur without formation of a transcription bubble if both DNA strands are only displaced and not rejoined after the transcribing polymerase (42). To analyze the impact of template-bound Nsi1 on Pol I elongation under more physiological conditions, a promoter-dependent in vitro transcription assay was developed. In a minimal Pol I transcription system, two additional initiation factors, Rrn3 and core factor (CF), are required in addition to purified Pol I (43, 44). Recombinant CF was purified from insect cells (Fig. 3A), and Rrn3 was purified in complex with Pol I from a yeast strain overexpressing Rrn3 (Fig. 3B). Addition of affinity-purified Rrn3-Pol I complex to purified CF in promoter-dependent transcription reaction mixtures produced a strongly labeled promoter-dependent runoff transcript (Fig. 3C, lane 3). Omission of either Rrn3-Pol I or CF resulted in a nearly complete loss of transcriptional activity (Fig. 3C, lanes 1 and 2). The promoter-dependent assay resulted in a single round of transcription, as determined by addition of heparin to the promoter-bound Pol I complex before addition of nucleotides (see Fig. S4 in the supplemental material). Pol I could be significantly blocked at the Nsi1 binding site upon addition of Nsi1 (Fig. 3C, lane 4). In the presence of Nsi1, 49% ± 2.8% of the transcripts were aborted at the Nsi1 binding site. In contrast, pausing efficiency was significantly reduced when the T-rich 1 element was missing. Only about 16% ± 3.1% of the shorter transcripts were detected in the presence of Nsi1 (Fig. 3D). Since transcription reactions were performed using templates attached to magnetic beads, the amount of released transcripts could be estimated. Whereas most (more than 80%) of the full-length transcript remained associated with the beads (Fig. 3C, lanes 5 and 6), the presence of Nsi1 caused the release of 55% ± 2.6% of the shorter transcript into the supernatant (Fig. 3C, compare lanes 5 to 8). Interestingly, without the T-rich 1 element, about 66% ± 7.9% of the aborted transcripts were released to the supernatant, suggesting that the T-rich stretch supports pausing rather than the release efficiency in this in vitro transcription assay (Fig. 3D, lanes 1 to 4). In summary, these experiments indicated that a significant proportion of elongating Pol I aborts in vitro transcription and releases the nascent RNA when it encounters DNA-bound Nsi1.
FIG 3.
Association of Nsi1 at its cognate binding site is sufficient to terminate Pol I transcription in a minimal promoter-dependent transcription assay. (A) Coomassie-stained gel of one-step affinity-purified core factor (CF). Core factor subunits Rrn6, Rrn7, and Rrn11 were coexpressed in Sf21 insect cells infected with recombinant baculovirus. FLAG-Rrn7 was purified with anti-FLAG affinity matrix according to Materials and Methods. Then, 2.5% of the FLAG eluate derived from lysates of 50 × 106 infected cells was separated by SDS-PAGE. CF subunits were identified by mass spectrometry. (B) Coomassie-stained SDS-PAGE of purified yeast Pol I-Rrn3 complex. Pol I-Rrn3 complex was purified from yeast strain y2183 which expresses His- and ProtA-tagged Rrn3 under the control of an inducible GAL1/10 promoter (see Materials and Methods). After 3 h of induction with galactose, a 20-liter culture was harvested at an OD600 of 1.5 and lysed. Thirty percent of affinity-purified Pol I/Rrn3 was loaded on an SDS gradient gel. Pol I subunits (all Pol I subunits including A12.2) and Rrn3 were identified by mass spectrometry. (C and D) Binding of Nsi1 to its cognate binding site is sufficient to halt transcription and release transcripts in a promoter-dependent termination assay. 3′ biotinylated templates which carry the biotinylation downstream of the Pol I promoter were immobilized on streptavidin-coupled magnetic beads. Preincubation of Nsi1 with the template DNA was done as described in the legend to Fig. 1. Transcription was started by addition of 2 μl of RNA Pol I-Rrn3 complex and 0.5 pmol of core factor and allowed to proceed for 30 min at room temperature. Samples were processed as a whole or fractionated in pellet (P) and supernatant (S) before RNA extraction. Presence of Nsi1, core factor, and Pol I in the reactions is indicated. A cartoon visualizes the identity of the transcripts. For quantification of termination efficiency, the ratio of terminated and the sum of full-length and terminated transcripts was calculated. For quantification of release efficiency, the ratio of terminated transcripts in the supernatant and the sum of terminated transcripts was calculated. The C content of each template was included in the calculation.
The Nsi1 binding site is sufficient for Nsi1-dependent termination using an in vivo reporter system.
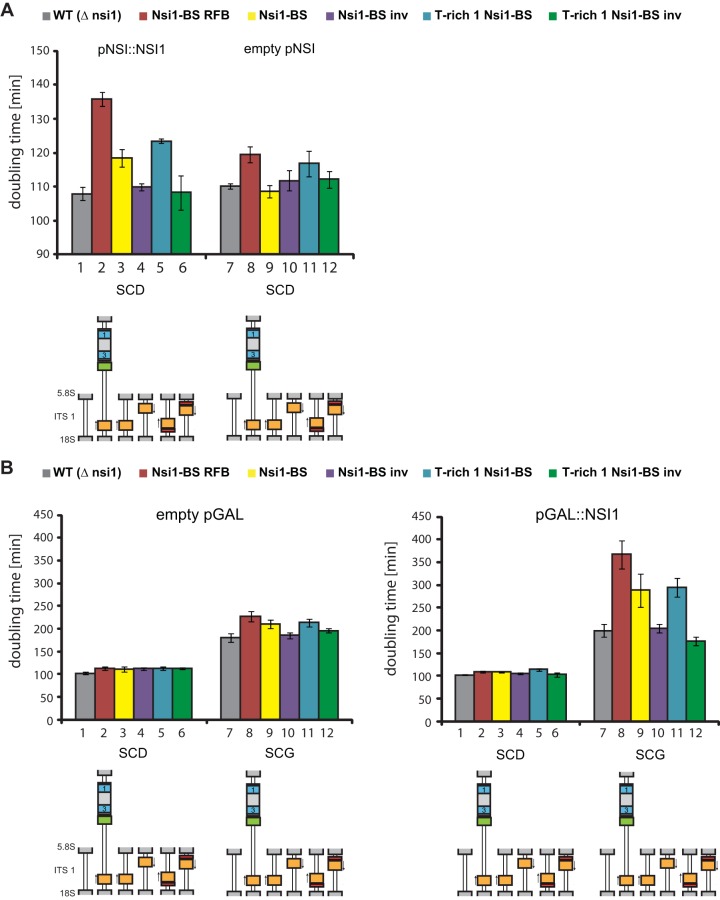
A recently published in vivo reporter assay (26) was used to compare the results obtained from the in vitro experiments with in vivo termination. In this system growth reduction of genetically modified yeast strains served as a readout for transcription termination. DNA elements were integrated in the internal transcribed spacer 1 (ITS1) elements of all of the 150 to 200 rDNA copies to search for sequences interfering with elongating Pol I. If the integrated DNA elements were involved in Pol I termination, synthesis of 5.8S and 25S rRNA encoded 3′ of the ITS1 sequence was inhibited, which led to growth defects (26). This reporter assay was used to analyze in more detail which combinations of cis elements of the terminator region affect cell growth, likely as a consequence of premature Pol I termination. We employed NSI1 deletion strains (see Table S3 in the supplemental material) which showed some, however not very pronounced, increase in generation time when the TER region or any other combination of cis elements contained in the TER region was integrated into ITS1 (Fig. 4, empty vector) (26). These mild effects (Fig. 4A and B, lanes 7 to 12) could be due to Nsi1-independent termination mechanisms or general toxic effects of the insertions.
FIG 4.
DNA cis element analysis in an artificial in vivo termination system and dependence on Nsi1 (over)expression. (A) Influence of terminator cis elements on doubling time is dependent on whether Nsi1 is ectopically expressed under the control of its own promoter. The indicated terminator cis elements TER, Nsi1 binding site (Nsi1-BS), inverted Nsi1 binding site (Nsi1-BS inv), T-rich Nsi1, and inverted T-rich Nsi1 (T-rich Nsi1 inv) were integrated into the ITS1 of all rDNA copies, resulting in strains y2281, y3097, y3098, y3099, and y3100, respectively, in which chromosomal NSI1 was deleted. As a control, strain y2276 was used, carrying an unmodified ITS1 but also lacking the chromosomal copy of NSI1. The strains were transformed with either plasmid 2172 (pNSI::NSI1), which expresses Nsi1 under the control of its own promoter (left panel), or with the empty vector 49 (pNSI, right panel). Doubling times (at least biological triplicates) of the different strains were determined from the growth curves obtained in a Tecan reader (see Materials and Methods). SCD, synthetic complete medium containing glucose. (B) Influence of terminator cis elements on growth rates and dependence on Nsi1 overexpression. Experiments were conducted as described for panel A but after transformation of yeast strains y2276, y2281, y3097, y3098, y3099, and y3100 with either plasmid 52 (pGAL::NSI1), which overexpresses Nsi1 under the control of a GAL1/10 promoter (right panel), or an empty vector 52 (pGAL) as a control. The transformed strains were cultivated in synthetic complete selection medium containing raffinose (SCR). Then, Nsi1 expression was induced by the addition of galactose to a 2% final concentration (synthetic complete medium containing galactose, SCG) or repressed in glucose-containing medium (SCD). Doubling times were determined at least in biological triplicates using a Tecan reader.
Generation times were compared after transformation with the control plasmids YCPlac33 (empty pNSI) and YCPlac181 (empty pGAL), a plasmid encoding Nsi1 under the control of its own promoter (pNSI1::NSI1) (Fig. 4A), or a plasmid encoding Nsi1 under the strong galactose-inducible GAL1/10 promoter (pGAL::NSI1) (Fig. 4B). When Nsi1 was expressed under the control of its own promoter, insertion of TER resulted in an extended doubling time (Fig. 4A, compare lane 2 with lanes 1 and 8). Omission of the RFB led to a substantial reduction of the doubling time, which was, however, still significantly higher than the doubling time of a strain carrying no insertion at ITS1 of the rDNA repeats (Fig. 4A, compare lane 3 with lanes 1 and 9). In contrast, removal of the T-rich 1 element led to only a minor reduction in generation time (Fig. 4A, compare lanes 3 and 5). Changing the orientation of the Nsi1 binding site resulted in similar generation times as observed in strains carrying no insertion at the ITS1 (Fig. 4A, compare lanes 4 and 6 with lane 1). These analyses suggest that the Nsi1 binding site in the correct orientation is sufficient for premature termination and that the RFB rather than the T-rich 1 element supports this effect in a strain expressing Nsi1 under the control of its endogenous promoter.
The observed growth defects were enhanced when GAL1/10 promoter-driven Nsi1 expression was induced by incubation of the respective strains in galactose-containing medium (Fig. 4B, right panel, lanes 7 to 12). Strikingly, under these conditions insertion of the Nsi1 binding site alone in correct orientation led to significant growth inhibition compared to a strain carrying no insertion in the ITS1 (Fig. 4B, right panel, compare lane 9 with lanes 7 and 10). The growth defect was further enhanced when the Nsi1 binding site was combined with the RFB element (lane 8) while addition of the T-rich 1 element alone resulted in no significant increase in generation times (Fig. 4B, compare lane 11 with lane 9). No extended generation times were observed if the Nsi1 binding site had the inverted orientation (Fig. 4B, right panel, lanes 10 and 12). In a control experiment no significant differences in doubling times were observed in glucose-containing medium with strains carrying the different ITS1 insertions and being transformed with an empty vector (Fig. 4B, left panel). Generation times of these strains were generally increased to a similar extent when galactose as opposed to glucose was used as a carbon source (compare Fig. 4B, empty vector control, lanes 1 to 12).
Altogether, these experiments suggest that a correctly oriented Nsi1 binding site is sufficient to promote Nsi1-dependent Pol I termination. Termination efficiency can be further enhanced by the T-rich 1 element, as indicated by the in vitro analyses shown above, and the RFB element, as seen in the in vivo reporter assay. Our in vivo data suggest that additional trans-acting factors could support Nsi1 to efficiently terminate Pol I transcription in vivo. This could be especially true for factors interacting with the RFB sequence since this element enhanced termination in vivo (Fig. 4) but not in vitro using purified Pol I and Nsi1 (Fig. 2).
DISCUSSION
Nsi1 was recently described as a nonessential nucleolar protein involved in silencing and preserving stability of the ribosomal DNA locus as well as in terminating Pol I transcription (26, 27). While Pol I-dependent termination was intensively studied in vitro using Reb1 as a termination factor, equivalent analyses were missing for the Reb1 homologue Nsi1. Such analyses became especially important since Nsi1 rather than Reb1 is associated with the terminator-proximal Reb1 binding site and since it was shown that Nsi1 expression is required for Pol I termination in vivo (26). The in vitro results employing tailed DNA templates and purified Pol I and Nsi1 match overall with previously published results describing Reb1 as a termination factor for Pol I transcription in yeast (15, 16, 45). Accordingly, the presence of a DNA-bound termination factor is sufficient to terminate Pol I transcription. In addition, the T-rich element upstream of the Nsi1 binding site supports factor-dependent termination. Nsi1 terminated RNA polymerase I transcription, whereas neither Pol II nor Pol III transcription termination was comparably affected in the employed in vitro assay. These results resemble those obtained in experiments investigating mammalian RNA polymerase I transcription termination by the transcription termination factor TTF1 (12, 38, 46). Hence, at least in such an artificial in vitro system, a specific molecular interplay between Pol I and Nsi1 is required to promote transcription termination.
Nsi1-dependent pausing efficiency was about 50% in both tailed-template and promoter-dependent transcription. In both assays the presence of the upstream T-rich element supported pausing efficiency. While the presence of the T-rich element increased the number of Nsi1-dependent shorter transcripts, it did not significantly influence the efficiency of transcript release in the promoter-dependent assay. This is in contrast to previous in vitro results using Reb1 as a termination factor in tailed-template assays, which suggested that the T-rich element functions especially as a release element (16, 47). These differences could be due to either specific features of the two homologous Myb-related proteins or the employed initiation-active Pol I fraction. Another explanation could be that possibly occuring DNA strand separation in tailed-template assays (39) provides different structural requirements to pause and release transcripts.
In the in vivo reporter system for Pol I termination, the T-rich 1 element did not significantly contribute to termination, especially when Nsi1 was overexpressed. In contrast, the presence of the RFB sequence led to a higher termination efficiency in vivo whether Nsi1 was overexpressed or not, whereas the sequence was not required for efficient transcription termination with purified factors in vitro. These in vivo data are in accordance with previously published experiments wherein in vivo termination was studied using reporter assays with rDNA minigenes. It was shown that deletion of the T-rich 1 element did not affect termination efficiency in WT strains (Fig. 4B) (25). Mapping of the pre-rRNA 3′ end in WT strains revealed that either one (17) or three (18) DNA sequences downstream of the Nsi1 binding site can behave as additional terminator sequences. In strains lacking the exonuclease Rat1, Pol I could transcribe even through these DNA elements, especially when the RFB binding protein Fob1 was lacking (20). These studies underline that the RFB and associated factors could contribute to Pol I termination in vivo. This could be achieved through facilitated recruitment of Nsi1 to its binding site through the replication fork binding protein Fob1, which was recently reported (27). On the other hand, Nsi1 binding to its 11-bp binding site in the vicinity of a random DNA sequence is sufficient to execute at least Pol I termination to some extent.
Our findings suggest that Pol I termination does not necessarily depend on additional factors (13, 14, 48) or upstream RNA secondary structures to release stalled transcripts as has been published for bacterial transcription termination (49) or recently for yeast Pol III termination (8). Since the sequence coding for the stem-loop that contains the Rnt1 cleavage site upstream of the T-rich 1 element (50–52) was absent from any of the templates used to analyze in vivo termination of transcription, our analyses also suggest that Rnt1 cleavage and the subsequent RNA degradation by Rat1 are not absolutely required to execute Pol I termination (20, 23). The same notion applies to the T-rich 1 element which was suggested to function as fail-safe cleavage site (25). However, secondary RNA structures and/or torpedo-like destabilizing mechanisms could certainly support Nsi1-triggered termination.
Fail-safe mechanisms might explain why Pol I transcription can still be terminated although the gene coding for Nsi1 is lacking and why Nsi1 is not essential for growth. However, in the absence of Nsi1, there appears to be readthrough by Pol I downstream of the Nsi1 binding site, which might interfere with binding of Fob1, the silent regulator protein Sir2, or Net1 (nucleolar silencing establishment factor and telophase regulator protein). This in turn could lead to increased rDNA recombination, reduced rDNA stability, and replicative life span (9, 27).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported through grants of the Deutsche Forschungsgemeinschaft (SFB 960). P. Merkl was partly supported by a fellowship of the German National Academic Foundation.
Footnotes
Published ahead of print 4 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00395-14.
REFERENCES
- 1.Peters JM, Vangeloff AD, Landick R. 2011. Bacterial transcription terminators: the RNA 3′-end chronicles. J. Mol. Biol. 412:793–813. 10.1016/j.jmb.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuehner JN, Pearson EL, Moore C. 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell. Biol. 12:283–294. 10.1038/nrm3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mischo HE, Proudfoot NJ. 2013. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim. Biophys. Acta 1829:174–185. 10.1016/j.bbagrm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porrua O, Libri D. 2013. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 20:884–891. 10.1038/nsmb.2592 [DOI] [PubMed] [Google Scholar]
- 5.Pearson EL, Moore CL. 2013. Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. J. Biol. Chem. 288:19750–19759. 10.1074/jbc.M112.434985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell FE, Jr, Setzer DR. 1992. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12:2260–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson MH, Greenleaf WJ, Landick R, Block SM. 2008. Applied force reveals mechanistic and energetic details of transcription termination. Cell 132:971–982. 10.1016/j.cell.2008.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen S, Yuzenkova Y, Zenkin N. 2013. Mechanism of eukaryotic RNA polymerase III transcription termination. Science 340:1577–1580. 10.1126/science.1237934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemeth A, Perez-Fernandez J, Merkl P, Hamperl S, Gerber J, Griesenbeck J, Tschochner H. 2013. RNA polymerase I termination: where is the end? Biochim. Biophys. Acta 1829:306–317. 10.1016/j.bbagrm.2012.10.007 [DOI] [PubMed] [Google Scholar]
- 10.Bartsch I, Schoneberg C, Grummt I. 1988. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol. Cell. Biol. 8:3891–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. 1995. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 14:1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn A, Normann A, Bartsch I, Grummt I. 1988. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 7:1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason SW, Sander EE, Grummt I. 1997. Identification of a transcript release activity acting on ternary transcription complexes containing murine RNA polymerase I. EMBO J. 16:163–172. 10.1093/emboj/16.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansa P, Mason SW, Hoffmann-Rohrer U, Grummt I. 1998. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J. 17:2855–2864. 10.1093/emboj/17.10.2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH. 1994. A model for transcription termination by RNA polymerase I. Cell 79:527–534. 10.1016/0092-8674(94)90261-5 [DOI] [PubMed] [Google Scholar]
- 16.Lang WH, Reeder RH. 1995. Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc. Natl. Acad. Sci. U. S. A. 92:9781–9785. 10.1073/pnas.92.21.9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeder RH, Guevara P, Roan JG. 1999. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 19:7369–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Sande CA, Kulkens T, Kramer AB, de Wijs IJ, van Heerikhuizen H, Klootwijk J, Planta RJ. 1989. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 17:9127–9146. 10.1093/nar/17.22.9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott EM, Osheim YN, Jones HS, Alen CM, Roan JG, Reeder RH, Beyer AL, Proudfoot NJ. 2004. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl. Acad. Sci. U. S. A. 101:6068–6073. 10.1073/pnas.0401393101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Hage A, Koper M, Kufel J, Tollervey D. 2008. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 22:1069–1081. 10.1101/gad.463708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 20:2887–2901. 10.1101/gad.1472706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeuchi Y, Horiuchi T, Kobayashi T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497–1506. 10.1101/gad.1085403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. 2008. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 22:1082–1092. 10.1101/gad.463408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braglia P, Heindl K, Schleiffer A, Martinez J, Proudfoot NJ. 2010. Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep. 11:758–764. 10.1038/embor.2010.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braglia P, Kawauchi J, Proudfoot NJ. 2011. Co-transcriptional RNA cleavage provides a failsafe termination mechanism for yeast RNA polymerase I. Nucleic Acids Res. 39:1439–1448. 10.1093/nar/gkq894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiter A, Hamperl S, Seitz H, Merkl P, Perez-Fernandez J, Williams L, Gerber J, Nemeth A, Leger I, Gadal O, Milkereit P, Griesenbeck J, Tschochner H. 2012. The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J. 31:3480–3493. 10.1038/emboj.2012.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha CW, Sung MK, Huh WK. 2012. Nsi1 plays a significant role in the silencing of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 40:4892–4903. 10.1093/nar/gks188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104. 10.1038/nature02800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30.Burke D, Dawson D, Stearns T. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Schiestl RH, Gietz RD. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339–346. 10.1007/BF00340712 [DOI] [PubMed] [Google Scholar]
- 32.Tschochner H. 1996. A novel RNA polymerase-I dependent RNase activity that shortens nascent transcripts from the 3′end. Proc. Natl. Acad. Sci. U. S. A. 93:12914–12919. 10.1073/pnas.93.23.12914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger I, Fitzgerald DJ, Richmond TJ. 2004. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22:1583–1587. 10.1038/nbt1036 [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. 2006. Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3:1021–1032. 10.1038/nmeth983 [DOI] [PubMed] [Google Scholar]
- 35.Hierlmeier T, Merl J, Sauert M, Perez-Fernandez J, Schultz P, Bruckmann A, Hamperl S, Ohmayer U, Rachel R, Jacob A, Hergert K, Deutzmann R, Griesenbeck J, Hurt E, Milkereit P, Bassler J, Tschochner H. 2013. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res. 41:1191–1210. 10.1093/nar/gks1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinkle DC, Ring J, Chamberlin MJ. 1972. Studies of the binding of Escherichia coli RNA polymerase to DNA. 3. Tight binding of RNA polymerase holoenzyme to single-strand breaks in T7 DNA. J. Mol. Biol. 70:197–207 [DOI] [PubMed] [Google Scholar]
- 37.Dezelee S, Sentenac A, Fromageot P. 1974. Role of deoxyribonucleic acid-ribonucleic acid hybrids in eukaryotes. Study of the template requirements of yeast ribonucleic acid polymerases and nature of the ribonucleic acid product. J. Biol. Chem. 249:5971–5977 [PubMed] [Google Scholar]
- 38.Kuhn A, Bartsch I, Grummt I. 1990. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature 344:559–562. 10.1038/344559a0 [DOI] [PubMed] [Google Scholar]
- 39.Coulter DE, Greenleaf AL. 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J. Biol. Chem. 260:13190–13198 [PubMed] [Google Scholar]
- 40.Matsuzaki H, Kassavetis GA, Geiduschek EP. 1994. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 235:1173–1192. 10.1006/jmbi.1994.1072 [DOI] [PubMed] [Google Scholar]
- 41.Bogenhagen DF, Brown DD. 1981. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24:261–270. 10.1016/0092-8674(81)90522-5 [DOI] [PubMed] [Google Scholar]
- 42.Dedrick RL, Chamberlin MJ. 1985. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry 24:2245–2253 [DOI] [PubMed] [Google Scholar]
- 43.Keener J, Josaitis C, Dodd J, Nomura M. 1998. Reconstitution of yeast RNA polymerase I transcription in vitro from purified factors. J. Biol. Chem. 273:33795–33802. 10.1074/jbc.273.50.33795 [DOI] [PubMed] [Google Scholar]
- 44.Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. 2012. Efficient transcription by RNA polymerase I using recombinant core factor. Gene 492:94–99. 10.1016/j.gene.2011.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang WH, Reeder RH. 1993. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mason SW, Wallisch M, Grummt I. 1997. RNA polymerase I transcription termination: similar mechanisms are employed by yeast and mammals. J. Mol. Biol. 268:229–234. 10.1006/jmbi.1997.0976 [DOI] [PubMed] [Google Scholar]
- 47.Jeong SW, Lang WH, Reeder RH. 1995. The release element of the yeast polymerase I transcription terminator can function independently of Reb1p. Mol. Cell. Biol. 15:5929–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschochner H, Milkereit P. 1997. RNA polymerase I from S. cerevisiae depends on an additional factor to release terminated transcripts from the template. FEBS Lett. 410:461–466. 10.1016/S0014-5793(97)00636-4 [DOI] [PubMed] [Google Scholar]
- 49.Yarnell WS, Roberts JW. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611–615. 10.1126/science.284.5414.611 [DOI] [PubMed] [Google Scholar]
- 50.Elela SA, Igel H, Ares M., Jr 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85:115–124. 10.1016/S0092-8674(00)81087-9 [DOI] [PubMed] [Google Scholar]
- 51.Kufel J, Dichtl B, Tollervey D. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909–917. 10.1017/S135583829999026X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allmang C, Tollervey D. 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278:67–78. 10.1006/jmbi.1998.1693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.