Abstract
Sporadic basal-like cancers (BLCs) are a common subtype of breast cancer that share multiple biological properties with BRCA1-mutated breast tumors. Despite being BRCA1+/+, sporadic BLCs are widely viewed as phenocopies of BRCA1-mutated breast cancers, because they are hypothesized to manifest a BRCA1 functional defect or breakdown of a pathway(s) in which BRCA1 plays a major role. The role of BRCA1 in the repair of double-strand DNA breaks by homologous recombination (HR) is its best understood function and the function most often implicated in BRCA1 breast cancer suppression. Therefore, it is suspected that sporadic BLCs exhibit a defect in HR. To test this hypothesis, multiple DNA damage repair assays focused on several types of repair were performed on a group of cell lines classified as sporadic BLCs and on controls. The sporadic BLC cell lines failed to exhibit an overt HR defect. Rather, they exhibited defects in the repair of stalled replication forks, another BRCA1 function. These results provide insight into why clinical trials of poly(ADP-ribose) polymerase (PARP) inhibitors, which require an HR defect for efficacy, have been unsuccessful in sporadic BLCs, unlike cisplatin, which elicits DNA damage that requires stalled fork repair and has shown efficacy in sporadic BLCs.
INTRODUCTION
Gene expression profiling of breast cancers has led to the identification of five subtypes: luminal A, luminal B, Her2 amplified, basal like, and normal breast like (1, 2). The basal-like subtype is of particular interest due to the lack of relevant targeted therapies as well as its phenotypic similarity to BRCA1−/− tumors. BRCA1−/− tumors segregate with the basal-like cancer (BLC) subtype by gene expression profiling (3, 4). These tumor species exhibit multiple other biological similarities. For example, both commonly fail to express estrogen receptor (ER), progesterone receptor (PR), and Her2 and are mutant for p53 (5–9). Moreover, both are associated with early relapse following clinically active breast cancer chemotherapy and exhibit similar patterns of metastasis (10). Given these similarities, it is widely speculated that sporadic BLCs manifest a defect(s) in a pathway(s) that is dependent upon BRCA1 function.
The BRCA1 gene encodes at least three known proteins: full-length p220, Δ11b, and IRIS (11). Much of the Δ11b protein sequence is shared with that of p220. However, it lacks most of the sequence encoded by the largest p220-coding exon, exon 11. There is limited knowledge regarding the function of Δ11b, despite the fact that it is the most conserved of all the known isoforms (12). Little is known of the IRIS function other than that the endogenous protein normally stimulates DNA replication, can modulate certain transcriptional events, and, when endogenously overexpressed, exhibits certain properties of an oncoprotein (13, 14).
Much more is known of the functions of p220, which, unlike the other known BRCA1 gene-encoded proteins, manifests breast and ovarian cancer suppression activity (15–18). p220 (also known as BRCA1) also performs multiple genome integrity maintenance functions together with its heterodimeric binding partner, BARD1 (19, 20). These include leadership in the performance of homologous recombination (HR) (21, 22), involvement in the repair of stalled or collapsed replication forks (23, 24), aiding in FANCD2 localization during interstrand cross-link repair (25–27), mitotic spindle pole formation (28), suppression of base mutagenesis and translesional synthesis (23, 24), maintenance of normal centrosome number (29, 30), and the suppression of satellite RNA expression (31).
Shortly after the induction of double-strand breaks (DSBs) by gamma irradiation (IR), BRCA1 becomes hyperphosphorylated and concentrates in focal areas of double-strand break-containing DNA damage (20). At these IR-induced nuclear foci (IRIF), BRCA1 participates in the repair of DSBs by HR (21, 22), and it does so as a member of multiple protein complexes, each of which is composed of unique protein binding partners, such as BRCA2, Rad51, NBS1, MRE11, BACH1, CtIP, and PALB2, among others (32, 33).
HR is one function through which BRCA1 is suspected of participating in breast cancer suppression (16–18). In keeping with this view, BRCA1 mutant cell lines and tumors are generally defective in HR (21, 22). Thus, a major goal of this study was to determine whether sporadic BLC cells, like BRCA1 mutant tumor cells, are also defective in HR repair of DSBs and/or exhibit defects in other BRCA1-dependent DNA damage repair pathways. The answers to these questions might influence the application of mechanism-based approaches to sporadic BLC therapy.
MATERIALS AND METHODS
Cell culture.
All cell lines were cultured as described by Neve et al. (34). For cell lines into which a single copy of the DR-GFP reporter (35) had been integrated, puromycin (1 μg/ml) was added to the culture medium to select for the constant presence of the integrated sequence.
IP and Western blotting.
Cell lines were grown to approximately 80% confluence, pelleted, and lysed in buffer containing 300 mM NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, 0.5% NP-40, 10% glycerol, and a protease inhibitor (catalog number 11836170001; Roche Diagnostics). Lysates containing equivalent amounts of protein were incubated overnight with either the C-terminal BRCA1 antibody sc6954 (Santa Cruz) or a mouse IgG control (antibody sc2025; Santa Cruz). On the next day, these lysates were incubated with protein A beads for 1 h at 4°C. The beads were washed three times in the above-noted lysis buffer, and equal amounts of Laemmli buffer (catalog number BP-110NR; Boston BioProducts) containing 2.5% beta-mercaptoethanol (BME; catalog number M6250; Sigma) were added to each sample. Equivalent amounts of protein from each cell extract were immunoprecipitated, and equivalent amounts of protein from each immunoprecipitation (IP) were electrophoresed in 4 to 12% Tris-glycine gels (catalog number EC60385BOX; Life Technologies), which were then transferred to 0.45-μm-pore-size nitrocellulose membranes. These were incubated in the N-terminal BRCA1 monoclonal antibody MS110 (catalog number ab16780; Abcam) overnight and incubated with horseradish peroxidase-conjugated anti-mouse light chain secondary antibody (catalog number 559751; BD Biosciences) for 1 h on the next day. The bands in each blot were developed using standard enhanced chemiluminescence solution (catalog number NEL105001EA; PerkinElmer) and visualized by exposing the film to the blot for various amounts of time. For the hemagglutinin (HA)-tagged I-SceI blots, the lysates were harvested from the cells in the lysis buffer described above, and appropriate amounts of Laemmli sample buffer containing 2.5% BME were added to each sample to normalize the concentrations. Equivalent amounts of cell extract were electrophoresed in 4 to 12% bis-Tris gels (catalog number NP0336BOX; Life Technologies), which were then transferred to 0.45-μm-pore-size nitrocellulose membranes. The blotting assays were carried out as described above, except that the primary antibodies were a monoclonal HA antibody (catalog number MMS-101P; Covance) and a tubulin antibody (catalog number T-5168; Sigma).
Immunofluorescence.
Cells were plated on coverslips in 6- or 12-well plates and allowed to settle overnight. For gamma irradiation experiments, on the next day one set was treated with 5 Gy using a cesium source, while a second control set was not. Cells were allowed to recover at 37°C for 8 h, fixed in 3% paraformaldehyde (dissolved in phosphate-buffered saline [PBS]), permeabilized with 0.5% Triton (0.5% Triton X-100, 20 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose, all of which were dissolved in double-distilled H2O), and then stained with primary and secondary antibodies. For the Rad51 counting experiments, a minimum of 150 cells was counted for each treatment for each cell line in each individual repetition of the experiment.
For UV treatment, cells on coverslips were irradiated at 30 J/m2 (the dose was measured using a UVX radiometer [UVP Inc., Upland, CA]) using a 254-nm UV-C lamp (UVP Inc., Upland, CA) through 3-mm-pore-size isopore/micropore polycarbonate filters (catalog number TSTP02500; Millipore), as described by Polo et al. (36). They were then allowed to recover for 4 h at 37°C and were then fixed in 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and stained for cyclobutane pyrimidine dimers (CPDs) as described by Polo et al. (36).
If the cells were to be transfected with small interfering RNAs (siRNAs), the cells were plated on coverslips in a 6-cm plate on day 1, transfected with 10 pmol of the appropriate siRNAs on day 2, transfected again with 10 pmol of the appropriate siRNAs on day 3, treated with DNA-damaging agents, fixed, permeabilized, and stained on day 4.
Primary antibodies in these assays included monoclonal BRCA1 C-terminal antibody sc6954 (Santa Cruz); a polyclonal BRCA1 C-terminal antibody (antibody 07-434; Upstate); a purified monoclonal antibody targeting exon 11 of BRCA1, SD118 (37); a monoclonal anti-CPD antibody, clone TDM-2 (catalog number CAC-NM-DND-001; Cosmo Bio); two different polyclonal γH2AX antibodies (antibodies 07-164 [Upstate] and ab2893 [Abcam]); a monoclonal γH2AX antibody (antibody 05-636; Millipore); two different monoclonal 53BP1 antibodies (antibodies 612523 [BD] and MAB3802 [Millipore]); and a polyclonal Rad51 (H-92) antibody (antibody sc8349; Santa Cruz). Secondary antibodies were from Jackson Labs (fluorescein isothiocyanate and rhodamine conjugated) and Abcam (Alexa Fluor 488 and 594 conjugated).
Colony formation assays.
All BLC cell lines and controls were tested for colony-forming efficiency. Enough cells of each line were plated in triplicate to form between 80 and 300 colonies. The cells were allowed to settle overnight. The plates were then treated with various doses of each of a series of DNA-damaging agents and allowed to recover at 37°C until colonies became visible (7 to 14 days, depending on the cell line).
For gamma irradiation, cells were exposed to 0 Gy, 1 Gy, 2 Gy, 3 Gy, 4 Gy, or 5 Gy using a cesium source.
For UV irradiation, cells were exposed to 0 J/m2 (cells had an air exposure time equal to the longest UV exposure time), 1 J/m2, 2 J/m2, 4 J/m2, 5 J/m2, 10 J/m2, 20 J/m2, or 50 J/m2 using a 254-nm UV-C lamp (UVP Inc., Upland, CA). UV doses were measured with a UVX radiometer (UVP Inc., Upland, CA).
For mitomycin C (MMC) treatment, the medium was removed and replaced with medium containing 0 μM (the volume of ethanol added to medium with 0 μM MMC was equivalent to that added with the highest MMC dose), 0.1 μM, 0.2 μM, 0.3 μM, 0.4 μM, or 0.5 μM MMC (catalog number M4287; Sigma) dissolved in ethanol for 4 h. The cells were then washed with PBS, and medium containing no drug was added to allow the cells to recover.
For methyl methanesulfonate (MMS) experiments, the medium was removed after the initial cell plating and replaced with medium containing 0 mM, 1 mM, 2 mM, 3 mM, 4 mM, or 5 mM MMS (catalog number 129925; Sigma) for 4 h. The cells were then washed once with PBS, and drug-free medium was added to allow the cells to recover.
For olaparib (AZD2281) exposure experiments, cells were seeded at a suitable density for transfection on day 1, transfected with 10 pmol of various siRNAs on days 2 and 3, and plated at a suitable density for colony formation on day 4. At 4 h after plating of the cells, the medium was replaced with medium containing 0 μM (which contained a volume of dimethyl sulfoxide [DMSO] equal to that contained in the highest dose), 0.01 μM, 0.05 μM, 0.1 μM, 0.5 μM, 0.75 μM, 1 μM, 2.5 μM, 5 μM, 10 μM, or 25 μM olaparib (catalog number S1060; Selleck) dissolved in DMSO. The cells were incubated in this medium until colonies of the appropriate size had grown.
For all of the treatments described above, after colonies of the appropriate size had grown, the cells were stained with crystal violet and counted with a Microbiology International ProtoCOL colony counter. The average number of colonies for each cell line at each treatment level was calculated, and from these values, the percentage of cells that survived at each level of treatment was determined by comparison of the number of surviving cells to the number of untreated control cells. A nonlinear regression curve was fit to these values using GraphPad Prism software to estimate a 50% inhibitory concentration (IC50) for each cell line in each individual experiment. Cell lines were tested with each damaging agent 2 to 3 times, and the IC50s were estimated in this way each time. The averages of these calculated IC50s from multiple experiments are shown in the bar graphs in the figures, with the error bars representing the standard deviations between separate experiments.
Southern blotting.
Genomic DNA was prepared from each clone after phenol-chloroform extraction. Fifty micrograms of DNA was digested with either HindIII or StuI overnight. The digested DNA was electrophoresed through a 0.8% agarose gel and transferred to a nylon membrane (catalog number 10416282; Whatman) by neutral transfer. Membranes were then probed with an 812-bp green fluorescent protein (GFP) gene sequence-containing fragment derived from pDR-GFP puromycin by HindIII digestion (35). The probe was labeled using a Ladderman labeling kit by TaKaRa (catalog number 6046), and blotting was performed using a modification of the method of Church and Gilbert (38).
siRNAs.
An siRNA against GL2 (siGL2) was purchased from Dharmacon (catalog number D-001100-01-20). The sequence of the siRNA against BRCA1 exon 13 (siBRCA1 exon 13) was GGGAUACCAUGCAACAUAA (catalog number D-003461-06-0020; Dharmacon). The sequence of siBRCA1 exon 11 no. 1 was CCAAAUCAGUAGAGAGUAAUU. The sequence of siBRCA1 exon 11 no. 2 was GUUAGAUGAUGGUGAAAUA. The siRNA against the Δ11b junction sequence (siΔ11b junction) was GUAUCAGGGUGAAGCAGCAUU.
Transfection.
Lipofectamine RNAiMax (catalog number 13778150; Life Technologies) was used for the transfection of all siRNAs. Lipofectamine 2000 (catalog number 11668019; Life Technologies) was used for the transfection of I-SceI.
HR reporter assay.
Plating efficiencies were determined for each cell line such that the cells could reach the end of the assay without becoming completely confluent. Cells were plated in a 6-cm plate at an appropriate density on day 1. On day 2, the cells were transfected with 10 pmol of each siRNA. On day 3, the cells were transfected with 2 μg of the I-SceI plasmid (pCBAS). The cells were then allowed to recover for 72 h. At that point, one half of the cells was harvested for an I-SceI–HA Western blot and the other half was fixed with 3% paraformaldehyde and assayed for GFP-positive cells by fluorescence-activated cell sorting (FACS).
For the IP-Western blot assays performed to test the effects of the various siRNAs introduced into each cell line, the transfection sequence and quantities of siRNA used were different from those used in the HR assays because these IP-Western blot analyses were performed before the HR assays in an effort to test the efficacy of the relevant siRNAs. It was later apparent that less siRNA could be used to achieve a reproducible HR phenotype without eliciting aberrant cell cycle phenotypes. The cells were plated on day 1, transfected with 75 pmol of each siRNA on day 2, transfected with 75 pmol of each siRNA on day 3, transfected with 2 μg I-SceI on day 4, and harvested and extracted for IP 72 h after I-SceI transfection.
RESULTS
BRCA1 expression in sporadic BLC cell lines.
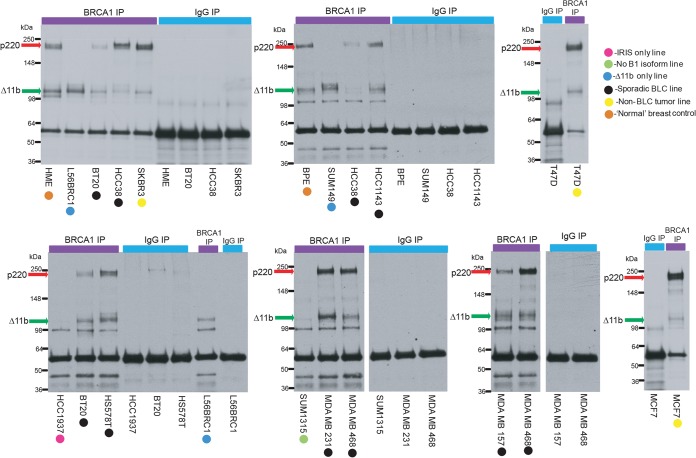
A collection of sporadic, BRCA1+/+ human breast cancer cell lines shown by expression profiling to represent the BLC subtype (34, 39, 40) was analyzed, along with controls. The controls consisted of two independently derived, telomerase-immortalized, normal breast epithelial cell lines, BPE and HME (41). Before performing assays of DNA repair proficiency in these cells, we asked whether these sporadic BLC and control cell lines express full-length BRCA1 using nonquantitative, standardized BRCA1 IP-Western blot analysis performed on asynchronous, undamaged cells (Fig. 1). IP was performed with a C-terminal BRCA1 antibody, and blots were developed with an N-terminal BRCA1 antibody. All sporadic BLC and control cell lines expressed readily detectable full-length p220 and Δ11b (Fig. 1). Thus, any HR defect observed in them cannot be attributed to a loss of BRCA1 p220 or Δ11b expression. IRIS expression was not tested in these lines, as IRIS does not bind BARD1 and has not been shown to be important in HR (14). No comparisons of the abundance of the two primary isoforms observed among the relevant cell lines from these IP-Western blot analyses could be made, as these assays were performed on asynchronous cells and, being IPs, present an amplification of the signal that might be observed in a direct Western blot. Bands that appear as doublets in these blots represent the phosphorylated versions (slower-migrating bands) and nonphosphorylated versions of the different isoforms.
FIG 1.
Sporadic BLC cell lines express p220 and Δ11b. Full-length BRCA1 and the Δ11b BRCA1 isoform were immunoprecipitated from lysates of the sporadic BLC cell lines, normal controls, and BRCA1 mutant lines, using a C-terminal epitope BRCA1 antibody (sc6954). Blots were performed with an N-terminal epitope BRCA1 antibody (MS110). p220 migrates between the 148- and 250-kDa markers and is marked by a red arrow in the molecular mass marker lane. Δ11b migrates just above the 98-kDa marker and is marked by a green arrow in the molecular mass marker lane. Different colored circles mark the type of line that was tested in each blot, and the key for the circles is shown. Several BLC cell lines were blotted more than once to confirm the results.
In addition, four BRCA1 mutant lines served as positive controls in all experiments. Two of them (SUM149 and L56BRC1) have sustained nonsense mutations in BRCA1 exon 11 which prevent them from expressing intact p220 but allow them to express Δ11b (Fig. 1). HCC1937 cells carry a classical, disease-producing, germ line BRCA1 mutation, an insertion of a C nucleotide at position 5383 (5382insC), in the sequence that encodes one of its BRCT motifs. SUM1315 cells contain another classical disease-producing mutation, a deletion of A and G nucleotides at position 185 (185delAG), which results in a severe truncation of p220. Importantly, no readily detected BRCA1 protein was observed in these two lines under the conditions that were used (Fig. 1).
Evidence of postdamage DNA repair responses in nuclear foci.
The repair of DSBs by HR and the repair of stalled or collapsed replication forks are dependent upon BRCA1-containing complexes concentrating at sites of DNA damage in a defined temporal order (24, 32, 33). Thus, assessing the postdamage concentration of specific members of these protein complexes in IRIF by immunofluorescent (IF) staining can be used as a surrogate reporter of HR, at least up to the point of the arrival of the assayed proteins.
(i) HR repair of DSBs.
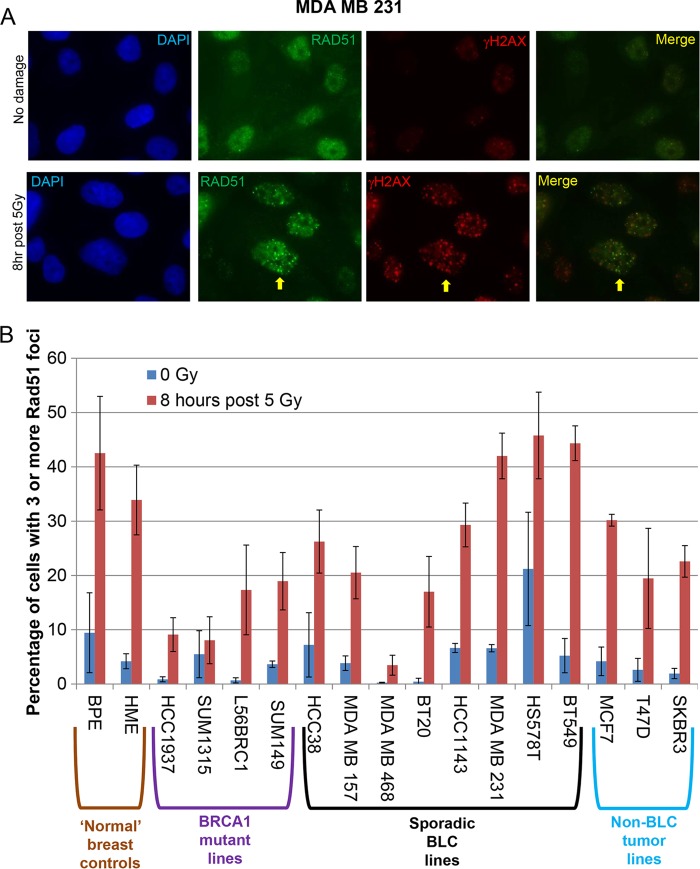
To study HR, we assayed the formation or absence of post-gamma-irradiation Rad51, BRCA1, and 53BP1 nuclear foci (Fig. 2 and 3A and B; Table 1), and all of the sporadic BLC cell lines and controls revealed post-gamma-irradiation BRCA1- and 53BP1-containing foci. While variable in prevalence, Rad51 foci were also regularly observed in all of these tumor cell lines.
FIG 2.
Postdamage Rad51 foci in sporadic BLC cells and controls. (A) MDA MB 231 cells treated or not treated with 5 Gy irradiation and stained with γH2AX and Rad51 antibodies 8 h after treatment. An arrow has been placed next to a representative cell in which there is colocalization. The brightness was increased by 20% and the contrast was increased by 20% using PowerPoint in every panel to alleviate difficulties with the conversion of the images to PDF. DAPI, 4′,6-diamidino-2-phenylindole. (B) All sporadic BLC cell lines, normal controls, BRCA1 mutant lines, and non-BLC cell lines were treated with 5 Gy or mock treated (0 Gy), allowed to recover for 8 h, and then fixed and stained for Rad51. The percentage of cells containing three or more Rad51 foci was calculated for each cell line under each condition. This experiment was repeated 2 to 3 times for each cell line, and the bars in the bar graph represent the average percentage of cells containing 3 or more Rad51 foci, as determined in these experiments. The error bars represent the standard deviations between the experiments.
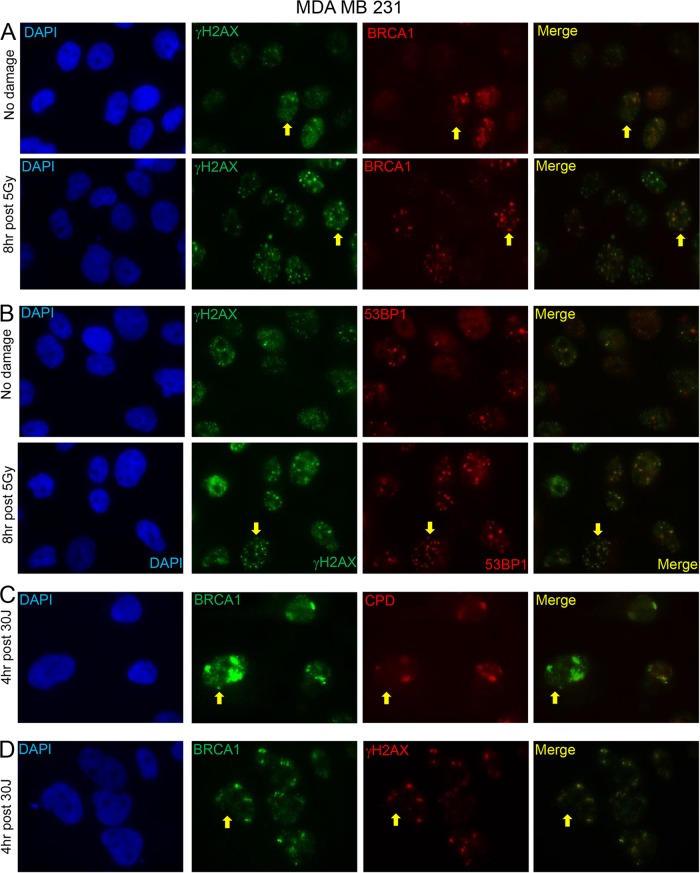
FIG 3.
Representative immunofluorescence results after exposure to different DNA-damaging agents. (A) MDA MB 231 cells treated or not treated with 5 Gy irradiation and stained with γH2AX and BRCA1 antibodies 8 h after treatment. (B) MDA MB 231 cells treated or not treated with 5 Gy irradiation and stained with γH2AX and 53BP1 antibodies 8 h after treatment. (C and D) MDA MB 231 cells 4 h after 30-J UV irradiation through micropores stained with BRCA1 and CPD antibodies (C) or BRCA1 and γH2AX antibodies (D). CPDs are known to mark sites of UV damage. Arrows are placed next to a few representative cells in which there is colocalization. The brightness was been increased by 20% and the contrast was increased by 20% using PowerPoint in every panel to alleviate difficulties with the conversion of the images to PDF.
TABLE 1.
Postdamage IF results in all BLC cell lines and controlsa
| Cell type and cell line | Result |
|||
|---|---|---|---|---|
| Post-IR BRCA1 foci | Post-IR Rad51 foci | Post-IR 53BP1 foci | Post-UV BRCA1-CPD colocalization | |
| Normal breast cells | ||||
| BPE | + | + | + | + |
| HME | + | + | + | + |
| BLC cells | ||||
| HCC38 | + | + | + | + |
| HCC1143 | + | + | + | + |
| MDA MB 157 | + | + | + | + |
| MDA MB 231 | + | + | + | + |
| MDA MB 468 | + | + | + | + |
| BT20 | + | + | + | + |
| BT549 | + | + | + | + |
| HS578T | + | + | + | + |
| BRCA1 mutant cells | ||||
| HCC1937 | − | + | + | − |
| SUM1315 | − | + | + | − |
| SUM149 | + | + | + | + |
| L56BRC1 | + | + | + | + |
| ER-positive tumor cells | ||||
| MCF7 | + | + | + | + |
| T47D | + | + | + | + |
| Her2-amplified tumor cell line, SKBR3 | + | + | + | + |
Cell lines were exposed to various DNA-damaging agents on coverslips and then stained for different DNA damage markers. For gamma irradiation (IR), one set was treated with 5 Gy IR, while a second, control set was not. The cells were allowed to recover at 37°C for 8 to 9 h. For UV treatment, cells were treated with 30 J through a UV micropore filter and then allowed to recover for 4 h at 37°C.
Because of the variability in the prevalence of Rad51 foci and because Rad51 foci are a potential biomarker for sensitivity to chemotherapeutic agents and targeted therapies that generate DSBs (42–44), we quantified the number of cells containing three or more Rad51 foci 8 h after treatment with 5 Gy of IR or mock treatment for all cell lines in this study (Fig. 2B). More than half of the sporadic BLC cell lines exhibited an abundance of Rad51 foci similar to that of the normal breast epithelial cell controls (Fig. 2B), implying that these lines do not harbor an HR defect or that, if they do, it is due to a malfunction downstream of Rad51 action. A subset of the sporadic BLC cell lines tested revealed fewer post-gamma irradiation Rad51 foci than the normal breast control cells, but detectable amounts were still present, indicating a potential HR defect that either is due to inefficient Rad51 loading or is manifest downstream of the Rad51 function (Fig. 2B).
We also analyzed 53BP1 foci in the sporadic BLC cell lines, because loss of 53BP1 expression in BRCA1 mutant cells led to the restoration of HR in these cells (45). In cells deficient for both 53BP1 and BRCA1, DSB end excision can occur in an ATM-dependent manner in such a way that it results in HR-mediated DSB repair, despite the loss of BRCA1 (46). However, against the possibility that some or all of the sporadic BLC cell lines manifest a BRCA1-related defect in HR that is rescued by 53BP1 deficiency (46), 53BP1 foci were readily detected in all sporadic BLC cell lines (Table 1). This implies that BRCA1-driven HR function and its regulation by 53BP1 are intact in these sporadic BLC cell lines, at least up to the point of Rad51 loading.
IRIF formation was also analyzed in the BRCA1 mutant lines. 53BP1 foci were observed in all four BRCA1 mutant lines (Table 1; see also Fig. S1 in the supplemental material), implying that any HR phenotypes observed in them are not a result of 53BP1 loss. In addition, BRCA1 foci were observed in both the SUM149 and L56BRC1 cell lines, the two lines that still express Δ11b (Table 1). These foci were absent when these lines were analyzed by IF using a BRCA1 antibody that recognizes an epitope in exon 11 (see Fig. S2 to S4 in the supplemental material) and disappeared upon exposure to Δ11b-specific but not p220-specific siRNAs (see Fig. S5 in the supplemental material). Therefore, it is likely that the BRCA1 foci observed in these two lines contain Δ11b and lack p220.
Finally, Rad51 foci were observed in all of the BRCA1 mutant lines (Table 1; Fig. 2B). SUM1315 cells, which contain a greatly truncated form of BRCA1 that cannot localize at breaks or interact with any HR repair proteins, and HCC1937 cells, which contain a truncated BRCA1 lacking one of the BRCT motifs that cannot efficiently localize to DSBs, revealed the lowest percentages of Rad51 foci post-gamma irradiation. This was expected, given the poor ability of the former to localize at breaks and to bind important BRCA1 interactors and of the latter to localize to DSBs. SUM149 and L56BRC1 cells, which produce the Δ11b isoform that can localize to DSBs, as demonstrated in Fig. S2 to S5 in the supplemental material, exhibited post-gamma-irradiation Rad51 foci that were less abundant than they were in the normal controls but more abundant than they were in BRCA1 mutant cells that contained a truncated BRCA1 species that could not localize to DSBs. Conceivably, SUM149 and L56BRC1 cells recruit Rad51 through Δ11b, or potentially, all four lines recruit it by a less efficient process that may be independent of BRCA1 (47). The localization of Δ11b to sites of DSBs, combined with percentages of post-gamma-irradiation Rad51 foci higher than the percentages observed in lines containing truncated BRCA1 polypeptides, hints at a limited HR function for this isoform, and this possibility needs to be explored further.
(ii) Stalled replication forks.
We also assayed these cell lines for their ability to repair stalled or collapsed replication forks, another BRCA1 function (23, 24). This was undertaken by performing UV micropore analysis (36, 48, 49) (Fig. 3C and D; Table 1). In these assays, BRCA1 concentrates in UV-irradiated micropore territories approximately 30 min after UV-C exposure in a replication-dependent manner (24). There it participates in the repair of stalled/collapsed replication forks (24).
In all sporadic BLC cell lines and controls that were tested, BRCA1 was readily attracted to micropore territories marked by both cyclobutane pyrimidine dimers (CPDs) and γH2AX (Fig. 3C and D and Table 1). This implies that the role of BRCA1 in stalled fork repair is intact in these cell lines up to the point of its recruitment to these territories. This does not mean that the function of some other BRCA1 partner in this pathway acting at or after BRCA1 recruitment to these areas of DNA damage is not altered. In such a scenario, wild-type (wt) BRCA1 would concentrate at genomic sites where stalled forks are located, but it might still not contribute to the repair of stalled or collapsed forks due to the loss of function of a relevant binding partner.
In contrast, no BRCA1 was observed in micropore territories in the BRCA1 mutant lines HCC1937 and SUM1315, and only faint BRCA1 micropore staining (suspected of reflecting the presence of Δ11b) was observed in the SUM149 and L56BRC1 cell lines, the two p220-negative, Δ11b-producing lines (Table 1).
DNA repair analysis: repair of mutagen-induced DNA damage.
Having accumulated these results, we searched for defects in the repair of the DNA damage elicited by various mutagens. We first assayed all BLC cell lines for proficiency in the response to the damage elicited by four different mutagens by assessing colony formation after exposure to a range of doses (Fig. 4).
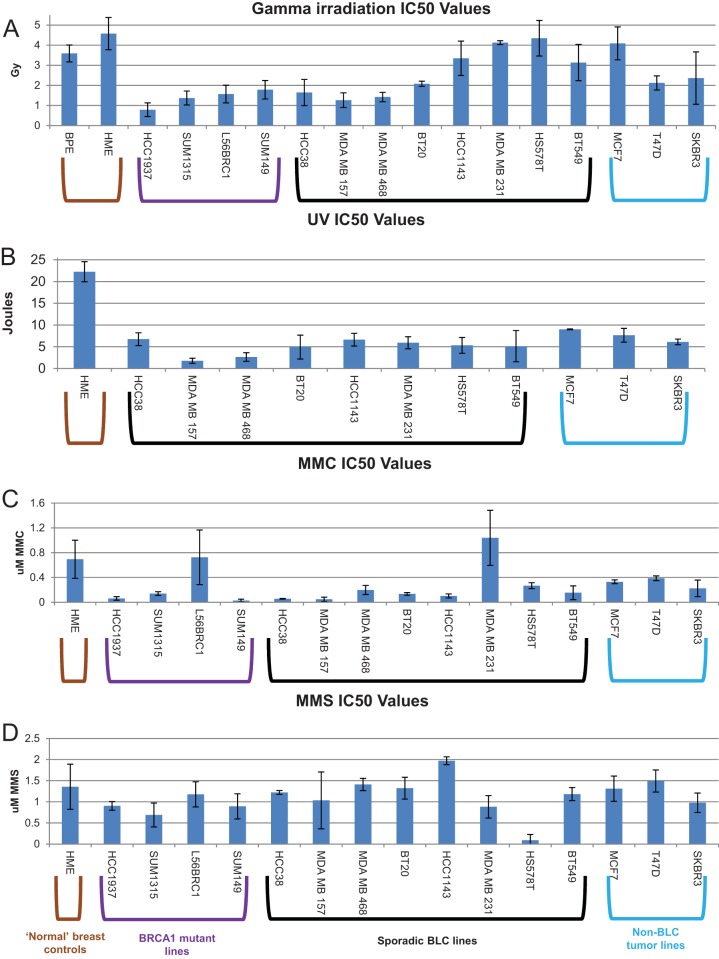
FIG 4.
Sensitivity of cell lines to various DNA-damaging agents. Cells of each line were plated in triplicate at a density suitable for colony formation, allowed to settle, treated with various doses of different DNA-damaging agents, and then allowed to recover and grow at 37°C until colonies became visible. After colonies of the appropriate size had grown, the cells were stained with crystal violet staining solution and counted with a Microbiology International ProtoCOL colony counter. IC50s for each cell line for each treatment were calculated on the basis of the dose-response curves generated from these counts. The bars in the bar graphs represent the average IC50s for each cell line from 2 to 3 experiments after the following: gamma irradiation (A), UV irradiation (B), MMC treatment (C), and MMS treatment (D). The error bars represent the standard deviations between experiments. The color of the brackets grouping certain cell lines represents the cell line type, as depicted in the key at the bottom.
To test DSB repair responses, we exposed the cell lines to gamma irradiation (IR) (Fig. 4A). Half of the sporadic BLC cell lines were supersensitive to gamma irradiation (meaning that they were significantly more sensitive than the normal breast cell controls), like all BRCA1 mutant cell lines, suggesting the possibility of a functional defect in gamma irradiation-induced DNA damage repair. For example, a BRCA1 partner protein(s) that operates in DSB repair in these cells might be depleted or otherwise functionally defective. However, half of the sporadic BLC cell lines were gamma irradiation resistant, like the controls. Since HR-incompetent cells are expected to be supersensitive to gamma irradiation, these results suggested that not all of the sporadic BLC cell lines are defective in HR, in keeping with the above-noted results of IRIF analyses of these cells. It is particularly interesting to note that the four sporadic BLC cell lines that were the most gamma irradiation resistant exhibited an abundance of post-gamma-irradiation Rad51 foci very similar to that of the controls (Fig. 2B). However, one of the gamma irradiation-sensitive lines also revealed focus formation similar to that of the controls (HCC38 cells) (Fig. 2B and 4A). Given all of these results, it is unclear how well the abundance of Rad51 foci and gamma irradiation sensitivity correlate with HR proficiency in these tumor cell lines. A more definitive test of HR was, therefore, needed and was undertaken (see below).
To test the responses to interstrand cross-link-associated stalled and collapsed replication forks, we exposed the cell lines to mitomycin C (MMC). All but one of the sporadic BLC cell lines and all but one of the BRCA1 mutant cell lines were supersensitive to MMC (Fig. 4C). This suggests that these sporadic BLC cell lines are phenocopies of the BRCA1 mutant lines in manifesting a defect in interstrand cross-link repair and/or the subsequent replication fork stalling that it elicits. BRCA1 contributes to the repair of this mode of cross-linking by multiple mechanisms (23–27).
The responses to stalled and collapsed replication forks were independently tested by exposing the cells to UV irradiation (a source of intrastrand cross-linking). Here all sporadic BLC cell lines were supersensitive to UV irradiation compared to the controls (Fig. 4B). The BRCA1 mutant lines had previously been tested for sensitivity to UV, and all were found to be supersensitive, with IC50s being similar to or less than those of all the sporadic BLC cell lines shown here (24).
When taken together, these and the MMC sensitivity results suggest that the sporadic BLC cell lines are phenocopies of BRCA1 mutant lines in their deficient responses to stalled replication fork development. When normally executed, stalled fork repair and the suppression of replication stress are known BRCA1 functions (23–27). Since the sporadic BLC cell lines that were tested lack BRCA1 mutations (34, 39, 40), failed suppression of replication stalling and failed repair of collapsed forks in these cells may be a product of the defective function of a protein(s) that is active in these types of repair or of a nongenetically derived alteration in BRCA1 function itself. These MMC and UV data suggest that stalled replication fork repair is a commonly encountered problem in sporadic BLC cell lines. This is in keeping with the clinical findings that certain cases of BLC are cisplatin sensitive (50–55).
Finally, in testing the cell lines for methyl methanesulfonate (MMS) responsiveness and, thus, an ability to perform base excision repair (BER) (Fig. 4D), BRCA1 mutants and nearly all sporadic BLC cell lines revealed drug sensitivity similar to that of the controls. In fact, only one sporadic BLC cell line exhibited supersensitivity. This suggests that most of these lines are competent for base excision repair of MMS damage.
In contrast, most of these sporadic BLC cell lines and BRCA1 mutant controls are known to be supersensitive to hydrogen peroxide-induced oxidative damage, which can also be repaired by BER, among other pathways (56). In addition, using an oxidative damage repair reporter, a decrease in oxidative damage repair efficiency has been observed in three of the sporadic BLC cell lines, one of the BRCA1 mutant lines, and a BRCA1-depleted line (56). Thus, loss of BRCA1 expression may result in a defect in oxidative damage-induced BER, as opposed to base alkylation-induced BER (56). Moreover, there may be a defect in this form of BER in certain sporadic BLCs.
Sporadic BLC cell lines are proficient in HR.
Given the mixed gamma irradiation sensitivity and post-gamma-irradiation Rad51 focus-forming results (Fig. 2B and 4A), experiments were performed to test whether or not the above-noted sporadic BLC cell lines can perform HR, using a well-established HR reporter that allows an assessment, by FACS analysis, of the ability of a cell line in which it is integrated to perform HR (35).
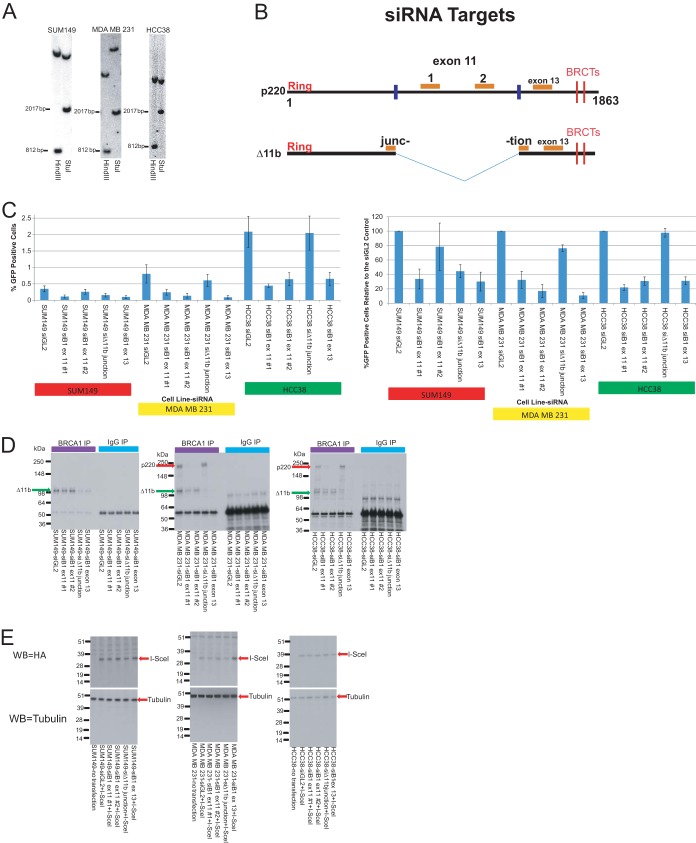
Stably transfected derivatives of three of the cell lines analyzed in the above-noted experiments were generated, with each bearing a single, integrated copy of the above-noted HR reporter. One was a BRCA1 mutant line (SUM149), one was a gamma irradiation-resistant sporadic BLC cell line (MDA MB 231), and one was a gamma irradiation-sensitive sporadic BLC cell line (HCC38). Multiple clones of each were tested by Southern blotting in a search for any that contained a single, integrated copy of the HR reporter. One clone of each was identified (Fig. 5A). Each was then studied for its ability to perform HR.
FIG 5.
BLC cell lines are HR proficient regardless of gamma irradiation sensitivity status. (A) Southern blots were performed on multiple clones of the three cell lines used in the HR reporter experiments in this figure to identify a clone from each line that carries a single, integrated copy of the I-SceI–GFP HR reporter. Separate blots for the three clones studied in the HR reporter experiments are shown here. Based on the sequence of the reporter, we digested clonal genomic DNA separately with two different enzymes to assess the reporter copy number. We digested with HindIII, which should give rise to a band of 812 bp and another single band of various sizes in clones bearing a single copy of the reporter. We also digested with StuI, which should give rise to a band of 2,017 bp and another single band of various sizes in clones bearing a single copy of the reporter. (B) Map of the target sites of the four BRCA1 siRNAs used in the HR experiments on p220 and Δ11b. (C) (Left) The average percentage of GFP-positive cells in the HR reporter assay for each cell line with each siRNA is represented in the bar graph. The error bars represent the standard deviations from four experiments. (Right) The HR reporter results have been normalized to those for the siGL2 control for each respective day in each respective cell line, and the bars in the graph represent the average percentage of GFP-positive cells relative to the number of GFP-positive siGL2-transfected control cells for that cell line, with the error bars representing the standard deviations from the four experiments. (D) IP-Western blotting was performed on extracts of each cell line transfected with each siRNA to demonstrate the efficacy of depletion of the specific isoform by each siRNA. IPs were performed with a C-terminal BRCA1 antibody, and the blots were developed with an N-terminal BRCA1 antibody so that both the p220 and Δ11b isoforms could be detected. Green arrow, Δ11b; red arrow, p220. (E) Western blotting (WB) assays were performed on half of the cells from one round of the HR reporter assay to demonstrate that the I-SceI protein levels were the same in each cell line after each siRNA treatment. The I-SceI that was transfected was tagged with HA. After loading of equivalent amounts of protein on the gel from each cell line transfected with each siRNA, the blots were stained with HA antibody to detect I-SceI. Tubulin blotting was performed to show that equivalent amounts of protein were loaded in each lane. B1, BRCA1; ex, exon.
To test these cell lines for HR proficiency, we performed the above-noted HR reporter assay in cells transfected with either a firefly luciferase control siRNA (siGL2) or one of four different BRCA1-specific siRNAs (Fig. 5C; see Table S1 in the supplemental material). To target Δ11b, we generated an siRNA targeting the unique junction sequence between the early part of exon 11, which is present in the Δ11b mRNA, and the 5′ region of exon 12 (Fig. 5B). An siRNA that targets exon 13 of BRCA1 was also employed (Fig. 5B). It should deplete both full-length p220 and Δ11b (Fig. 5B). Finally, we employed two exon 11-specific siRNAs that depleted p220 but not Δ11b (Fig. 5B).
IP-Western blot analyses performed with extracts of each of these single-copy HR reporter-containing clones showed that these siRNAs deplete the predicted BRCA1 isoforms (Fig. 5D). The slight depletion of Δ11b in the MDA MB 231 clone observed with exon 11 siRNA 1 is likely due to slowing of the cell cycle caused by depletion of p220 and not an off-target effect of the siRNA. Thus, with this possible exception, the siRNAs deplete the appropriate BRCA1 isoforms.
BRCA1 siRNAs were employed in these experiments for multiple reasons. First, BRCA1 is a physiological contributor to normal HR function (16–18). Hence, its depletion should result in a loss of HR capacity, if a given cell line produces the relevant BRCA1 isoform (e.g., p220) and can perform this function (21, 22). Thus, in these experiments, BRCA1 depletion served as a positive control employed to determine whether any GFP signals that were observed were a product of BRCA1-driven HR function. Second, the use of BRCA1 siRNAs made it possible to determine whether any HR activity that was measured in a reporter cell line was BRCA1 dependent. Finally, four different BRCA1 siRNAs, each targeting a different region of the BRCA1 mRNA, were employed (Fig. 5B) to determine which isoform—p220, Δ11b, or both—was responsible for any HR activity that was observed in a given cell line. This question was particularly relevant for SUM149 cells, which express the Δ11b isoform but not the full-length p220 isoform. IRIS does not concentrate in post-gamma irradiation foci or bind BARD1, both of which are HR-associated properties (14). Therefore, it is not believed to be involved in HR and was not tested here.
In the HR analyses that were performed, it was not possible to compare, even semiquantitatively, the HR functionality of the three lines that were tested. This is because the I-SceI-driven HR reporter was integrated at different genomic sites in each cell line and because the cell lines are not isogenic. However, these considerations should not nullify the validity of an internal comparison within each cell line of I-SceI-driven GFP production measured before and after BRCA1 depletion.
The results of the HR assay were presented as both the percentage of GFP-positive cells for each cell line and the percentage of GFP-positive cells relative to the number of GFP-positive siGL2-transfected control cells for that line to facilitate their interpretation (Fig. 5C; see Table S1 in the supplemental material).
To control for variability in the transfection efficiency of I-SceI, standardized Western blotting for I-SceI was performed in conjunction with the HR assay (Fig. 5E). I-SceI levels were relatively uniform in each cell line and were unaffected by the various BRCA1-directed siRNA treatments (Fig. 5E). Thus, any differences in the percentage of GFP-positive cells within a single cell line among the different siRNA transfections were not a product of various transfection efficiencies and/or I-SceI expression levels. In initial experiments, we also cotransfected mCherry with I-SceI and scored only mCherry-positive cells in the FACS, also to control for transfection variability. There, too, the results were similar to those noted above, regardless of mCherry cotransfection (data not shown).
As shown in Fig. 5C (for other related results, see Table S1 in the supplemental material), the SUM149 HR reporter-containing clone, which synthesizes Δ11b and not p220, exhibited measurable BRCA1-dependent HR function. When the line was transfected with the exon 13-specific siRNA, which depletes both p220 and the Δ11b BRCA1 isoform, or with the Δ11b-specific junction siRNA, the percentage of GFP-positive cells decreased, reflecting the presence of BRCA1 gene product-dependent HR in this line. As expected, one of the exon 11 siRNAs (exon 11 siRNA 2) had no effect on HR activity in this line, while the other one (exon 11 siRNA 1) caused a decrease. In light of the junction siRNA effect and the lack of effect of exon 11 siRNA 2, one can hypothesize that SUM149 cell HR activity is Δ11b dependent. Given the HR-suppressing effect of the other exon 11 siRNA (exon 11 siRNA 1), it is conceivable that a yet undetected, alternatively spliced BRCA1 mRNA is expressed in these cells and its product contributes to the observed HR function. Alternatively, HR suppression by this siRNA species represents an off-target effect. These results for SUM149 cells correlate with the localization of Δ11b to DSBs observed in Fig. S2 to S5 in the supplemental material and by others (57), as well as the slight increase in post-gamma-irradiation Rad51 foci in this line compared to the BRCA1 mutant lines containing mutations that produce greatly truncated proteins. Thus, they further support the hypothesis that Δ11b exhibits a limited HR capacity.
Among sporadic BLC cell HR reporter-containing lines, MDA MB 231, which was relatively gamma irradiation resistant (Fig. 4A) and revealed an abundance of post-gamma-irradiation Rad51 foci similar to that for the normal breast cell controls (Fig. 2B), appeared to be HR proficient. When this line was transfected with each of the exon 11-specific siRNAs or the exon 13-specific siRNA that depletes both isoforms, HR proficiency decreased significantly (Fig. 5C; see Table S1 in the supplemental material). In contrast, the Δ11b-specific junction siRNA had no effect (Fig. 5C; see Table S1 in the supplemental material). Thus, this line appears to perform HR in a p220-dependent manner.
HCC38 cells were supersensitive to gamma irradiation (Fig. 4A) and also revealed an abundance of post-gamma-irradiation Rad51 foci slightly reduced compared to that of the controls (Fig. 2B). However, HCC38 cells yielded a significant HR signal after siGL2 transfection which decreased significantly after exposure to either of the exon 11 siRNAs or the exon 13 siRNA (Fig. 5C; see Table S1 in the supplemental material). Like MDA MB 231 cells, its HR signal was unaffected by the Δ11b junction-specific siRNA (Fig. 5C; see Table S1 in the supplemental material). Thus, although it was supersensitive to gamma irradiation, HCC38 cells are HR proficient, and they, too, perform HR in a p220-dependent manner. This finding is interesting with regard to post-gamma-irradiation Rad51 focus formation. The HCC38 cell line is the one gamma irradiation-sensitive line that exhibited a post-gamma irradiation Rad51 focus-forming effect close to that of the control cell lines (Fig. 2B).
Taken together, these results strongly suggest that in cells lacking a BRCA1 mutation (39, 40), p220 is responsible for HR, but in cells that express only Δ11b, the Δ11b isoform is itself capable of supporting this function. In addition, these results and those described earlier collectively show, or otherwise strongly suggest, that the sporadic BLC cell lines tested, regardless of their gamma irradiation sensitivity and post-gamma-irradiation Rad51 focus-forming activity, are HR proficient and, where tested, this DNA repair function is BRCA1 gene product dependent.
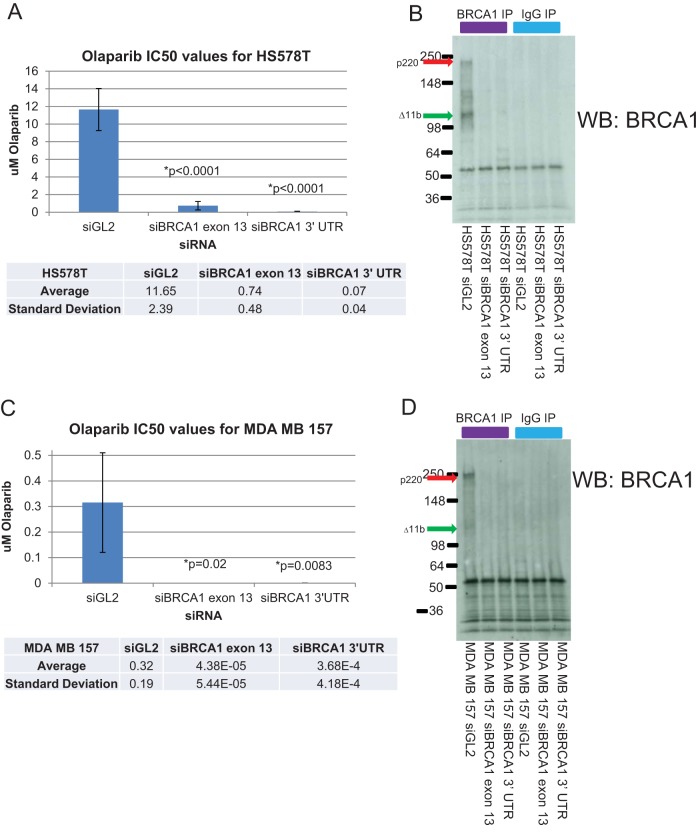
The SUM149, HCC38, and MDA MB 231 cell lines were the only lines used in these studies into which we were able to successfully integrate a single copy of the HR reporter. However, we were particularly interested in attempting to perform a more definitive test of the HR proficiency of the HS578T and MDA MB 157 sporadic BLC cell lines. These two lines have been shown to express large quantities of the microRNAs miR-146a and miR-146b-5p, which have been shown to downregulate the expression of BRCA1 and to decrease HR when their precursors are transfected into an unrelated cell line (RG37) that contains an HR reporter (58). The HR proficiency of the MDA MB 157 and HS578T cell lines was not definitively tested in the study, demonstrating that these lines overexpress these BRCA1-regulating microRNAs. However, it is possible that one or both of these cell lines are defective in HR, given the HR findings obtained when the above-noted microRNAs were expressed ectopically in an unrelated cell line (58).
Our IP-Western blot analysis of BRCA1 protein expression revealed that both of these cell lines express both p220 and Δ11b (Fig. 1). However, these studies were not quantitative, so it is not possible to know what levels were expressed compared to the levels expressed by the other cell lines tested. The results of surrogate HR assays in these lines were mixed. HS578T cells exhibited a post-gamma-irradiation Rad51 focus response similar to that of the normal breast cell controls and were gamma irradiation resistant, while MDA MB 157 cells revealed a post-gamma-irradiation Rad51 response below that of the normal breast cell controls but still greater than that of the two least-effective Rad51 focus-producing BRCA1 mutant lines (Fig. 2B and 4A). MDA MB 157 cells were gamma irradiation sensitive (Fig. 4A). Based on these results, one might have suspected that HS578T cells would be HR proficient and MDA MB 157 cells would be HR defective.
Therefore, a more definitive test was needed. Despite numerous attempts, we were unable to generate clones of these lines bearing a single copy of the HR reporter. Thus, we assayed their sensitivity to the poly(ADP-ribose) polymerase (PARP) inhibitor olaparib with and without BRCA1 depletion. In this type of assay, a cell line would be deemed HR proficient if it were more sensitive to the PARP inhibitor in the setting of BRCA1 depletion than in the wt setting (59). Hence, we transfected each cell line with a control siRNA and two different BRCA1 siRNAs and then tested its sensitivity to a range of concentrations of olaparib by colony formation assay (Fig. 6). Though their baseline levels of sensitivity to olaparib were different, each line became much more sensitive to the PARP inhibitor upon BRCA1 depletion, indicating that both lines are HR proficient and perform this HR in a BRCA1-dependent manner (Fig. 6). Determining the amplitude of HR proficiency is not possible in this assay, since one is comparing entirely different nonisogenic cell lines. However, an internal comparison of the results of different siRNA exposures within a cell line remains a valid method of assessing whether or not the line can perform HR.
FIG 6.
HR capacity of HS578T and MDA MB 157 cell lines. The HR capacity of the sporadic BLC cell lines HS578T and MDA MB157 was tested by transfecting these lines with a control siRNA or two different BRCA1 siRNAs, seeding the cells at a density suitable for colony formation, and exposing the cells to various concentrations of the PARP inhibitor olaparib. The cells were grown in PARP inhibitor-containing medium until colonies of suitable size had formed. Colonies were stained and counted, and IC50s were estimated from the dose response curves generated by these colony counts. (A and C) The average IC50 for each siRNA from at least three separate experiments is shown in the bar graphs for HS578T (A) and MDA MB 157 (C) cells. Since the siBRCA1 IC50s are so low compared to those for the controls, a table with the relevant values is introduced below each bar graph. The error bars in the bar graphs represent the standard deviations between the averages for multiple experiments. The P values comparing each set of siBRCA1 results to the siGL2 results were calculated using a two-tailed t test in GraphPad Prism software. (B and D) BRCA1 IP-Western blots of HS578T and MDA MB 157 cells transfected with the siRNAs used for the assays whose results are presented in panels A and C demonstrate the ability of these siRNAs to deplete BRCA1 and are shown for HS578T (B) and MDA MB 157 (D) cells. In these blots, the p220 and Δ11b isoforms are indicated by arrows of different colors. UTR, untranslated region.
Overall, these results indicate that these two cell lines are HR proficient and perform HR in a BRCA1-dependent manner, despite overexpressing microRNAs which downregulate BRCA1 expression (58). It also again demonstrates that gamma irradiation sensitivity and post-gamma-irradiation Rad51 focus formation, though capable of reflecting differences in HR capacity between BRCA1+/+ and BRCA1−/− cells, are not able to show definitively whether or not a given non-BRCA1 mutant cell is capable of performing HR.
DISCUSSION
This work was performed on a subset of human breast cancer cell lines that collectively represent the BLC subtype (34). The results fail to support the hypothesis that, as a general matter, sporadic BLCs exhibit an HR defect and, therefore, are valid phenocopies of BRCA1 mutant tumors in this regard. Rather, they suggest that some other BRCA1-related DNA repair defect(s) exists in significant numbers of sporadic, BRCA1 wt BLCs. Specifically, these defects affect the responses of cells to stalled and collapsed replication forks (23–27). These data are supported by the results of recent clinical trials and retrospective studies of this tumor subtype (50–55), and it suggests that therapeutic strategies aimed at inducing replication stress in tumor cells may be useful in this tumor type.
In keeping with this notion, PARP inhibitors, the toxicity of which relies on tumor cells exhibiting an HR defect (59–61), have been of particular interest. These agents have been tested in a wide variety of tumors, but they have been most promising in the clinic in the therapy of both BRCA1 and BRCA2 mutant breast and ovarian tumors (62–64), as well as in certain sporadic ovarian cancers (62, 63). The notion that sporadic BLCs manifest a defect in HR and the success of these agents in BRCA1 mutant tumors have spurred clinical trials with PARP inhibitors in sporadic BLCs. However, sporadic BLCs have failed after treatment with single agents (62, 64) and PARP inhibitor/antiangiogenic therapy combinations (63). We did not test our entire cell line panel for PARP inhibitor sensitivity, because most of the cell lines, along with others representing different subtypes, have already been tested for sensitivity to two different PARP inhibitors (65). Five of the sporadic BLC cell lines (MDA MB 231, MDA MB 468, BT20, HCC1143, and HS578T), one of the BRCA1 mutant lines (HCC1937), and the three non-BLC cell lines (MCF7, SKBR3, and T47D) tested in our work were assayed in the PARP study (65). The results are difficult to interpret without comparing the lines to a normal breast control cell line, but there was no uniformity in PARP inhibitor sensitivity among the different subtypes from these data (65). Thus, on the basis of that work, it also appears that not all BRCA1 mutant genotypes and breast tumor subtype designations are effective predictors of sensitivity to PARP inhibitors. These results, taken together with the lack of an HR defect in sporadic BLC cell lines observed in our results, may help to explain why PARP inhibitors have failed in the therapy of sporadic BLCs. In comparison, we did not test any ovarian cancer cell lines for HR proficiency, but on the basis of the PARP inhibitor trial results, this may be informative (62–64).
In contrast, clinical trials and retrospective studies examining the efficacy of the interstrand cross-link-inducing and subsequent replication fork-stalling drug cisplatin have been successful in a fraction of BLCs (50–55), and results presented here suggest a mechanism that underlies this success. The data reveal that all of the sporadic BLC cell lines tested were supersensitive to an interstrand cross-link-inducing agent and UV irradiation (Fig. 4), both of which ultimately give rise to stalled and collapsed replication forks. Moreover, BRCA1 participates in the repair of interstrand cross-links and stalled and collapsed replication forks, in some cases operating in conjunction with the Fanconi pathway (23–27). In addition, BRCA1 mutant cell lines proved to be supersensitive to agents that cause stalled replication forks, such as MMC, cisplatin, and UV irradiation (24, 66).
Given these findings, we propose that sporadic BLCs are phenocopies of BRCA1 tumors in manifesting a defect in various forms of stalled and collapsed replication fork repair (23–27). However, they do not appear to be uniformly defective in HR. These considerations suggest that pursuing agents which elicit replication fork stalling through a mechanism that threatens BLC survival represents a rational therapeutic strategy in at least some cases of sporadic BLC.
In addition, there may be a role for Δ11b in HR. We have confirmed that Δ11b is recruited to post-gamma-irradiation damage foci (Table 1; see Fig. S2 to S5 in the supplemental material) (57), which suggests that Δ11b concentrates in a suitable nuclear location to participate in HR function. Moreover, the BRCA1 mutant cell lines that expressed Δ11b but not p220 revealed post-gamma irradiation Rad51 focus formation levels that were lower than those of normal breast cell controls but higher than those of BRCA1 mutant cell lines that produced truncated versions of p220 that cannot localize to DSBs (Fig. 2B). This implies the existence of some HR function in these lines. In addition, a BRCA1 mutant cell line that expresses Δ11b and not p220 exhibited a measurable level of Δ11b-driven HR in an HR reporter assay (Fig. 5C).
These data are supported by work in model organisms. Δ11b is the only BRCA1 isoform expressed in Caenorhabditis elegans, and it and the C. elegans BARD1 ortholog appeared to support DNA damage repair in that organism (12). In addition, in HR reporter-bearing mouse ES cells in which p220 but not Δ11b was absent, HR levels were still detectable, suggesting that Δ11b is an HR contributor in this setting, an observation that has also been made by others (22, 67, 68).
The importance of this finding is highlighted by observations in patients and model organisms. Mice that express only Δ11b in a p53 heterozygous setting developed normally but exhibited early-onset breast tumors (69). Similarly, a patient who had a biallelic mutation at the BRCA1 locus but who still expressed intact Δ11b but not p220 exhibited multiple developmental abnormalities and was diagnosed with early-onset ovarian cancer (70).
Taken together, these results suggest a scenario in which Δ11b expression, in the absence of p220, generates enough BRCA1 DNA repair function, manifest at least in part through HR, to prevent embryonic lethality. However, it does not generate enough other BRCA1 function to prevent developmental defects and BRCA1-related tumorigenesis. If validated, this would further imply that, even if it is required to suppress breast and ovarian cancer development, its HR function is insufficient to do so alone.
In summary, the results of the studies reported here demonstrate that tumor cell lines derived from multiple sporadic BLCs (34) are BRCA1 mutant breast cancer cell phenocopies with respect to a subset of BRCA1 functions that direct the repair of stalled and collapsed replication forks (23–27). These findings are consistent with successes observed in clinical trials (50, 52, 55) and retrospective studies (51, 53, 54) of cisplatin in sporadic BLC and support efforts to adapt knowledge of a state of increased, BLC-specific replication stress to its therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kristine McKinney for valuable experimental advice, Christian Colin for advice on Southern blotting, and Kristine McKinney and other members of the D. M. Livingston lab for helpful discussions.
S.J.H. was initially funded by DOD BCRP fellowship W81XWH-08-1-0748 and subsequently by NCI fellowship 1F30CA167895-01. This work was supported by grants from the Susan G. Komen Foundation for the Cure (SAC110022), the Breast Cancer Research Foundation, an NCI SPORE grant (P50CA089393) in breast cancer research to the Dana-Farber/Harvard Cancer Center, and RO1 (R01CA136512) and P01 (P01CA080111) grants from NCI to D.M.L.
S.J.H., D.M.L., and D.P.S. conceived of the project, and D.M.L. and D.P.S. mentored S.J.H. in its execution. A.P.C. performed the Southern blotting assays whose results are shown in Fig. 5A. S.J.H. performed all other experiments and wrote the manuscript. S.J.H., D.M.L., A.P.C., and D.P.S. collectively edited the manuscript.
Footnotes
Published ahead of print 4 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01646-13.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. 2000. Molecular portraits of human breast tumours. Nature 406:747–752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 98:10869–10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100:8418–8423. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- 5.Breast Cancer Linkage Consortium. 1997. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Lancet 349:1505–1510. 10.1016/S0140-6736(96)10109-4 [DOI] [PubMed] [Google Scholar]
- 6.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. 2002. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 20:2310–2318. 10.1200/JCO.2002.09.023 [DOI] [PubMed] [Google Scholar]
- 7.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF. 2005. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin. Cancer Res. 11:5175–5180. 10.1158/1078-0432.CCR-04-2424 [DOI] [PubMed] [Google Scholar]
- 8.Crook T, Brooks LA, Crossland S, Osin P, Barker KT, Waller J, Philp E, Smith PD, Yulug I, Peto J, Parker G, Allday MJ, Crompton MR, Gusterson BA. 1998. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene 17:1681–1689. 10.1038/sj.onc.1202106 [DOI] [PubMed] [Google Scholar]
- 9.Manie E, Vincent-Salomon A, Lehmann-Che J, Pierron G, Turpin E, Warcoin M, Gruel N, Lebigot I, Sastre-Garau X, Lidereau R, Remenieras A, Feunteun J, Delattre O, de The H, Stoppa-Lyonnet D, Stern MH. 2009. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res. 69:663–671. 10.1158/0008-5472.CAN-08-1560 [DOI] [PubMed] [Google Scholar]
- 10.Valentin MD, da Silva SD, Privat M, Alaoui-Jamali M, Bignon YJ. 2012. Molecular insights on basal-like breast cancer. Breast Cancer Res. Treat. 134:21–30. 10.1007/s10549-011-1934-z [DOI] [PubMed] [Google Scholar]
- 11.Tammaro C, Raponi M, Wilson DI, Baralle D. 2012. BRCA1 exon 11 alternative splicing, multiple functions and the association with cancer. Biochem. Soc. Trans. 40:768–772. 10.1042/BST20120140 [DOI] [PubMed] [Google Scholar]
- 12.Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M. 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14:33–39. 10.1016/j.cub.2003.11.029 [DOI] [PubMed] [Google Scholar]
- 13.Nakuci E, Mahner S, Direnzo J, ElShamy WM. 2006. BRCA1-IRIS regulates cyclin D1 expression in breast cancer cells. Exp. Cell Res. 312:3120–3131. 10.1016/j.yexcr.2006.06.021 [DOI] [PubMed] [Google Scholar]
- 14.ElShamy WM, Livingston DM. 2004. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat. Cell Biol. 6:954–967. 10.1038/ncb1171 [DOI] [PubMed] [Google Scholar]
- 15.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71. 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 16.Silver DP, Livingston DM. 2012. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2:679–684. 10.1158/2159-8290.CD-12-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li ML, Greenberg RA. 2012. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 37:418–424. 10.1016/j.tibs.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh T, King MC. 2007. Ten genes for inherited breast cancer. Cancer Cell 11:103–105. 10.1016/j.ccr.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 19.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14:430–440. 10.1038/ng1296-430 [DOI] [PubMed] [Google Scholar]
- 20.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. 1997. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90:425–435. 10.1016/S0092-8674(00)80503-6 [DOI] [PubMed] [Google Scholar]
- 21.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell 4:1093–1099. 10.1016/S1097-2765(00)80238-5 [DOI] [PubMed] [Google Scholar]
- 22.Moynahan ME, Chiu JW, Koller BH, Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4:511–518. 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- 23.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, Livingston DM. 2011. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 44:235–251. 10.1016/j.molcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, Chen HT, Fernandez-Capetillo O, D'Andrea A, Nussenzweig A. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 46:125–135. 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249–262. 10.1016/S1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- 27.Vandenberg CJ, Gergely F, Ong CY, Pace P, Mallery DL, Hiom K, Patel KJ. 2003. BRCA1-independent ubiquitination of FANCD2. Mol. Cell 12:247–254. 10.1016/S1097-2765(03)00281-8 [DOI] [PubMed] [Google Scholar]
- 28.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. 2006. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 127:539–552. 10.1016/j.cell.2006.08.053 [DOI] [PubMed] [Google Scholar]
- 29.Hsu LC, Doan TP, White RL. 2001. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 61:7713–7718 [PubMed] [Google Scholar]
- 30.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389–395. 10.1016/S1097-2765(00)80466-9 [DOI] [PubMed] [Google Scholar]
- 31.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. 2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477:179–184. 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberg RA. 2008. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma 117:305–317. 10.1007/s00412-008-0154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huen MS, Sy SM, Chen J. 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11:138–148. 10.1038/nrm2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633–2638. 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polo SE, Roche D, Almouzni G. 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127:481–493. 10.1016/j.cell.2006.08.049 [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317–328. 10.1016/S1097-2765(00)80276-2 [DOI] [PubMed] [Google Scholar]
- 38.Church GM, Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995. 10.1073/pnas.81.7.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, Merajver SD, Ethier SP, Schutte M. 2006. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 66:41–45. 10.1158/0008-5472.CAN-05-2853 [DOI] [PubMed] [Google Scholar]
- 40.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R. 2004. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91:355–358. 10.1038/sj.bjc.6601894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. 2007. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell 12:160–170. 10.1016/j.ccr.2007.06.013 [DOI] [PubMed] [Google Scholar]
- 42.Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, Lenobel R, Bartkova J, Lau A, O'Connor MJ, Lukas J, Bartek J. 2012. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle 11:3837–3850. 10.4161/cc.22026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, Orr N, Parton M, Smith IE, Reis-Filho JS, Dowsett M, Ashworth A, Turner NC. 2010. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin. Cancer Res. 16:6159–6168. 10.1158/1078-0432.CCR-10-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willers H, Taghian AG, Luo CM, Treszezamsky A, Sgroi DC, Powell SN. 2009. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol. Cancer Res. 7:1304–1309. 10.1158/1541-7786.MCR-09-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakada S, Yonamine RM, Matsuo K. 2012. RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res. 72:4974–4983. 10.1158/0008-5472.CAN-12-1057 [DOI] [PubMed] [Google Scholar]
- 48.Mone MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2:1013–1017. 10.1093/embo-reports/kve224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213–224. 10.1016/S1097-2765(01)00281-7 [DOI] [PubMed] [Google Scholar]
- 50.Chew HK, Doroshow JH, Frankel P, Margolin KA, Somlo G, Lenz HJ, Gordon M, Zhang W, Yang D, Russell C, Spicer D, Synold T, Bayer R, Hantel A, Stiff PJ, Tetef ML, Gandara DR, Albain KS. 2009. Phase II studies of gemcitabine and cisplatin in heavily and minimally pretreated metastatic breast cancer. J. Clin. Oncol. 27:2163–2169. 10.1200/JCO.2008.17.4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurley J, Reis IM, Rodgers SE, Gomez-Fernandez C, Wright J, Leone JP, Larrieu R, Pegram MD. 2013. The use of neoadjuvant platinum-based chemotherapy in locally advanced breast cancer that is triple negative: retrospective analysis of 144 patients. Breast Cancer Res. Treat. 138:783–794. 10.1007/s10549-013-2497-y [DOI] [PubMed] [Google Scholar]
- 52.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. 2010. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28:1145–1153. 10.1200/JCO.2009.22.4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sirohi B, Arnedos M, Popat S, Ashley S, Nerurkar A, Walsh G, Johnston S, Smith IE. 2008. Platinum-based chemotherapy in triple-negative breast cancer. Ann. Oncol. 19:1847–1852. 10.1093/annonc/mdn395 [DOI] [PubMed] [Google Scholar]
- 54.Staudacher L, Cottu PH, Dieras V, Vincent-Salomon A, Guilhaume MN, Escalup L, Dorval T, Beuzeboc P, Mignot L, Pierga JY. 2011. Platinum-based chemotherapy in metastatic triple-negative breast cancer: the Institut Curie experience. Ann. Oncol. 22:848–856. 10.1093/annonc/mdq461 [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Wang Z, Hu X, Wang B, Wang L, Yang W, Liu Y, Liu G, Di G, Hu Z, Wu J, Shao Z. 13 May 2014. Cisplatin and gemcitabine as the first line therapy in metastatic triple negative breast cancer. Int. J. Cancer 10.1002/ijc.28966 [DOI] [PubMed] [Google Scholar]
- 56.Alli E, Sharma VB, Sunderesakumar P, Ford JM. 2009. Defective repair of oxidative DNA damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 69:3589–3596. 10.1158/0008-5472.CAN-08-4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber LJ, Yang TW, Sarkisian CJ, Master SR, Deng CX, Chodosh LA. 2001. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell. Biol. 21:4005–4015. 10.1128/MCB.21.12.4005-4015.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaelian I, Mazoyer S. 2011. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 3:279–290. 10.1002/emmm.201100136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 60.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. 2012. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72:5588–5599. 10.1158/0008-5472.CAN-12-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helleday T. 2011. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 5:387–393. 10.1016/j.molonc.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, Hirte H, Huntsman D, Clemons M, Gilks B, Yerushalmi R, Macpherson E, Carmichael J, Oza A. 2011. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12:852–861. 10.1016/S1470-2045(11)70214-5 [DOI] [PubMed] [Google Scholar]
- 63.Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, Lee H, Whalen C, Tyburski K, Winer E, Ivy P, Matulonis UA. 2013. A phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur. J. Cancer 49:2972–2978. 10.1016/j.ejca.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361:123–134. 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]
- 65.Pierce A, McGowan PM, Cotter M, Mullooly M, O'Donovan N, Rani S, O'Driscoll L, Crown J, Duffy MJ. 2013. Comparative antiproliferative effects of iniparib and olaparib on a panel of triple-negative and non-triple-negative breast cancer cell lines. Cancer Biol. Ther. 14:537–545. 10.4161/cbt.24349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899–23903. 10.1074/jbc.C000276200 [DOI] [PubMed] [Google Scholar]
- 67.DelloRusso C, Welcsh PL, Wang W, Garcia RL, King MC, Swisher EM. 2007. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 5:35–45. 10.1158/1541-7786.MCR-06-0234 [DOI] [PubMed] [Google Scholar]
- 68.Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME. 2003. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 23:7926–7936. 10.1128/MCB.23.21.7926-7936.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. 2001. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 28:266–271. 10.1038/90108 [DOI] [PubMed] [Google Scholar]
- 70.Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker A, Couch FJ, Goldgar DE, Davidson HR, Nathanson KL, Foulkes WD, Greenberg RA. 2013. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 3:399–405. 10.1158/2159-8290.CD-12-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.