Abstract
Human UTX, a member of the Jumonji C family of proteins, associates with mixed-lineage leukemia 3/4 complexes. Stimulation with retinoic acid leads to the recruitment of UTX-containing complexes to HOX genes, which results in demethylation of histone H3 lysine 27 and concomitant methylation of histone H3 lysine 4. Here, we show that UTX interacts with the retinoic acid receptor α (RARα) and that this interaction is essential for proper differentiation of leukemic U937 cells in response to retinoic acid. UTX occupies the promoters of several RAR target genes and regulates their transcriptional output by modulating ASH2L complex recruitment. Overexpression of UTX in promyelocytic NB4 cells results in enhanced cellular differentiation upon retinoic acid treatment. Our results show that UTX is important for RAR-mediated transcription and provide insight into the critical role of cross talk between histone H3 lysine 4 methylation and histone H3 lysine 27 demethylation during cellular differentiation.
INTRODUCTION
UTX, a JmjC domain-containing histone demethylase, can specifically demethylate di- and trimethylated histone 3 lysine 27 (H3K27) (1–3). The posttranslational methylation of H3K27 is highly correlated with genomic silencing, and H3K27me3 demethylation, which is regulated by the UTX protein, is essential for proper development, differentiation, and proliferation (4–6). It has been reported that UTX has a role in establishing and maintaining differentiation programs during stem and progenitor cell commitment to differentiated lineages (7). Aberrant methylation of core histone tails and deregulation of the corresponding enzymes have been implicated in leukemia as well as other types of cancers (8). Together, these observations suggest that UTX might contribute to epigenetic mechanisms of initiation or progression of leukemia. Acute promyelocytic leukemia (APL) is phenotypically characterized by the accumulation of clonal hematopoietic precursors blocked at the stage of promyelocytic cells, and it represents a paradigm for cell transformations that are caused by an aberrant epigenetic status that leads to gene silencing (9). The oncogenic PML-retinoic acid receptor α (PML-RARα) fusion protein generated by chromosomal translocation t(15;17) behaves as a constitutive and potent transcriptional repressor by recruiting different chromatin-modifying enzymes, including histone deacetylases, Polycomb complexes, and DNA methyltransferases (10–12). In contrast to the wild-type RARα, PML-RARα is insensitive to physiological concentrations of retinoic acid (RA) that would usually trigger transcriptional activation. However, pharmacological doses of RA, which are used for patients in early phases of the disease, can lead to partial derepression of PML-RAR target genes (13).
Interestingly, UTX was identified as a component of known histone H3 lysine 4 (H3K4)-specific MLL/SET methyltransferase complexes (14). ASH2L protein is considered one of the integral members of these complexes (15) and is required for trimethylation of global H3K4 as well as of specific MLL target genes in human cells (16). During RA signaling events, the coordinated removal of repressive marks, Polycomb group displacement, and concomitant deposition of activating marks are important for the stringent regulation of transcription during cellular differentiation. In human embryonic teratocarcinoma NT2/D1 cells, RA-induced differentiation results in a concerted mechanism for the recruitment and/or activation of a complex containing trimethyl H3K27 demethylase activity and trimethyl H3K4 methyltransferase activity that mediates transcriptional activation of HOX genes (1).
To better understand the molecular mechanisms underlying UTX involvement in RA-mediated gene regulation in leukemia, we studied the effects of UTX silencing in a human myeloid precursor leukemic cell line (U937) and of its overexpression in a human APL cell line expressing PML-RAR (NB4). Here, we show that UTX interacts with RARα and is necessary for proper cellular differentiation. UTX contributes to the activation of different target genes during RA-induced differentiation of U937 cells. Moreover, we demonstrated that UTX protein mediates ASH2L association with RARα. Strikingly, UTX overexpression facilitates cellular differentiation of APL cells. Together these results identify UTX as a critical mediator of gene activation during RA-induced differentiation in leukemic cells.
MATERIALS AND METHODS
Plasmids and antibodies.
pRS-derived vectors (shRandom, shUTX, shASH2L, and shNCoA6) were generated by ligating synthetic oligonucleotides against the target sequence into pRetro-Super (17). The retroviral expression vector was generated by subcloning the cDNA of UTX into pBabe, and the RARα expression vector was previously described (12).
Antibodies were obtained from the following suppliers: anti-trimethyl H3K4 (07473) and anti-trimethyl H3K27 (05851), Upstate/Millipore; anti-RARα (sc-551), Santa Cruz; anti-ASH2L (A300-107A) and anti-NCoA6 (A300-411A), Bethyl Laboratories; antitubulin, Abcam; and anti-Flag (F3165), Sigma-Aldrich. Anti-UTX was previously described (1). For fluorescence-activated cell sorting (FACS) analysis, we used antibodies that recognize cell surface myeloid-specific antigens CD11c and CD11b (Becton Dickinson).
Cell lines, transfection, and retroviral infection.
U937 and NB4 cells were cultured at 37°C with 5% CO2 in RPMI medium supplemented with 10% fetal bovine serum (FBS). HEK 293T cells were cultured at 37°C with 5% of CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and transfected with standard techniques. Cell extracts were prepared as described previously (12). HEK 293T cells were transfected using the calcium phosphate coprecipitation method (10). The pRS-based retrovirus was produced by transfecting GP2-293 packaging cells (Clontech). The collected retrovirus was subsequently used to transduce U937 or NB4 cells by spinoculation (900 × g, 90 min, 32°C) in the presence of protamine sulfate followed by an additional overnight incubation at 37°C in 5% CO2. The protocol was repeated for two consecutive days. U937 and NB4 cells were treated with RA (100 nM and 10 nM, respectively) to induce differentiation and were harvested at 5, 24, and 48 h.
Differentiation assays.
U937 and NB4 cells were treated with RA (100 nM and 10 nM, respectively), rinsed twice with PBS, and incubated 30 min with specific antibodies. After a washing, the percentage of differentiated antigen-positive cells and the fluorescence levels were analyzed by flow cytometry on a Becton Dickinson FACSCanto analyzer. The nitroblue tetrazolium (NBT) assay was performed using commercially available NBT (Sigma-Aldrich). Two hundred microliters of cell suspension at a density of 2 × 105 cells in RPMI–5% FBS was mixed with 0.2 μl of filtered 0.2% NBT solution and 3 μl of TPA (1 μM) and then further incubated for 30 min at 37°C. Cytocentrifuge slides were then prepared (200 rpm, 4 min). NBT-positive cells were determined by scoring 400 cells under a light microscope.
Immunoprecipitation and ChIP.
For coimmunoprecipitation assays, cells that had been treated with RA (1 μM for 30 min) or not treated were washed in PBS and lysed in lysis buffer (10). Cell extracts were incubated overnight with specific antibodies at 4°C in the presence or absence of RA. Protein A-Sepharose beads (GE Life Sciences) saturated with bovine serum albumin (BSA) were added to the lysates for 2 h, and the immunoprecipitates were then washed and loaded on SDS-PAGE gels.
For chromatin immunoprecipitation, cells were cross-linked with 0.8% HCHO at room temperature for 6 min. ChIP assays were performed and analyzed as described previously (11). Equal amounts of unrelated antibody (IgG) were included as controls for the ChIP assays. Chromatin immunoprecipitates for proteins and methyl marks were amplified by real-time quantitative PCR (Roche LightCycler), normalized to input, and calculated as percentage of input. PCR primer sequences are available upon request.
RNA purification and RT-PCR analysis.
For quantitative RT-PCR, total RNA was isolated using the Qiagen RNeasy kit and reverse transcribed using Invitrogen's first-strand synthesis kit. Samples were amplified by real-time quantitative PCR (Roche LightCycler), and mRNA levels were normalized to PUM1 levels. The relative mRNA level represents the fold change over the control.
RESULTS
UTX interacts with RARα in vivo.
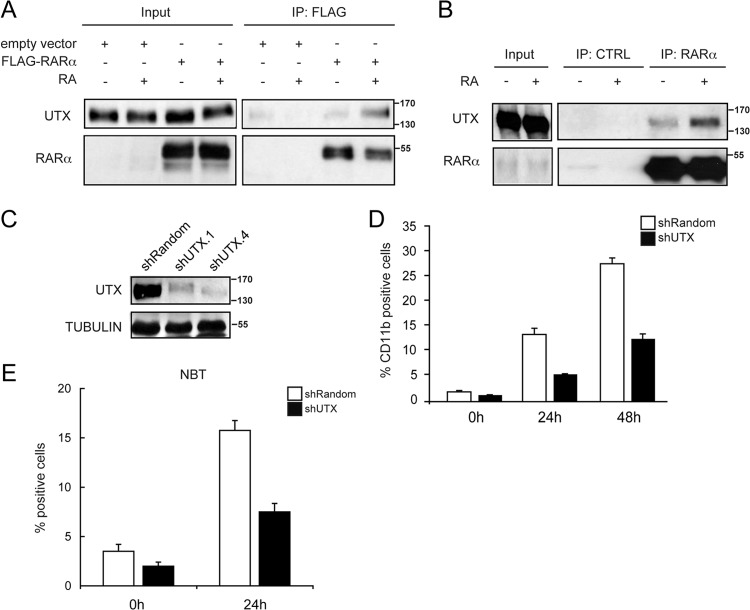
We have recently demonstrated that components of the MLL complex and UTX can be recruited to RA-responsive genes after RA stimulation, thereby promoting H3K4 methylation, H3K27 demethylation, and consequently, transcription activation (1). Based on these results, we explored the possibility that UTX interacts with RARα by treating 293T cells transiently transfected with a Flag-tagged RARα with RA. After immunoprecipitation with anti-Flag antibody, we found that RARα was specifically associated with UTX in the presence of RA (Fig. 1A).
FIG 1.
UTX specifically interacts with RARα and is important for cellular differentiation. (A) Interaction between UTX and RARα. 293T cells were transfected with a Flag-RARα expression vector, and extracts from cells that were untreated (−) or treated with RA (+) were immunoprecipitated with an anti-Flag antibody (IP: FLAG). Western blots with input lysate or immunoprecipitates were analyzed using antisera against UTX or RARα, as indicated. (B) Endogenous interaction between UTX and RARα. Extracts from U937 cells that were untreated (−) or treated with RA (+) were immunoprecipitated with IgG antibody (IP: CTRL) or with the anti-RARα antibody (IP: RARα). Western blots with input lysate or immunoprecipitates were analyzed using antisera against UTX or RARα. (C) Knockdown of UTX in the U937 cell line. Equal amounts of extracts from cells infected with a retroviral construct generating the scrambled control sequence (shRandom) or two UTX-specific small hairpin RNAs (shUTX.1 and shUTX.4) were analyzed by Western blotting with the indicated antibodies. (D and E) Downregulation of UTX affects U937 differentiation after treatment with RA. shRandom or shUTX U937 cells were treated with RA for 24 or 48 h or not treated. Cell differentiation was evaluated by quantitative expression of CD11b antigen (D) and NBT reduction (E). Error bars represent standard deviations from the means for triplicate experiments.
We then performed endogenous coimmunoprecipitation experiments using lysates from promyelocytic leukemia U937 cells. Immunoblot analysis of anti-RARα immunoprecipitates revealed a stable, endogenous complex between RARα and UTX that was present in the absence of stimulus and was stimulated by RA administration (Fig. 1B).
UTX is necessary for proper cellular differentiation.
Given the endogenous interaction between UTX and RARα in U937 cells, we wondered whether UTX could participate directly in RA-mediated gene regulation. We generated a stable UTX knockdown U937 cell line (shUTX) using a retroviral vector-based short hairpin RNA (shRNA) approach (17). A reduction of >75% of UTX protein was achieved under these conditions (using the shUTX.4 vectors), compared to the mock knockdown cells (shRandom) (Fig. 1C).
To investigate whether the UTX knockdown could affect global cell differentiation, we quantified the levels of CD11b differentiation marker by flow cytometry as an indicator of maturation of hematopoietic cells (18). The percentage of CD11b-positive cells was increased to 13.6% and 27.9% after 24 h and 48 h of RA administration, respectively, in shRandom control U937 cells. In contrast, shUTX cells showed a delayed onset of differentiation after RA stimulus, with 5.6% and 12.8% CD11b-positive cells at 24 h and 48 h of treatment, respectively (Fig. 1D). We also monitored U937 maturation by measuring the capacity of differentiated cells to reduce nitroblue tetrazolium (NBT). We found no evidence of spontaneous differentiation in untreated shRandom control and shUTX cells, while we observed a significantly reduced number of NBT-positive cells in response to RA in shUTX U937 cells compared to shRandom control cells (Fig. 1E).
UTX activates different target genes during RA-induced differentiation of U937 cells.
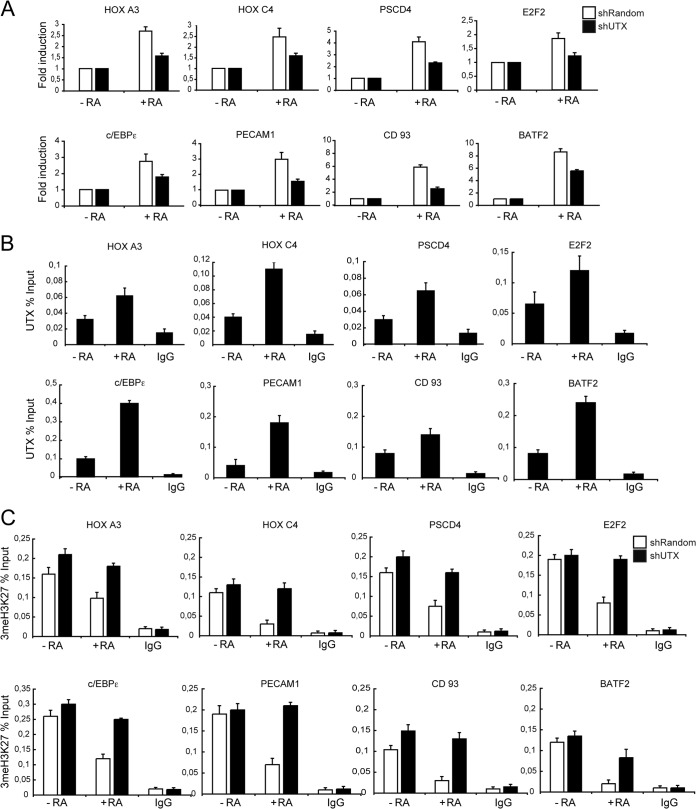
We next tested by quantitative reverse transcription-PCR (RT-qPCR) how UTX knockdown affects promoter activity of essential genes for RA-induced differentiation of leukemic cells (19–25). Quantitative analysis of mRNA levels indicated that genes reported to be key regulators of myeloid precursor differentiation are RA-dependent UTX targets (Fig. 2A, white bars). Specifically, the expression of HOXA3, HOXC4, PSCD4, E2F2, c/EBPε, PECAM1, CD93, and BATF2 was compromised upon UTX depletion in RA-treated cells (Fig. 2A, black bars).
FIG 2.
UTX is required for transcriptional activation of RAR target genes. (A) Knockdown of UTX affects RAR-target gene expression. Total RNA from shRandom or shUTX U937 cells that were untreated (−RA) or treated with RA (+RA) was prepared. mRNA levels of the indicated genes were analyzed relative to that in the PUM1 control by RT-qPCR. Results are presented as the means and standard errors of the means (SEM) from three independent experiments. (B and C) RA treatment results in recruitment of UTX concomitant with reduced levels of trimethyl H3K27 in control U937 cells. UTX occupancy in shRandom cells (B) and trimethyl H3K27 (3meH3K27) levels in shRandom and shUTX cells (C) at the RAR target gene promoters were analyzed by a qChIP assay. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments.
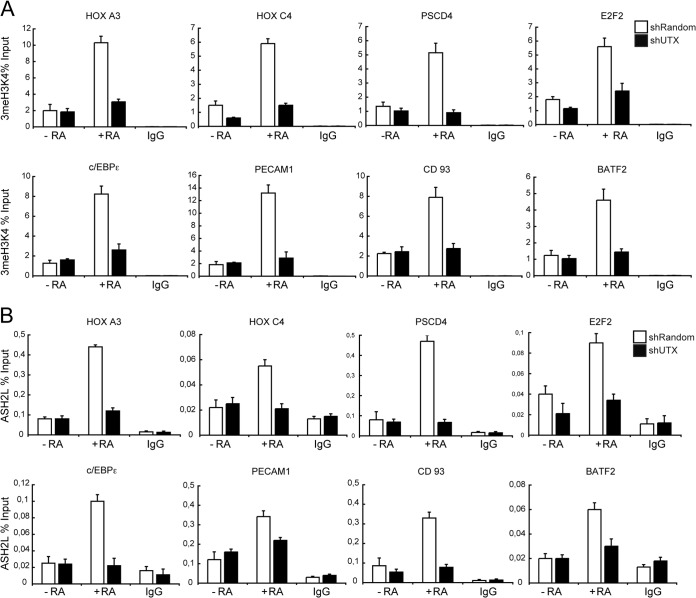
To demonstrate that UTX mediates such transcriptional effects on all these genes directly, we examined UTX occupancy on their promoters by quantitative chromatin immunoprecipitation (qChIP) assay. UTX was associated at those promoters, and its occupancy increased after control U937 cells were treated with RA (Fig. 2B). As expected, this increase was paralleled by a decrease in trimethyl H3K27 (Fig. 2C, white bars). The RA-induced transcriptional activation of these promoters was accompanied by increased levels of trimethyl H3K4 (Fig. 3A, white bars) and the recruitment of ASH2L, a critical component of the mixed-lineage leukemia (MLL)-containing complex (Fig. 3B, white bars). Importantly, the levels of these epigenetic marks were significantly affected in shUTX cells (Fig. 2C and 3A and B, black bars).
FIG 3.
UTX knockdown affects the MLL complex activity at RAR target genes. Downregulation of UTX impaired trimethyl H3K4 (3meH3K4) levels (A) and ASH2L occupancy (B) at RAR target genes. shRandom or shUTX U937 cells that were untreated (−RA) or treated with RA (+RA) were subjected to ChIP analysis, using the indicated antibodies. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments.
ASH2L knockdown confirms the interplay between the MLL complex and the UTX protein.
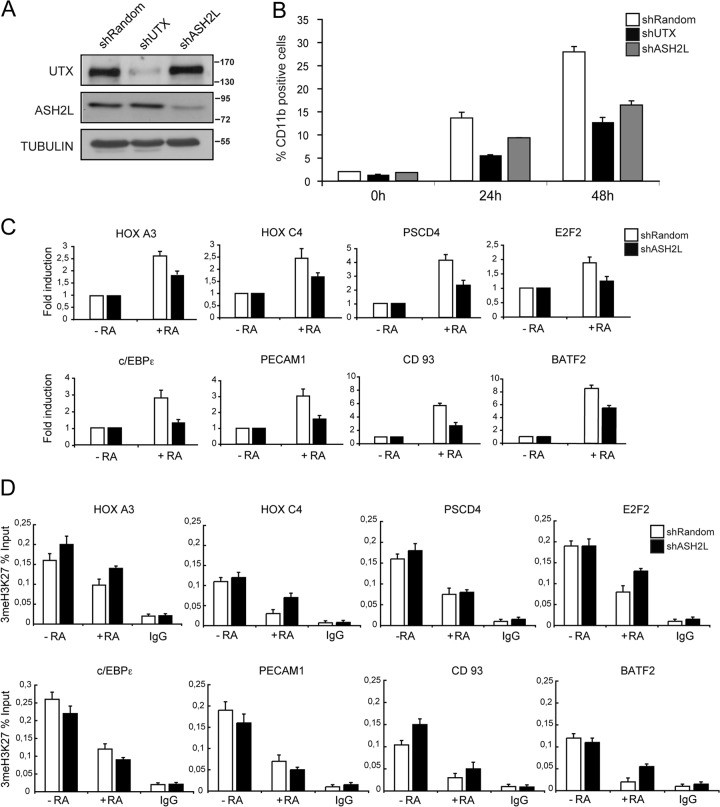
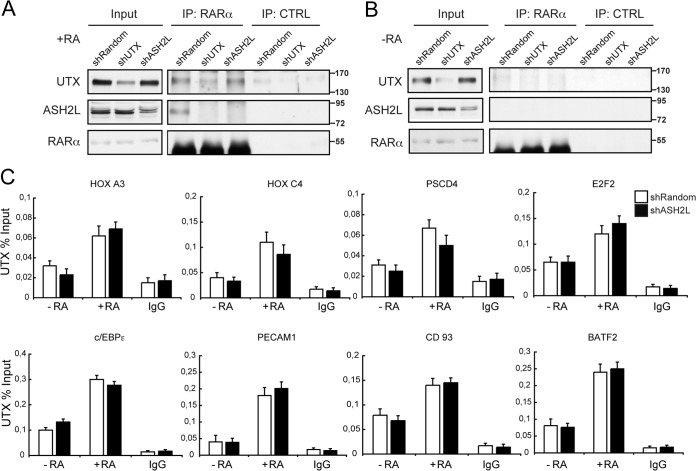
We next wanted to assess the interaction of UTX with the MLL complex (2, 14) and to investigate whether ASH2L is involved in cellular differentiation in response to RA, using a stable ASH2L knockdown U937 cell line (shASH2L) (Fig. 4A). To study the contribution of ASH2L to cell differentiation, we analyzed the CD11b differentiation marker by flow cytometry. Interestingly, shASH2L cells showed a delayed onset of differentiation upon RA treatment, with 9.3% and 16.4% of CD11b-positive cells at 24 h and 48 h of treatment, respectively (Fig. 4B, gray bars), which was, however, not as pronounced as for shUTX U937 cells (Fig. 4B, black bars); shRandom control U937 had 13.6% and 27.9% CD11b-positive cells, respectively, under the same conditions (Fig. 4B, white bars).
FIG 4.
Effects of ASH2L depletion in U937 leukemic cells. (A) Knockdown of ASH2L in U937 cells. Human U937 leukemic cells were infected and selected as indicated in the legend to Fig. 1C. Equal amounts of cell extracts from mock knockdown (shRandom) and interfering-RNA cells (shUTX or shASH2L) were analyzed by Western blotting with the indicated antibodies. Knockdown in shUTX U937 cells is shown as a control. (B) Downregulation of ASH2L affects U937 differentiation after treatment with RA. shRandom or shASH2L U937 cells were treated with RA for 24 and 48 h or untreated, and cell differentiation was evaluated as percent CD11b-positive cells, as indicated for Fig. 1D. Error bars represent standard deviations from the mean for triplicate experiments. Differentiation in shUTX U937 cells is shown for comparison. (C) Knockdown of ASH2L affects RAR target gene expression. Total RNA from shRandom or shASH2L U937 cells that were untreated (−RA) or treated with RA (+RA) was prepared. mRNA levels of the genes indicated were analyzed relative to a PUM1 control by RT-qPCR. Results are presented as the means and SEM from three independent experiments. (D) Downregulation of ASH2L did not impair trimethyl H3K27 (3meH3K27) levels at RAR target genes. shRandom or shASH2L U937 cells that were untreated (−RA) or treated with RA (+RA) were subjected to ChIP analysis using the indicated antibodies. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments.
To study how ASH2L knockdown affects the expression of the UTX target genes, we analyzed RA-treated shASH2L cells by RT-qPCR and found that they elicited a gene expression profile similar to that of UTX knockdown cells. The UTX target genes we studied (e.g., HOXA3, HOXC4, PSCD4, E2F2, c/EBPε, PECAM1, CD93, and BATF2) were poorly induced upon RA stimulus (Fig. 4C). Importantly, control ChIP experiments indicated that inhibition of ASH2L expression correlates with decreased levels of trimethyl H3K4 (data not shown). However, no significant changes in trimethyl H3K27 levels were observed upon inhibition of ASH2L expression (Fig. 4D).
We next investigated the possibility of a functional interplay between RARα, UTX, and ASH2L, speculating that a trimeric complex should assemble upon RA administration. We performed endogenous coimmunoprecipitation experiments using lysates from either shRandom control, shUTX, or shASH2L U937 cells that had been treated with RA or not treated. Immunoblot analysis of anti-RARα immunoprecipitates revealed endogenous complexes of RARα with UTX, as previously shown (Fig. 5A, top), and with ASH2L after RA treatment (Fig. 5A, middle, shRandom lanes). Interestingly, the RARα-UTX interaction was not affected after ASH2L depletion (Fig. 5A, top, shASH2L lanes). Only a weak interaction was observed in the absence of RA (Fig. 5B, top). To further support these findings, we examined the occupancy of UTX in ASH2L-depleted cells. Following RA treatment of U937 cells, UTX is recruited to its target genes (e.g., HOXA3, HOXC4, PSCD4, E2F2, c/EBPε, PECAM1, CD93, and BATF2) even in the absence of ASH2L (Fig. 5C), and this was paralleled by a decrease in trimethyl H3K27 similar to that observed in control cells (Fig. 4D). Additionally, coimmunoprecipitation experiments between RARα and ASH2L in both control and UTX-depleted cells indicate that this interaction was lost in UTX knockdown cells (Fig. 5A). No interactions were observed in the absence of RA (Fig. 5B). Thus, the association between ASH2L and RARα depends on the presence of UTX. This is in agreement with the ChIP analysis performed in UTX knockdown cells, indicating that UTX depletion resulted in a decreased recruitment of ASH2L (Fig. 3B). Together, these data suggest that UTX is a strong mediator of the RARα-ASH2L interaction and that it promotes their recruitment to target promoters through a RA-mediated mechanism.
FIG 5.
RA induces UTX bridging of RARα and ASH2L. (A and B) Endogenous interaction between RARα, UTX, and ASH2L. Extracts from shRandom, shUTX, and shASH2L U937 cells that were treated with RA (+RA) (A) or untreated (−RA) (B) were immunoprecipitated with the RARα antibody (IP: RARα) or with IgG antibody (IP: CTRL). Western blots with input lysate or immunoprecipitates were analyzed using antisera against UTX, ASH2L, or RARα. (C) Downregulation of ASH2L did not impair UTX recruitment to RAR target genes. shRandom or shASH2L U937 cells that were untreated (−RA) or treated with RA (+RA) were subjected to ChIP analysis. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments.
NCoA6/ASC-2 mediates UTX and ASH2L interaction with RARα.
The nuclear factor NCoA6, also known as ASC-2, stimulates transactivation activity of nuclear receptors, including RAR, and of numerous other transcription factors (26). We recently demonstrated that UTX associates with a 2-MDa, MLL3/4-containing complex that displayed H3K27 demethylase and H3K4 methyltransferase activities (1). Moreover, we showed that NCoA6 indeed belongs to this complex (1).
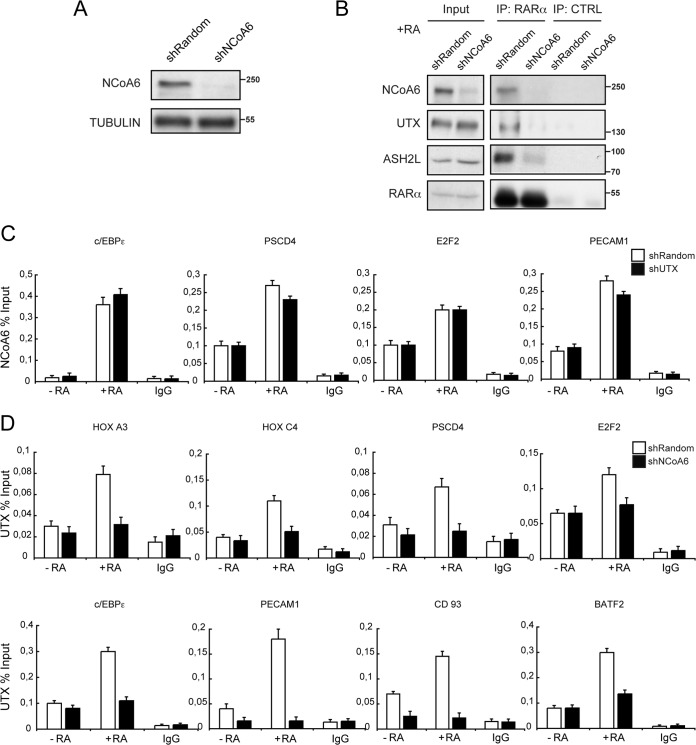
To assess whether NCoA6 plays a role in RA-mediated gene transcription, we developed a stable NCoA6 knockdown U937 cell line (shNCoA6), which has a reduction of >90% of NCoA6 protein compared to the mock knockdown cells (shRandom) (Fig. 6A). Coimmunoprecipitation experiments with an anti-RARα antibody on lysates from U937 control cells treated with RA confirmed that RARα formed an endogenous complex with NCoA6, UTX, and ASH2L (Fig. 6B), as previously reported (1). These interactions were lost in U937 NCoA6-depleted cells, suggesting that the coactivator NCoA6 is an important mediator of the association of UTX and ASH2L with RARα. These observations prompted us to investigate the interplay between NCoA6 and UTX protein at the promoter of the previously described target genes. UTX depletion had no significant effect on NCoA6 promoter occupancy, as measured by qChIP (Fig. 6C). Nevertheless, UTX occupancy at these genes in response to RA was severely affected after NCoA6 depletion (Fig. 6D, compare white and black bars). These results indicate that NCoA6 confers RAR target gene specificity for the recruitment of UTX and ASH2L, promoting H3K4 methylation, H3K27 demethylation, and, consequently, transcription activation, in leukemic U937 cells.
FIG 6.
NCoA6 regulates UTX recruitment to RAR target genes. (A) Knockdown of NCoA6 in the human U937 cell line. U937 leukemic cells were infected and selected as described for Fig. 1C. Equal amounts of cell extracts from mock (shRandom) and interfering-RNA (shNCoA6) cells were analyzed by Western blotting with the indicated antibodies. (B) NCoA6 mediates UTX and ASH2L endogenous association to RARα. Extracts from shRandom or shNCoA6 U937 cells treated with RA (+RA) were immunoprecipitated with the anti-RARα antibody (IP: RARα) or with IgG antibody (IP: CTRL). Western blots with input lysate and blots of immunoprecipitates were analyzed using antisera against NCoA6, UTX, ASH2L, or RARα. (C) NCoA6 occupancy at RAR target genes is not affected upon UTX knockdown. shRandom or shUTX U937 cells that were untreated (−RA) or treated with RA (+RA) were subjected to ChIP analysis. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments. (D) NCoA6 downregulation impairs UTX occupancy at RAR target genes. shRandom or shNCoA6 U937 cells, untreated (−RA) or treated with RA (+RA), were subjected to ChIP analysis. IgG was included as a control. Error bars indicate standard deviations obtained from three independent experiments.
Overexpression of UTX facilitates cellular differentiation in acute promyelocytic leukemia.
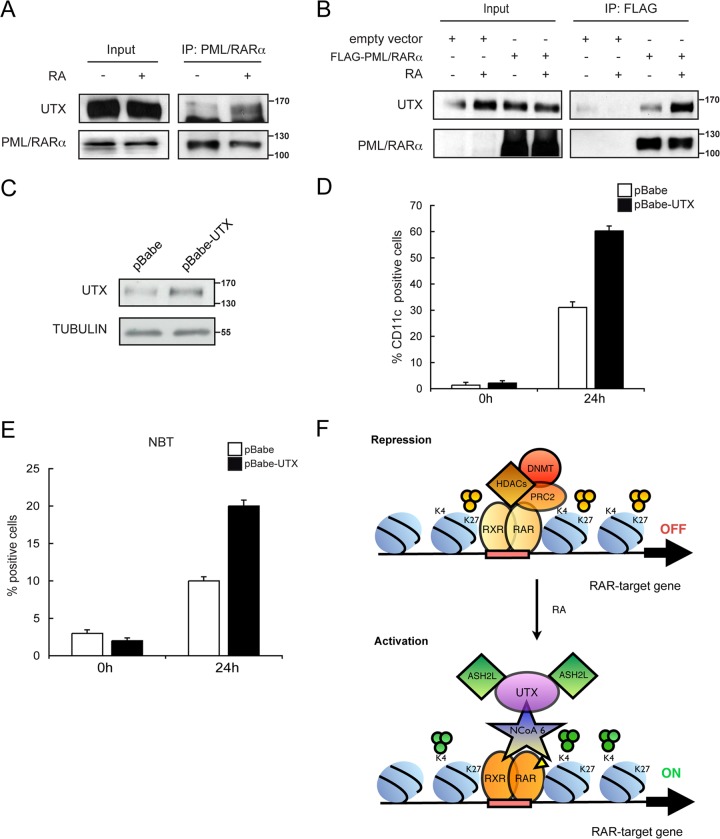
We expanded our analysis to fully established, patient-derived NB4 leukemic cells in order to further characterize the role of UTX on the epigenetic silencing imposed by the oncogenic transcription factor PML/RARα. We initially tested whether PML/RARα and UTX associate endogenously with coimmunoprecipitation experiments using anti-PML/RARα antibody (PGM3). Indeed, we observed an endogenous stable complex of PML/RARα with UTX in the presence of RA (Fig. 7A). We next transiently transfected 293T cells with a Flag-tagged PML/RARα and treated them with RA. After immunoprecipitation with anti-Flag antibody, we found that PML/RARα was specifically associated with UTX in the presence of RA (Fig. 7B).
FIG 7.
UTX overexpression facilitates the differentiation of NB4 leukemic cells. (A) Endogenous interaction between UTX and PML-RARα. Extracts from U937 cells that were untreated (−) or treated with RA (+) were immunoprecipitated with the PML antibody (IP: PML/RARα). Western blots with input lysate or immunoprecipitates were analyzed using antisera against UTX or RARα. (B) Interaction between UTX and PML-RARα. 293T cells were transfected with a Flag-PML/RARα expression vector, and extracts from cells that were untreated (−) or treated with RA (+) were immunoprecipitated with an antibody against Flag (IP: FLAG). Western blots with input lysate or immunoprecipitates were analyzed using antisera against UTX or RARα as indicated. (C) Overexpression of UTX in NB4 leukemic cells. Equal amounts of extracts from cells infected with a retroviral construct generating the scrambled control sequence (pBabe) or a stable UTX-overexpressing cell line (pBabe-UTX) were analyzed by Western blotting with the indicated antibodies. (D and E) Differentiation of UTX-overexpressing NB4 cells after treatment with suboptimal concentrations of RA (10 nM). pBabe control or pBabe-UTX NB4 cells were treated with RA for 24 h or were not treated. Cell differentiation was evaluated by quantitative expression of CD11c antigen (D) and NBT reduction (E). Error bars represent standard deviations from the means from triplicate experiments. (F) Model for the involvement of UTX in the transcriptional regulation of RAR target genes during differentiation of leukemic cells. Genes are silenced in the absence of stimulus (9). Administration of RA promotes gene activation by modulating NCoA6 recruitment, which in turn allows occupancy of UTX and ASH2L, concomitant with demethylation of H3K27 and trimethylation of H3K4.
Since the PML/RARα-induced differentiation block of NB4 cells is known to be reversed with high doses of RA (1 μM), we next tested whether UTX overexpression could affect global cell differentiation in the presence of lower concentrations of the stimulus (10 nM). For this purpose, we generated a stable UTX overexpressed NB4 cell line (pBabe-UTX) using a retroviral-vector-based approach. An increase of >60% of UTX protein was achieved under these conditions, compared to control transfected cells (pBabe) (Fig. 7C). As expected, PML/RARα blocked differentiation of control cells by 70% after 24 h of RA administration (Fig. 7D, white bars). Interestingly, under these conditions, overexpression of UTX induced cell differentiation by 60% as measured by flow cytometry analysis of surface differentiation marker CD11c (Fig. 7D, black bars). We also monitored NB4 maturation by the NBT assay and observed a significantly increased number of NBT-positive cells in UTX overexpressed NB4 cells (Fig. 7E). In summary, these studies reveal that UTX overexpressed NB4 cells are sensitized and thus more prone to differentiation upon RA administration.
DISCUSSION
Here, we show that UTX interacts endogenously with RARα and PML/RARα and is required for differentiation of leukemic cells (specifically, of U937 and NB4 cells) in response to RA. Our in vivo studies demonstrated critical roles for UTX in regulating transcription as well as the levels of H3K27 methylation at RAR target genes. We provide evidence that UTX regulates transcription of important genes for the differentiation process, such as HOXA3, HOXC4, PSCD4, E2F2, c/EBPε, PECAM1, CD93, and BATF2, by modulating the recruitment of ASH2L and the deposition of the trimethylation of H3K4. Moreover, recruitment of the RAR coactivator NCoA6 after RA treatment regulates the occupancy of the UTX and ASH2L complexes, thereby modulating the transcriptional output. Our results suggest a mechanism of RA-mediated gene activation that involves NCoA6 recruitment, H3K4 trimethylation brought about by ASH2L, and H3K27 demethylation mediated by UTX in U937 cells (Fig. 7F). Taken together, these findings reveal that UTX plays a key role during RA-induced differentiation of leukemic cells. On the basis of our studies, we could further support H3K27 demethylation and H3K4 methylation are interdependent processes in leukemia. We predict that NCoA6 is a central component of the complex and is required for UTX and ASH2L recruitment to RAR target genes. We cannot discard the possibility of a recruitment hierarchy for a sequential NCoA6, UTX, and ASH2L binding to chromatin. Furthermore, it is tempting to speculate that a NCoA6-UTX-ASH2L preformed complex, as previously described (1, 14), is involved in this scenario. Interestingly, an ordered cycle of H3K4 trimethylation followed by demethylation of H3K27 mediated by UTX has already been described during RA-induced transcriptional activation of HOX cluster genes in human embryonic teratocarcinoma NT2/D1 and ES cells (1, 7). Our data unveil an elegant mechanism in leukemia cells for cell differentiation in response to RA. Importantly, we demonstrated a novel endogenous interaction between UTX and RAR and determined that this confers target gene specificity and gives a physiological significance to the physical association between the H3K27 demethylase UTX and the MLL3/4-containing H3K4 methyltransferase complexes in the regulation of gene expression, as previously reported (1, 2). In particular, the trithorax protein ASH2L is considered one of the core subunits of all known H3K4-specific MLL/SET methyltransferase complexes (15), functions as an oncoprotein that is overexpressed in human tumors (27), and is essential for pluripotency in ES cells (28). NCoA6, which is essential for nuclear receptor function in vivo, is amplified and overexpressed in various human cancers (29). It has been reported that transactivation by RAR could involve ligand-dependent recruitment of NCoA6 complex to target DNA response elements (RAREs) and subsequent transient H3K4 methylation of the promoter region in vivo (26, 30). In this work, we found that NCoA6 bridges UTX and RARα and mediates UTX recruitment to RAR target genes during differentiation. These findings are of particular interest based on the presence of RAREs in the regulatory region of all the genes analyzed here, as we speculate that RARα can influence epigenetic regulation and consequently the expression of RA-responsive genes during differentiation of leukemic cells. Moreover, our findings that NCoA6 interacts with the UTX protein may shed light onto how RA-responsive genes are regulated upon cell differentiation, as NCoA6 is known to functionally associate with other classes of transcription factors, including AP-1, NF-κB, CBP/p300, c/EBPα, and ATF2 (31). Further characterization of the RARα-UTX complex could thus be instrumental in defining a general epigenetic mechanism for cell differentiation.
A major impediment to successfully treating APL patients is RA resistance, which is associated with a consequent relapse of the disease. Therefore, identifying protein complexes that can effectively synergize with or provide an alternative to RA is of paramount relevance (11). Our finding that UTX overexpression facilitates cellular differentiation of NB4 cells in the presence of physiological concentrations of RA suggests that specific HMT inhibitors for H3K27 methylation can be used in cancer therapy, making these attractive targets for therapeutic strategies.
ACKNOWLEDGMENTS
We are grateful to Veronica A. Raker for critical reading of the manuscript and to the CRG Genomic Facility.
This work was supported by grants from the Spanish Ministerio de Educación y Ciencia, from AGAUR, from AICR (10-0177), and from the European Commission's 7th Framework Program 4DCellFate (grant 277899 to L.D.C.).
Footnotes
Published ahead of print 28 July 2014
REFERENCES
- 1.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318:447–450. 10.1126/science.1149042 [DOI] [PubMed] [Google Scholar]
- 2.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449:731–734. 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- 3.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U. S. A. 104:18439–18444. 10.1073/pnas.0707292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morales Torres C, Laugesen A, Helin K. 2013. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 8:e60020. 10.1371/journal.pone.0060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. 2010. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 29:1401–1411. 10.1038/emboj.2010.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Mercher T, Scholl C, Brumme K, Gilliland DG, Zhu N. 2012. A functional role for the histone demethylase UTX in normal and malignant hematopoietic cells. Exp. Hematol. 40:487–498.e3. 10.1016/j.exphem.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 7.Shahhoseini M, Taghizadeh Z, Hatami M, Baharvand H. 2013. Retinoic acid dependent histone 3 demethylation of the clustered HOX genes during neural differentiation of human embryonic stem cells. Biochem. Cell Biol. 91:116–122. 10.1139/bcb-2012-0049 [DOI] [PubMed] [Google Scholar]
- 8.de The H, Chen Z. 2010. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat. Rev. Cancer 10:775–783. 10.1038/nrc2943 [DOI] [PubMed] [Google Scholar]
- 9.Di Croce L. 2005. Chromatin modifying activity of leukaemia associated fusion proteins. Hum. Mol. Genet. 14(Spec No 1):R77–R84. 10.1093/hmg/ddi109 [DOI] [PubMed] [Google Scholar]
- 10.Villa R, Morey L, Raker VA, Buschbeck M, Gutierrez A, De Santis F, Corsaro M, Varas F, Bossi D, Minucci S, Pelicci PG, Di Croce L. 2006. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc. Natl. Acad. Sci. U. S. A. 103:1400–1405. 10.1073/pnas.0509343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, Fuks F, Helin K, Di Croce L. 2007. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell 11:513–525. 10.1016/j.ccr.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 12.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, Minucci S, Pelicci PG. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295:1079–1082. 10.1126/science.1065173 [DOI] [PubMed] [Google Scholar]
- 13.Pandolfi PP. 2001. Oncogenes and tumor suppressors in the molecular pathogenesis of acute promyelocytic leukemia. Hum. Mol. Genet. 10:769–775. 10.1093/hmg/10.7.769 [DOI] [PubMed] [Google Scholar]
- 14.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395–20406. 10.1074/jbc.M701574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13:713–719. 10.1038/nsmb1128 [DOI] [PubMed] [Google Scholar]
- 16.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. 2006. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 13:852–854. 10.1038/nsmb1131 [DOI] [PubMed] [Google Scholar]
- 17.Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 18.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. 1991. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 77:1080–1086 [PubMed] [Google Scholar]
- 19.Bijl JJ, van Oostveen JW, Walboomers JM, Brink AT, Vos W, Ossenkoppele GJ, Meijer CJ. 1998. Differentiation and cell-type-restricted expression of HOXC4, HOXC5 and HOXC6 in myeloid leukemias and normal myeloid cells. Leukemia 12:1724–1732. 10.1038/sj.leu.2401106 [DOI] [PubMed] [Google Scholar]
- 20.Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, Nervi C, Minucci S, Fuks F, Di Croce L. 2008. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol. Cell. Biol. 28:5912–5923. 10.1128/MCB.00467-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenbauer F, Tenen DG. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117. 10.1038/nri2024 [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Ichikawa H, Tagata Y, Katsumoto T, Ohnishi K, Akao Y, Naoe T, Pandolfi PP, Kitabayashi I. 2007. PML-retinoic acid receptor alpha inhibits PML IV enhancement of PU. 1-induced C/EBPepsilon expression in myeloid differentiation. Mol. Cell. Biol. 27:5819–5834. 10.1128/MCB.02422-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park DJ, Vuong PT, de Vos S, Douer D, Koeffler HP. 2003. Comparative analysis of genes regulated by PML/RAR alpha and PLZF/RAR alpha in response to retinoic acid using oligonucleotide arrays. Blood 102:3727–3736. 10.1182/blood-2003-02-0412 [DOI] [PubMed] [Google Scholar]
- 24.Hofmann A, Gerrits B, Schmidt A, Bock T, Bausch-Fluck D, Aebersold R, Wollscheid B. 2010. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood 116:e26–34. 10.1182/blood-2010-02-271270 [DOI] [PubMed] [Google Scholar]
- 25.Liao J, Humphrey SE, Poston S, Taparowsky EJ. 2011. Batf promotes growth arrest and terminal differentiation of mouse myeloid leukemia cells. Mol. Cancer Res. 9:350–363. 10.1158/1541-7786.MCR-10-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, Roeder RG, Azorsa DO, Meltzer PS, Suh PG, Song EJ, Lee KJ, Lee YC, Lee JW. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140–149. 10.1128/MCB.23.1.140-149.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luscher-Firzlaff J, Gawlista I, Vervoorts J, Kapelle K, Braunschweig T, Walsemann G, Rodgarkia-Schamberger C, Schuchlautz H, Dreschers S, Kremmer E, Lilischkis R, Cerni C, Wellmann A, Luscher B. 2008. The human trithorax protein hASH2 functions as an oncoprotein. Cancer Res. 68:749–758. 10.1158/0008-5472.CAN-07-3158 [DOI] [PubMed] [Google Scholar]
- 28.Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, He Q, Yang D, Chen R, Liu D, Barton M, Songyang Z. 2013. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J. Biol. Chem. 288:5039–5048. 10.1074/jbc.M112.424515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SK, Anzick SL, Choi JE, Bubendorf L, Guan XY, Jung YK, Kallioniemi OP, Kononen J, Trent JM, Azorsa D, Jhun BH, Cheong JH, Lee YC, Meltzer PS, Lee JW. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283–34293. 10.1074/jbc.274.48.34283 [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. U. S. A. 103:15392–15397. 10.1073/pnas.0607313103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S, Choi HM, Park MJ, Kim YH, Choi YH, Kim HH, Cheong J. 2004. Activation and interaction of ATF2 with the coactivator ASC-2 are responsive for granulocytic differentiation by retinoic acid. J. Biol. Chem. 279:16996–17003. 10.1074/jbc.M311752200 [DOI] [PubMed] [Google Scholar]