Abstract
The acetyltransferase Gcn5 is critical for embryogenesis and shows partial functional redundancy with its homolog PCAF. However, the tissue- and cell lineage-specific functions of Gcn5 and PCAF are still not well defined. Here we probe the functions of Gcn5 and PCAF in adipogenesis. We found that the double knockout (DKO) of Gcn5/PCAF inhibits expression of the master adipogenic transcription factor gene PPARγ, thereby preventing adipocyte differentiation. The adipogenesis defects in Gcn5/PCAF DKO cells are rescued by ectopic expression of peroxisome proliferator-activated receptor γ (PPARγ), suggesting Gcn5/PCAF act upstream of PPARγ to facilitate adipogenesis. The requirement of Gcn5/PCAF for PPARγ expression was unexpectedly bypassed by prolonged treatment with an adipogenic inducer, 3-isobutyl-1-methylxanthine (IBMX). However, neither PPARγ ectopic expression nor prolonged IBMX treatment rescued defects in Prdm16 expression in DKO cells, indicating that Gcn5/PCAF are essential for normal Prdm16 expression. Gcn5/PCAF regulate PPARγ and Prdm16 expression at different steps in the transcription process, facilitating RNA polymerase II recruitment to Prdm16 and elongation of PPARγ transcripts. Overall, our study reveals that Gcn5/PCAF facilitate adipogenesis through regulation of PPARγ expression and regulate brown adipogenesis by influencing Prdm16 expression.
INTRODUCTION
White adipose tissue (WAT) and brown adipose tissue (BAT) provide different functions in energy metabolism. WAT stores excess energy as fat and has hallmark features such as a large unilocular lipid droplet and sparse mitochondria. In contrast, BAT dispenses energy in the form of heat and features small multilocular lipid droplets and abundant mitochondria. Adipogenesis in WAT and BAT is controlled by a similar adipogenic transcriptional cascade (1, 2), in which peroxisome proliferator-activated receptor γ (PPARγ) serves as a master regulator. PPARγ is a member of the nuclear receptor superfamily of ligand-activated transcription factors, and it is both necessary and sufficient for adipocyte differentiation. Two PPARγ isoforms exist: PPARγ1 and PPARγ2. PPARγ1 is expressed in multiple cell types, whereas PPARγ2 is highly adipocyte specific. Both isoforms are strongly induced during adipogenesis. BAT-specific features are driven by additional factors, in particular Prdm16 (3). However, how Prdm16 expression is regulated is not well defined.
PPARγ expression is activated in the adipogenic transcriptional cascade by two transcription factors, C/EBPβ and C/EBPδ, which are immediately induced upon initiation of adipogenesis by cyclic AMP (cAMP)-activated cAMP response element-binding protein (CREB) and glucocorticoid-activated glucocorticoid receptor, respectively. C/EBPβ binds directly to the PPARγ2 promoter to activate PPARγ2 expression (4). C/EBPα is also important to adipogenesis, and it is also induced by C/EBPβ and C/EBPδ. PPARγ and C/EBPα positively regulate themselves and each other to form a feed-forward loop to promote terminal differentiation of adipocytes (5–8).
PPARγ expression during adipogenesis is regulated by alterations in histone modification patterns (7, 9). Mono- and dimethylation of histone H3 lysine 4 (H3K4me1/2) on PPARγ gene locus by MLL3/4 complexes facilitates, whereas dimethylation of histone H3 lysine 9 (H3K9me2) by G9a represses, PPARγ expression (10, 11). Adipogenesis also induces acetylation of histone H3 lysines 9 (H3K9ac) and 27 (H3K27ac) at the PPARγ gene locus (10, 12, 13). H3K27ac marks active enhancers and is catalyzed by histone acetyltransferases (HATs) CBP and p300 (14). Both CBP and p300 have been shown to be required for adipogenesis (15). However, it is still unclear whether the HATs responsible for H3K9ac regulate PPARγ expression and adipogenesis.
One HAT that acetylates histone H3 is Gcn5 (also known as Kat2a). Gcn5 is highly conserved across evolution in structure and enzymatic specificity (16), and mammals contain two highly homologous Gcn5-like paralogues, Gcn5 and PCAF (also known as Kat2b). Gcn5 regulates transcription as part of large multisubunit complexes, such as the SAGA complex, which is recruited to target gene loci by interactions with transcription factors, such as p53, c-myc, and E2F (17–19). In mouse embryonic fibroblasts (MEFs), Gcn5 and PCAF act redundantly for robust acetylation of H3K9. DKO of these HATs eliminates H3K9ac in MEFs (14).
Since Gcn5/PCAF-mediated H3K9ac is induced and correlates with chromatin opening and gene activation during adipogenesis (12, 20), we established immortalized Gcn5/PCAF DKO brown preadipocytes to probe the function of Gcn5/PCAF in adipogenesis. Our findings indicate that Gcn5/PCAF act redundantly during adipogenesis to regulate expression of both PPARγ and Prdm16. Interestingly, the requirement for Gcn5/PCAF for PPARγ expression and adipogenesis was bypassed by prolonged IBMX treatment. In contrast, Prdm16 expression during adipogenesis was highly dependent on Gcn5/PCAF and was not bypassed by prolonged IBMX treatment. Mechanistically, Gcn5/PCAF modulate transcription elongation to regulate PPARγ expression, and modulate polymerase II (Pol II) recruitment to regulate Prdm16 expression. Together, these results indicate that Gcn5/PCAF act at multiple steps to regulate adipocyte differentiation.
MATERIALS AND METHODS
Plasmids and antibodies.
Retroviral plasmids MSCVhygro-Cre and MSCVpuro-PPARγ2 have been described previously (21). The simian virus 40 (SV40) large T antigen-expressing retroviral plasmid pBABEneo-large T was obtained from Addgene (catalog no. 1780). Gcn5 and Gcn5 D608A mutant cDNAs were generated by PCR and inserted into MSCVpuro to generate MSCVpuro-Gcn5 and MSCVpuro-Gcn5-D608A. Primer sequences are available upon request.
Antibodies against C/EBPα (sc-61), C/EBPβ (sc-150X), C/EBPδ (sc-151), PPARγ (sc-7273), and β-actin (sc-47778) were from Santa Cruz. The Gcn5 (catalog no. 3305) and PCAF (catalog no. 3378) antibodies were from Cell Signaling. Antibodies to RNA Pol II (ab5408), S5P Pol II (ab5131), and S2P Pol II (ab5095) were from Abcam. The anti-H3K9ac (1328-1) antibody was from Epitomics. All chemicals were from Sigma unless otherwise indicated.
Isolation of primary preadipocytes, immortalization, virus infection, and adipogenesis assays.
These experiments have been described previously (11). Briefly, primary brown preadipocytes were isolated from interscapular brown adipose tissues of newborn pups and immortalized with retrovirus pBABEneo expressing SV40 large T antigen. The immortalized cells were routinely cultured in Dulbecco modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS). For adipogenesis assays, brown preadipocytes were cultured in differentiation medium (DMEM plus 10% FBS, 0.1 μM insulin, and 1 nM T3) for 4 days before induction of adipogenesis. At day 0, overconfluent brown preadipocytes were treated with differentiation medium supplemented with an adipogenic cocktail consisting of 0.125 mM indomethacin, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), and 2 μg/ml dexamethasone (DEX). Two days later, cells were changed to the differentiation medium alone. At day 6 postinduction, cells were analyzed by Oil Red O staining or quantitative reverse transcription-PCR (qRT-PCR) analysis of gene expression.
Immunoblotting, qRT-PCR, ChIP, and quantitative PCR.
Immunoblotting, qRT-PCR using SYBR green assays, and chromatin immunoprecipitation (ChIP) assays were performed as previously described (14). To determine the Gcn5 KO efficiency in cells with ectopic Gcn5, genomic DNA was extracted. The Gcn5 genomic DNA was quantified with SYBR green assays using standard curve and relative quantitation with GAPDH genomic DNA as control. The data are presented as means ± the standard deviations.
Histology and immunofluorescence staining.
E16.5 embryos dissected by Cesarean section were analyzed by hematoxylin and eosin (H&E) and immunofluorescence staining on paraffin sections as described previously (10). All mouse works were approved by the Animal Care and Use Committee of National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
RESULTS
Gcn5 and PCAF function redundantly to regulate adipogenesis.
To investigate the function of Gcn5 and PCAF in adipogenesis, primary white preadipocytes were isolated from Gcn5flox/flox or Gcn5flox/flox; PCAF−/− mice, and Gcn5 was deleted by adenovirus expressing Cre to generate Gcn5 single-knockout (KO) or Gcn5/PCAF DKO cells (see Fig. S1A in the supplemental material). The cells were then induced to undergo adipogenesis by treatment with an adipogenic cocktail that included insulin, IBMX, and DEX. Individual deletion of either Gcn5 or PCAF had little effect on adipogenesis. However, Gcn5/PCAF DKO inhibited lipid droplet formation and expression of adipogenic genes, such as PPARγ and aP2 (see Fig. S1B and C in the supplemental material).
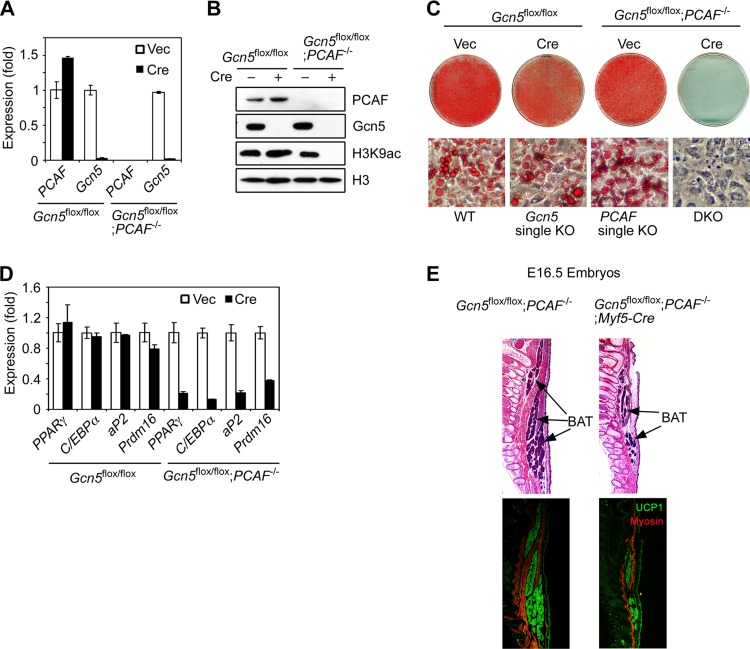
To obtain sufficient amounts of cells for mechanistic studies, brown preadipocytes from Gcn5flox/flox or Gcn5flox/flox; PCAF−/− mice were immortalized with SV40 large T antigen (SV40T). The immortalized cells were infected with the retrovirus expressing Cre (retroviral Cre) to delete Gcn5 (Fig. 1A). Gcn5 deletion in Gcn5flox/flox; PCAF−/− cells did not change cell morphology but did retard cell growth (see Fig. S1D and E in the supplemental material). Consistent with previous findings in MEFs (14), Gcn5/PCAF DKO dramatically reduced global levels of H3K9ac (Fig. 1B), indicating that Gcn5 and PCAF act redundantly as the major drivers of H3K9ac in brown preadipocytes. Similar to our observations in white preadipocytes, deletion of both PCAF and Gcn, but not Gcn5 or PCAF alone strongly prevented lipid accumulation and expression of adipogenic genes, including PPARγ, C/EBPα, and aP2, as well as the BAT master regulator, Prdm16 (Fig. 1C and D).
FIG 1.
Gcn5 and PCAF function redundantly to regulate adipogenesis. (A to D) Gcn5 and PCAF function redundantly for H3K9ac and adipogenesis in brown preadipocytes. Brown preadipocytes were isolated from Gcn5flox/flox and Gcn5flox/flox; PCAF−/− mice and immortalized with SV40T prior to Gcn5 deletion by retroviral Cre. Subconfluent cells were subjected to qRT-PCR analysis of Gcn5 and PCAF mRNA (A) and immunoblotting with the antibodies indicated on the right (B). The cells were induced to undergo adipogenesis for 6 days, followed by Oil Red O staining (C) and qRT-PCR analysis of gene expression (D). (E) Gcn5 and PCAF are required for development of BAT in vivo. Gcn5flox/flox; PCAF−/− males were crossed with Gcn5flox/+; PCAF−/−; Myf5-Cre females to generate Gcn5flox/flox; PCAF−/−; Myf5-Cre pups. Sagittal sections of E16.5 embryos were subjected to H&E staining (upper panels) and immunofluorescence staining with antibodies against myosin and UCP1 (lower panels). Myosin (red) and UCP1 (green) are specifically expressed in skeletal muscle and BAT, respectively.
Although PCAF KO mice do not show any obvious, abnormal phenotype, Gcn5 KO leads to early embryonic lethality (22). DKO embryos die even earlier (22), preventing analysis of BAT development in either Gcn5 single-KO or DKO mice. To circumvent this lethality, we introduced a Myf5-driven Cre transgene into Gcn5flox/flox; PCAF−/− mice to specifically delete Gcn5 in precursor cells that develop to both brown fat and skeletal muscle tissues in the back (3). Gcn5flox/flox; PCAF−/−; Myf5-Cre pups died right after birth, likely due to skeletal muscle defects (data not shown). Importantly, these pups had much less BAT compared to their littermates (see Fig. S1F in the supplemental material). To further confirm defects in BAT development in these mice, sagittal sections of E16.5 embryos were subjected to H&E and immunofluorescence staining of BAT- and skeletal muscle-specific markers, UCP1 and myosin, respectively. These results again reveal that BAT was reduced substantially in Gcn5flox/flox; PCAF−/−; Myf5-Cre pups compared to their littermate controls (Fig. 1E), indicating that Gcn5/PCAF are required for BAT development in vivo. Collectively, these results indicate that, Gcn5 and PCAF function redundantly to regulate adipocyte differentiation.
Gcn5 and PCAF act upstream of PPARγ to facilitate adipogenesis.
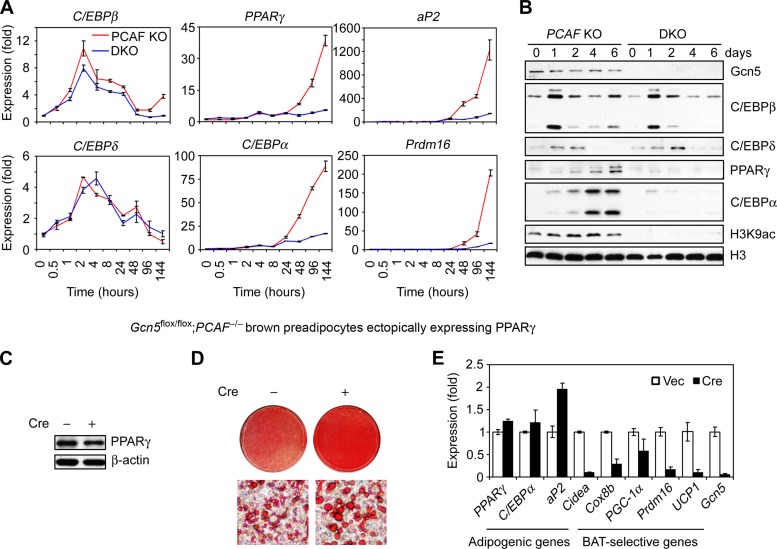
To understand the mechanism by which Gcn5 and PCAF facilitate adipogenesis, we monitored induction of the adipogenic transcriptional cascade in DKO brown preadipocytes. We found that C/EBPβ and C/EBPδ were induced normally in DKO cells, whereas the expression of PPARγ and C/EBPα, as well as that of their downstream target gene aP2, was severely impaired by Gcn5/PCAF double deletion (Fig. 2A and B). Prdm16 induction was also impaired in the DKO cells (Fig. 2A). Gcn5 protein levels were not significantly altered during adipogenesis in PCAF KO cells (Fig. 2B). These results suggest that Gcn5 and PCAF function upstream of PPARγ. To test this, we ectopically expressed PPARγ from a retroviral vector in Gcn5flox/flox; PCAF−/− brown preadipocytes prior to Gcn5 deletion by Cre (Fig. 2C). As shown in Fig. 2D and E, ectopic PPARγ stimulated adipogenesis well in both control and DKO cells, indicating that Gcn5/PCAF work upstream of PPARγ to facilitate adipogenesis.
FIG 2.
Gcn5 and PCAF act upstream of PPARγ during adipogenesis. (A and B) Gcn5 and PCAF act downstream of C/EBPβ/δ but upstream of PPARγ during adipogenesis. Retroviral Vec- or Cre-infected Gcn5flox/flox; PCAF−/− brown preadipocytes were induced to undergo adipogenesis, and samples were collected at the indicated times for qRT-PCR analysis of gene expression (A) and immunoblotting with the antibodies indicated on the right (B). (C to E) Gcn5 and PCAF are dispensable for ectopic PPARγ-induced adipogenesis. Gcn5flox/flox; PCAF−/− brown preadipocytes were sequentially infected with retroviral PPARγ2 and retroviral Cre. Ectopic PPARγ2 proteins in subconfluent cells were detected by immunoblotting (C). The cells were induced to undergo adipogenesis. Six days later, the cells were subjected to Oil Red O staining (D) and qRT-PCR analysis of gene expression (E).
Gcn5 catalytic activity is required for efficient adipogenesis.
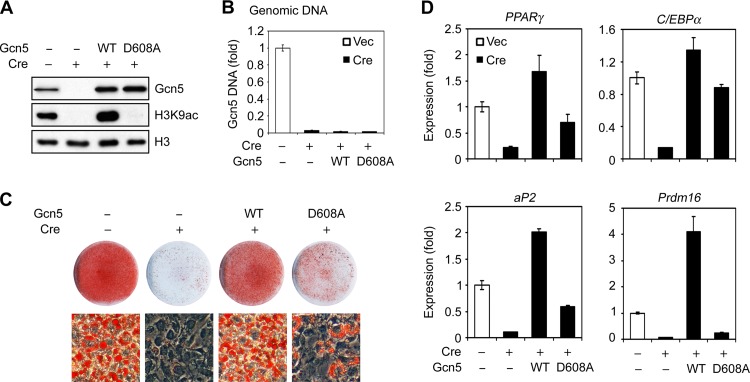
Gcn5 has both HAT activity-dependent and -independent functions (23, 24). To investigate the role of Gcn5 HAT activity in adipogenesis, wild-type (WT) Gcn5 or an enzymatically dead Gcn5 mutant (D608A) was transduced into Gcn5flox/flox; PCAF−/− brown preadipocytes using retroviral vectors, followed by deletion of endogenous Gcn5 by retroviral Cre (Fig. 3A and B). As expected, WT Gcn5 and PCAF, but not the Gcn5 D608A mutant, prevented loss of global H3K9ac after deletion of endogenous Gcn5 (Fig. 3A; see Fig. S2A in the supplemental material). Upon induction of adipogenesis, ectopic expression of either WT Gcn5 or PCAF prevented defective adipogenesis in the DKO cells (Fig. 3C and D; see Fig. S2B and C in the supplemental material), confirming these HATs serve redundant functions during adipogenesis. Introduction of the Gcn5 D608A was less effective than wild-type Gcn5 in restoring lipid accumulation and adipogenic gene expression (Fig. 3C and D). Strikingly, ectopic overexpression of WT Gcn5 elevated Prdm16 expression ∼4-fold relative to expression seen in the presence of the wild-type endogenous protein, and ∼16-fold relative to equally expressed ectopic Gcn5 D608A (Fig. 3A and D). These data suggest that Gcn5 has both HAT-dependent and -independent functions in adipogenesis and that Prdm16 expression is highly dependent on Gcn5 HAT activity.
FIG 3.
Gcn5 catalytic activity is required for efficient adipogenesis. Gcn5flox/flox; PCAF−/− brown preadipocytes were first infected with retroviruses expressing wild-type (WT) Gcn5 or an enzymatically dead Gcn5 mutant D608A prior to endogenous Gcn5 deletion by Cre. In subconfluent cells, Gcn5, histone H3, and H3K9ac were analyzed by immunoblotting (A), and Gcn5 knockout was confirmed by qPCR analysis of Gcn5 genomic DNA (B). The cells were induced to undergo adipogenesis for 6 days, followed by Oil Red O staining (C) and qRT-PCR analysis of gene expression (D).
Gcn5 and PCAF are required for Prdm16 expression during brown adipogenesis.
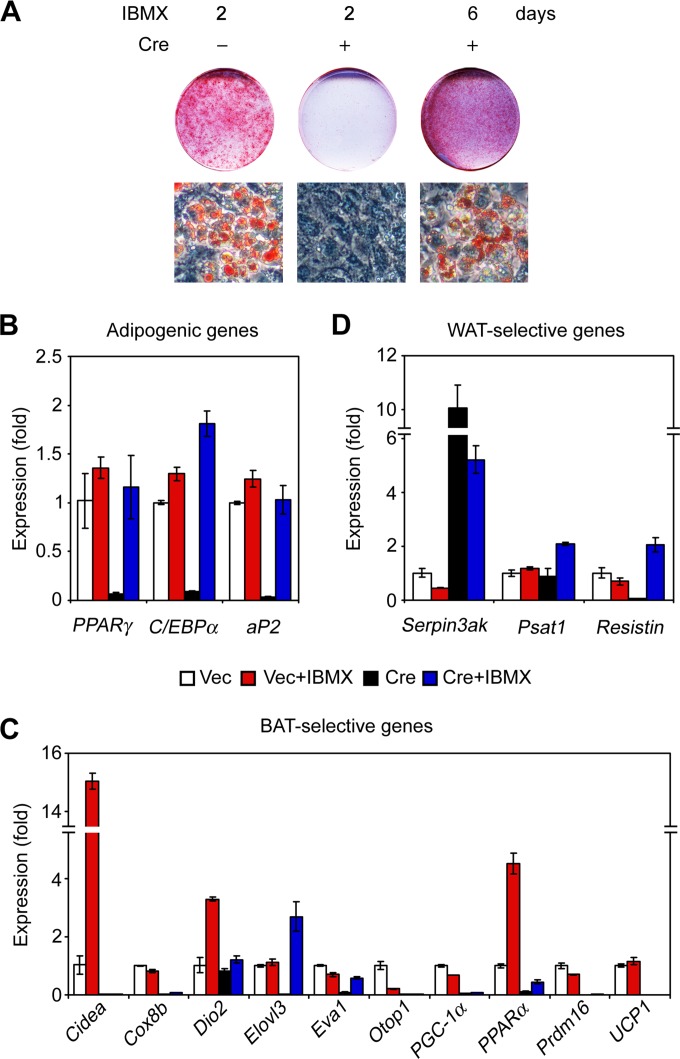
Ectopic expression of PPARγ in Gcn5/PCAF DKO brown preadipocytes rescued adipogenesis but not Prdm16 expression (Fig. 2E), indicating that Gcn5/PCAF are also involved in regulating Prdm16 expression during brown adipogenesis. Unexpectedly, we found that prolonged treatment of DKO cells with IBMX rescued adipogenesis and expression of adipogenic genes such as PPARγ, C/EBPα, and aP2 (Fig. 4A and B; see Fig. S3A and B in the supplemental material). However, this treatment did not rescue expression of Prdm16 (Fig. 4C).
FIG 4.
Gcn5 and PCAF are required for Prdm16 expression during brown adipogenesis. Retroviral Vec- or Cre-infected Gcn5flox/flox; PCAF−/− brown preadipocytes were treated with the adipogenic cocktail to induce adipogenesis at day 0. At day 2, the adipogenic cocktail was removed and cells were treated with differentiation medium supplemented with or without 0.5 mM IBMX for 4 days. At day 6, cells were subjected to Oil Red O staining (A), and qRT-PCR analysis of adipogenic genes (B), BAT-selective genes (C), and WAT-selective genes (D).
Since Prdm16 is pivotal for development of brown adipocytes, we analyzed the effects of Gcn5/PCAF double deletion on expression of BAT- and WAT-specific genes (25) in differentiated cells after prolonged IBMX treatment (Fig. 4C and D). Prolonged IBMX treatment increased several BAT-selective genes in control cells, including PPARα, Cidea, and Dio2, but rescued expression of only a few BAT-specific genes, such as Eva1 and Elovl3, in DKO cells. Consistent with the requirement of Gcn5/PCAF for Prdm16 expression, prolonged IBMX treatment of the DKO cells failed to rescue expression of Prdm16-dependent BAT-selective genes, including Cidea, Cox8b, Dio2, Otop1, PGC-1α, PPARα, and UCP1 (25–27) (Fig. 4C). The expression of the WAT-selective genes, Psat1, Resistin, and particularly Serpin3ak, was increased in DKO cells after prolonged IBMX treatment (Fig. 4D). Collectively, these data indicate that Gcn5 and PCAF are required for Prdm16 expression during brown adipogenesis.
Gcn5 and PCAF regulate different steps of PPARγ and Prdm16 transcription.
Both PCAF and H3K9ac are enriched around transcription start sites (TSSs) of active genes (28, 29). PCAF and H3K9ac were gradually enriched at the PPARγ2 and Prdm16 genes during adipogenesis (Fig. 5A to D), and the increase in H3K9ac at these two genes was completely blocked by Gcn5/PCAF double deletion (Fig. 5B and D), a finding consistent with loss of the global H3K9ac in DKO cells. These results suggest PCAF and Gcn5 are set on PPARγ2 and Prdm16 genes to regulate their expression.
FIG 5.
Gcn5 and PCAF regulate different steps of PPARγ and Prdm16 transcription. (A to D) Gcn5/PCAF acetylate H3K9 at PPARγ2 and Prdm16 genes during adipogenesis. Gcn5flox/flox brown preadipocytes (A and C) or retroviral Vec- or Cre-infected Gcn5flox/flox; PCAF−/− brown preadipocytes (B and D) were induced for adipogenesis. ChIP assays were performed to analyze PCAF (A and C) and H3K9ac (B and D) at the indicated sites in the PPARγ2 and Prdm16 genes. (E and F) Gcn5/PCAF regulate different steps of PPARγ and Prdm16 transcription. Retroviral Vec- or Cre-infected Gcn5flox/flox; PCAF−/− brown preadipocytes were induced to undergo adipogenesis, and cells were collected at the indicated times for ChIP analysis of C/EBPβ, Pol II, Pol II (S5P), and Pol II (S2P) association at the indicated sites in the PPARγ2 (E) and Prdm16 (F) genes. (G and H) Prolonged IBMX treatment increases C/EBPβ and Pol II recruitment and reactivates transcription elongation of the PPARγ2 gene in Gcn5/PCAF DKO cells but does not promote Pol II recruitment at the Prdm16 gene. Retroviral Vec- or Cre-infected Gcn5flox/flox; PCAF−/− brown preadipocytes were induced to adipogenesis. After removal of the adipogenic cocktail, the cells were treated with or without 0.5 mM IBMX for 1 more day and then collected for ChIP assays of C/EBPβ, Pol II, and Pol II (S2P) enrichments at indicated sites of the PPARγ2 (G) and Prdm16 (H) genes.
Gcn5/PCAF loss did not impair C/EBPβ induction during adipogenesis (Fig. 2A). To determine whether loss of these HATs affects C/EBPβ recruitment onto the PPARγ2 gene, we performed ChIP assays for this factor to monitor its association with a C/EBPβ binding site located at kb −0.3 upstream of PPARγ2 TSS (4). We found that C/EBPβ was greatly enriched at this site at day 2 of adipogenesis and that this enrichment decreased at days 4 and 6 (Fig. 5E). This pattern of C/EBPβ binding was not affected in the DKO cells. We next examined RNA Pol II association with PPARγ2. Phosphorylation on Serine 5 (S5P) and Serine 2 (S2P) of the Pol II C-terminal domain are markers of transcription initiation and elongation, respectively. Peaks of S2P Pol II are observed commonly at TSSs and at the 3′ ends of genes (30). Adipogenesis-induced enrichment of Pol II, S5P Pol II, and S2P Pol II had similar trends of enrichment as C/EBPβ at PPARγ2 (Fig. 5E). Gcn5/PCAF double deletion did not change Pol II and S5P Pol II levels at the PPARγ2 gene at days 2 and 4 but lowered enrichment of these factors at day 6 (Fig. 5E). S2P Pol II enrichment at the PPARγ2 gene was not changed in the DKO cells at day 2 but was lowered at days 4 and 6, suggesting defective transcription elongation of PPARγ2 at these time points. These data indicate that Gcn5/PCAF are required for efficient and sustained elongation of PPARγ2 transcription.
In contrast, the enrichment of Pol II, S5P Pol II, and S2P Pol II near the TSS of the Prdm16 gene (at kb +0.1), which increased first at day 4 in control cells with further increase at day 6, was diminished in the DKO cells (Fig. 5F). These results suggest that Gcn5/PCAF are required for Pol II recruitment onto the Prdm16 gene.
Next, we investigated whether prolonged IBMX treatment stimulates transcription elongation of the PPARγ2 gene in DKO cells, since it rescued PPARγ2 expression in DKO cells. Prolonged treatment of IBMX enhanced C/EBPβ and Pol II recruitment at the PPARγ promoter in both control and DKO cells, and importantly, IBMX stimulated S2P Pol II enrichment at the PPARγ gene in DKO cells (Fig. 5G), which is consistent with elevated PPARγ expression in response to prolonged IBMX treatment (see Fig. S3C in the supplemental material). IBMX also enhanced Pol II enrichment on the Prdm16 gene locus in control cells but not in DKO cells (Fig. 5H). These results indicate prolonged IBMX treatment rescues PPARγ expression in DKO cells by reactivating transcription elongation of this gene.
DISCUSSION
Our data suggest a model for Gcn5 and PCAF facilitation of brown adipogenesis via regulation of PPARγ and Prdm16 expression (Fig. 6). PPARγ is known to be a master transcription factor that governs adipogenesis, whereas Prdm16 is a master regulator for development of brown adipocyte-specific features. Upon adipogenic stimulation, Pol II is recruited onto the PPARγ promoter by C/EBPβ and onto the Prdm16 promoter by unknown transcription factors to activate expression of these genes. PPARγ and Prdm16 then drive cells differentiation into mature brown adipocytes. In the absence of Gcn5/PCAF, Pol II is still recruited to the PPARγ promoter via C/EBPβ, but Pol II elongation is inhibited, leading to inhibition of PPARγ transcription. Loss of these HATs prevents Pol II recruitment to the Prdm16 promoter. Thus, adipogenesis is inhibited in DKO cells. In the absence of Gcn5/PCAF, prolonged IBMX treatment enhances C/EBPβ and Pol II recruitment to PPARγ, stimulating transcript elongation, but IBMX does not promote Pol II recruitment onto the Prdm16 promoter. Consequently, IBMX restores expression of PPARγ but not Prdm16 in DKO cells, resulting in their development into adipocytes that lack brown adipocyte molecular phenotype.
FIG 6.
Model for Gcn5 and PCAF regulating PPARγ and Prdm16 expression to facilitate brown adipogenesis.
Gcn5/PCAF double deletion in preadipocytes caused severe defects in adipogenesis. These defects were rescued by ectopic expression of PPARγ, indicating these HATs act largely at the level of PPARγ expression. However, adipogenesis was also rescued by prolonged IBMX treatment of the DKO cells. The main function of IBMX in the adipogenic cocktail is to elevate intracellular cAMP concentrations, thereby activating CREB to induce C/EBPβ expression. cAMP also stimulates C/EBPβ phosphorylation (31), which is required for C/EBPβ binding to target genes (32). Therefore, prolonged exposure to IBMX likely stimulates C/EBPβ phosphorylation to enhance C/EBPβ binding to the PPARγ2 promoter, consequently increasing Pol II recruitment and bypassing the need for Gcn5 and PCAF.
Gcn5 functions in transcription elongation have been reported previously in yeast. Yeast Gcn5 is predominantly localized to coding regions of highly transcribed genes (33), and it enhances transcription elongation by facilitating nucleosome eviction or other histone acetylation-dependent processes (34, 35). Our study provides evidence that Gcn5/PCAF are also involved in transcription elongation in mammalian cells. Previous work reporting that PCAF is distributed on both promoters and transcribed regions of active genes is also consistent with a role for this HAT in transcription elongation (28). Gcn5/PCAF were reported to interact with transcription elongation factor p-TEFb (36), which may be recruited onto the PPARγ gene by Gcn5/PCAF to phosphorylate Pol II and trigger transcription elongation.
The transcriptional control of BAT development has received much attention over the last several years, since Prdm16 is the master transcription factor for BAT development, and furthermore, Prdm16 is required for browning of subcutaneous fat cells (27). Since Gcn5 and PCAF are important for Prdm16 expression, these HATs are likely required not only for BAT development but also for browning of white adipocytes. Increased Prdm16 expression provides a potential strategy in the fight against the obesity epidemic. Prdm16 expression is regulated at both the mRNA and protein levels. miR-133 directly targets and downregulates Prdm16 mRNA to inhibit brown adipocyte differentiation (37). The PPARγ agonist rosiglitazone stabilizes Prdm16 to induce a white-to-brown fat conversion (26). Before our study, little was known about how Prdm16 is regulated at the transcriptional level. Gcn5/PCAF might mediate acetylation of either H3K9 or transcription factors/regulators to activate Prdm16. Both C/EBPβ and PGC-1α, which are involved in inducing expression of BAT-selective genes (38), are acetylated by Gcn5 to change their transcriptional activity (39, 40). Our findings might provide additional options for treatment of obesity. Modulation of Gcn5 HAT activity by small molecules or other agents might allow modulation of Prdm16 expression, facilitating BAT development or white-to-brown fat conversion. Development of such therapeutic strategies could provide important steps forward in combating obesity and obesity-associated disorders.
Supplementary Material
ACKNOWLEDGMENTS
We thank Boyko Atanassov, Aimee T. Farria, and Elizabeth Mclvor for critical reading of the manuscript.
Mouse experiments were facilitated by the Science Park Research Animal facility, which is partly supported by a Cancer Center Support (CORE) grant (5 P30 CA16672-32, DePinho as the principal investigator) from the National Cancer Institute. This study was supported by NIGMS/NIH R01 067718 to S.Y.R.D.; by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, to K.G.; and by CAS/SAFEA International Partnership Program for Creative Research Teams to H.Y.
Footnotes
Published ahead of print 28 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00622-14.
REFERENCES
- 1.Farmer SR. 2008. Molecular determinants of brown adipocyte formation and function. Genes Dev. 22:1269–1275. 10.1101/gad.1681308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fruhbeck G, Sesma P, Burrell MA. 2009. PRDM16: the interconvertible adipo-myocyte switch. Trends Cell Biol. 19:141–146. 10.1016/j.tcb.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 3.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967. 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salma N, Xiao H, Imbalzano AN. 2006. Temporal recruitment of CCAAT/enhancer-binding proteins to early and late adipogenic promoters in vivo. J. Mol. Endocrinol. 36:139–151. 10.1677/jme.1.01918 [DOI] [PubMed] [Google Scholar]
- 5.Cristancho AG, Lazar MA. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell. Biol. 12:722–734. 10.1038/nrm3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4:263–273. 10.1016/j.cmet.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge K. 2012. Epigenetic regulation of adipogenesis by histone methylation. Biochim. Biophys. Acta 1819:727–732. 10.1016/j.bbagrm.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 7:885–896. 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- 9.Lee JE, Ge K. 2014. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 4:29. 10.1186/2045-3701-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2:e01503. 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Xu S, Lee J-E, Baldridge A, Grullon S, Peng W, Ge K. 2013. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 32:45–59. 10.1038/emboj.2012.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Jr, Lazar MA. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 24:1035–1044. 10.1101/gad.1907110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. 2010. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143:156–169. 10.1016/j.cell.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30:249–262. 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, Fushiki T. 2002. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 277:16906–16912. 10.1074/jbc.M200585200 [DOI] [PubMed] [Google Scholar]
- 16.Roth SY, Denu JM, Allis CD. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81–120. 10.1146/annurev.biochem.70.1.81 [DOI] [PubMed] [Google Scholar]
- 17.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243–1254. 10.1016/S1097-2765(01)00414-2 [DOI] [PubMed] [Google Scholar]
- 18.McMahon SB, Wood MA, Cole MD. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556–562. 10.1128/MCB.20.2.556-562.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang SE, McMahon SB, Cole MD, Hearing P. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276:32627–32634. 10.1074/jbc.M102067200 [DOI] [PubMed] [Google Scholar]
- 20.Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodeling and establishment of transcription factor “hot spots” during early adipogenesis. EMBO J. 30:1459–1472. 10.1038/emboj.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. 2009. Histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis. Cell Metab. 10:27–39. 10.1016/j.cmet.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229–232. 10.1038/79973 [DOI] [PubMed] [Google Scholar]
- 23.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 35:352–364. 10.1016/j.molcel.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E. 2009. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev. 23:849–861. 10.1101/gad.1748409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 6:38–54. 10.1016/j.cmet.2007.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. 2012. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15:395–404. 10.1016/j.cmet.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, Wu J, Gunawardana SC, Banks AS, Camporez JP, Jurczak MJ, Kajimura S, Piston DW, Mathis D, Cinti S, Shulman GI, Seale P, Spiegelman BM. 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156:304–316. 10.1016/j.cell.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40:897–903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mousavi K, Zare H, Wang AH, Sartorelli V. 2012. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell 45:255–262. 10.1016/j.molcel.2011.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metz R, Ziff E. 1991. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 5:1754–1766. 10.1101/gad.5.10.1754 [DOI] [PubMed] [Google Scholar]
- 32.Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. 2005. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 102:9766–9771. 10.1073/pnas.0503891102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. 2009. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 10:1009–1014. 10.1038/embor.2009.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25:31–42. 10.1016/j.molcel.2006.11.020 [DOI] [PubMed] [Google Scholar]
- 35.Kristjuhan A, Svejstrup JQ. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243–4252. 10.1038/sj.emboj.7600433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabo A, Lusic M, Cereseto A, Giacca M. 2008. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell. Biol. 28:2201–2212. 10.1128/MCB.01557-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trajkovski M, Ahmed K, Esau CC, Stoffel M. 2012. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 14:1330–1335. 10.1038/ncb2612 [DOI] [PubMed] [Google Scholar]
- 38.Kajimura S, Seale P, Spiegelman BM. 2010. Transcriptional control of brown fat development. Cell Metab. 11:257–262. 10.1016/j.cmet.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 3:429–438. 10.1016/j.cmet.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 40.Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ. 2007. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPβ by GCN5. Proc. Natl. Acad. Sci. U. S. A. 104:2703–2708. 10.1073/pnas.0607378104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.