Abstract
Aspergillus spp. are among the most common causes of opportunistic invasive fungal infections in tertiary care hospitals. Little is known about the prevalence and in vitro susceptibility of Aspergillus species in Latin America, because there are few medical centers able to perform accurate identification at the species level. The purpose of this study was to analyze the distribution of cryptic and rare Aspergillus species among clinical samples from 133 patients with suspected aspergillosis admitted in 12 medical centers in Brazil and to analyze the in vitro activity of different antifungal drugs. The identification of Aspergillus species was performed based on a polyphasic approach, as well as sequencing analysis of the internal transcribed spacer (ITS) region, calmodulin, and β-tubulin genes and phylogenetic analysis when necessary. The in vitro susceptibility tests with voriconazole, posaconazole, and itraconazole were performed according to the CLSI M38-A2 document (2008). We demonstrated a high prevalence of cryptic species causing human infection. Only three isolates, representing the species Aspergillus thermomutatus, A. ochraceus, and A. calidoustus, showed less in vitro susceptibility to at least one of the triazoles tested. Accurate identifications of Aspergillus at the species level and with in vitro susceptibility tests are important because some species may present unique resistance patterns against specific antifungal drugs.
INTRODUCTION
Aspergillus is a ubiquitous fungus that is responsible for a wide spectrum of infections. One of the most important clinical manifestations of Aspergillus is invasive aspergillosis (IA), which is associated with high morbidity and mortality rates (1, 2). The genus Aspergillus is divided into eight subgenera that in turn are subdivided into several sections that include a large variety of closely related species (3, 4). The most clinically relevant sections are Fumigati, Flavi, Terrei, Usti, Nigri, and Nidulantes (5). Molecular studies have revealed numerous cryptic species within the different sections of the genus Aspergillus (6).
Historically, Aspergillus has been identified in the laboratory by conventional methods such as colony morphology and microscopic characteristics. However, there is a consensus that morphological characteristics may not be reliable for distinguishing between Aspergillus species (7). Despite its clinical relevance and several comprehensive studies dealing with the taxonomy of Aspergillus in the last few years, the taxonomy of Aspergillus remains somewhat ill defined. For consistent species identification, analyses of morphological, physiological, and molecular characteristics are required (7, 8). As this process is not suitable for routine testing by clinical microbiological laboratories, identification of Aspergillus clinical isolates at the species level has been scarcely reported (9). The accurate identification of species is critical given that different species may present peculiarities in terms of reservoir, virulence factors, natural history of infection, and in vitro susceptibility to antifungal drugs (10, 11).
The aim of this study was to analyze the distribution of Aspergillus species among clinical samples isolated from 133 patients with suspected aspergillosis admitted to 12 medical centers in Brazil and to analyze the in vitro antifungal susceptibility profiles of rare and cryptic species within the genus.
MATERIALS AND METHODS
Fungal isolates.
We selected 133 isolates previously identified as Aspergillus spp. obtained from 133 different patients admitted to 12 medical centers in Brazil between 2006 and 2013. All isolates were interpreted as pathogens by the clinicians following the criteria suggested by the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) before being sent for further identification in our reference lab. The isolates were grown on slanted potato dextrose agar (PDA) (Difco Laboratories, Detroit, MI, USA) for 7 days at 25°C and were covered with mineral oil for long-term room temperature storage until analysis.
Morphological examination and thermotolerance.
The isolates were grown on PDA, malt extract agar (MEA) (Difco Laboratories, Detroit, MI, USA), and Czapek agar (CZK) (Difco Laboratories, Detroit, MI, USA). The fungi were inoculated at three points on duplicate plates of each medium and incubated at 15, 25, 37, 42, and 50°C for 14 days in the dark (12). Micromorphology observations were performed on microscopic mounts prepared in lactic acid from MEA colonies. The thermotolerance test involved assessment of the presence or absence of fungal growth at different temperatures (8).
Molecular identification: DNA extraction, amplification, and sequencing of ITS, calmodulin, and β-tubulin genes.
The isolates were grown on yeast extract sucrose agar (YES) (10 g yeast extract, 75 g sucrose, 10 g agar, and 500 ml distilled water). Then, DNA was extracted with the PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The DNA concentration and purity (relative to proteins and salts) were determined by optical density at 260 nm (OD260) and ratios of OD260/280 and OD260/230, respectively. Fragments of the calmodulin (CAL) and β-tubulin (BT2) genes and the internal transcribed spacer (ITS) region of rDNA were amplified with the primer pairs cl1/cl2a, bt2a/bt2b (13), and ITS1/ITS4, respectively. The reactions were performed with PCR master mix (Promega, Madison, WI, USA) according to the manufacturer's instructions. After amplification, the fragments were sequenced following the protocol provided with the BigDye reagent kit (Applied Biosystems, Foster City, CA, USA) in an ABI 3130 (Applied Biosystems, Foster City, CA, USA) automatic sequencer. PCR products were sequenced with the same primers used for amplification. Contig assembly and editing were performed with Sequencher DNA sequence assembly software 4.1.4 (Gene Codes Corporation, Ann Arbor, MI, USA). Successful assembly of the contigs required a minimum match percentage of ≥85 and a minimum overlap of 20.
BLAST analysis.
Complete CAL, BT2, and ITS consensus sequences were used to conduct BLAST search analysis (BLASTn) for species identification from the NCBI genomic database (http://blast.ncbi.nlm.nih.gov/). For all regions analyzed by BLAST search, the sequences that presented with high identity (≥99%), queries and E values of e10−5 were considered for the final species identification using the sequencing method.
The identification of Aspergillus species was performed based on macromorphology, micromorphology, and thermotolerance of the colonies, as well as on sequence comparisons of the ITS region and the calmodulin and β-tubulin genes with published sequences in genomic databases. Phylogenetic analyses using Bayesian inference and maximum parsimony methodologies were performed to characterize isolates with inconsistent identification by morphological and genotypic analysis.
Phylogenetic analysis.
Most-parsimonious analysis was carried out for all data sets using PAUP* version 4.0b10 (14). One hundred heuristic searches were conducted using random sequence addition and tree bisection-reconnection branch-swapping algorithms, collapsing zero-length branches, and saving all minimal-length trees (MulTrees) on different sets of data. Gaps were treated as missing data. Support for internal branches was assessed by a heuristic parsimony search of 1,000 bootstrapped sets of data. Other measures were also taken, including tree length, consistency index (CI), homoplasy index (HI), and retention index (RI). The combined data set was tested for incongruence with the partition homogeneity test as implemented in PAUP*. The alignments used in the phylogenetic analysis were deposited in TreeBASE (www.treebase.org).
Bayesian posterior probabilities were calculated using MrBayes 3.12 (15). A neighbor-joining (NJ) tree was analyzed in the ModelTest program to estimate the best model of nucleotide substitution for application on the phylogeny inference. A general time-reversible (GTR) model was used with a proportion of invariant sites and a gamma-shaped distribution of rates across the sites. Markov chain Monte Carlo (MCMC) analysis was conducted for up to 1 × 106 generations until the chain converged. Concordance analysis was based on the exclusionary principle of Baum and Shaw (16) and the genealogical concordance phylogenetic species recognition concepts of Taylor et al. (17). Clades were recognized as independent evolutionary lineages if a clade was strongly supported by both parsimony and Bayesian analysis in at least one locus and the result was not contradicted by another strongly supported locus. Strong support was assessed at >70% bootstrap and >0.95 Bayesian posterior probability (11, 18, 19).
Nucleotide sequence accession numbers.
The nucleotide sequence representatives of each identified species in this study were deposited in the GenBank database under the accession numbers given in Table 1.
TABLE 1.
GenBank accession numbers of the nucleotide sequences representatives of each identified species and reference strains included in this study
| Aspergillus species | Straina | GenBank accession no. for: |
|
|---|---|---|---|
| β-Tubulin | Calmodulin | ||
| A. flavus | CBS 100927NT | AY819992.1 | EF202063.1 |
| A. flavus | LEMI803 | KJ767726 | KJ766990 |
| A. flavus | LEMI925 | KJ767727 | KJ766989 |
| A. flavus | LEMI953 | KJ767728 | KJ766988 |
| A. flavus | LEMI896 | KJ767729 | KJ766987 |
| A. arachidicola | CBS 117610T | EF203158 | EF202049.1 |
| A. arachidicola | LEMI760 | KJ767720 | KJ767736 |
| A. minisclerotigenes | CBS 117620 | EF203150.1 | EF202073.1 |
| A. parasiticus | CBS 100926NT | EF203155.1 | EF202043.1 |
| A. sergii | MUM 10.219T | HM803082 | HM803029 |
| A. transmontanensis | MUM 10.214T | HM803101 | HM803020 |
| A. transmontanensis | LEMI800 | KJ766997 | KJ767732 |
| A. nomius | NRRL 13137T | EF661494.1 | EF661531.1 |
| A. nomius | LEMI878 | KJ767725 | KJ767731 |
| A. oryzae var. effusus | NRRL 506T | JN185446.1 | JN185447.1 |
| A. oryzae | NRRL 447T | EF661483.1 | EF661506.1 |
| A. sojae | CBS 100928T | EF203168.1 | EF202041.1 |
| A. tamarii | NRRL 20818T | EF661474.1 | EF661526.1 |
| A. tamarii | LEMI436 | KJ767721 | KJ767722 |
| A. tamarii | LEMI999 | KJ767733 | KJ767735 |
| A. caelatus | NRRL 25528T | EF661470.1 | AF255036.1 |
| A. tennesseensis | NRRL 13150T | JN853976.1 | JN854017.1 |
| A. tennesseensis | LEMI875 | KJ766999 | KJ766995 |
| A. tennesseensis | LEMI917 | KJ766998 | KJ766994 |
| A. tennesseensis | LEMI870 | KJ767000 | KJ766996 |
| A. puulaauensis | NRRL 35641T | JN853979.1 | JN854034.1 |
| A. puulaauensis | NRRL 58602 | JN853999.1 | JN854048.1 |
| A. cvjetkovicii | NRRL 227T | EF652264.1 | EF652352.1 |
| A. cvjetkovicii | NRRL 58593 | JN853998.1 | JN854044.1 |
| A. cvjetkovicii | NRRL 4642 | EF652291.1 | EF652379.1 |
| A. creber | NRRL 58592T | JN853980.1 | JN854043.1 |
| A. subversicolor | NRRL 58999T | JN853970.1 | JN854010.1 |
| A. asperescens | NRRL 4770T | EF652299.1 | EF652387.1 |
| A. multicolor | NRRL 4775T | EF652301.1 | EF652389.1 |
| A. awamori | CCF 4068T | HE661602.1 | FR751414.1 |
| A. awamori | LEMI1010/LEMI993 | KJ777804 | KJ777809 |
| A. foetidus | CBS564.65 | GU296697.1 | FN594547.1 |
| A. foetidus | LEMI891 | KJ777808 | KJ777811 |
| A. tubingensis | CBS 134.48T | FJ629305.1 | FN594558.1 |
| A. creber | NRRL 58673T | JN853993.1 | JN854056.1 |
| A. creber | NRRL 58670 | JN853991.1 | JN854053.1 |
| A. creber | NRRL 58672 | JN853992.1 | JN854055.1 |
| A. creber | LEMI984 | KJ767001 | KJ766991 |
| A. jensenii | NRRL 58600T | JN854007.1 | JN854046.1 |
| A. jensenii | NRRL 225 | JN854000.1 | JN854020.1 |
| A. jensenii | NRRL 235 | JN854001.1 | JN854027.1 |
| A. jensenii | NRRL 240 | JN854002.1 | JN854030.1 |
| A. venenatus | NRRL 13147T | JN854003.1 | JN854014.1 |
| A. venenatus | NRRL 13148 | JN854004.1 | JN854015.1 |
| A. venenatus | NRRL 13149 | JN854005.1 | JN854016.1 |
| A. sydowii | NRRL 250T | EF652274.1 | EF652362.1 |
| A. sydowii | NRRL 254 | EF652275.1 | EF652363.1 |
| A. sydowii | NRRL 4768 | EF652297.1 | EF652385.1 |
| A. sydowii | NRRL 5585 | JN853936.1 | JN854039.1 |
| A. austroafricanus | NRRL 233T | JN853963.1 | JN854025.1 |
| A. protuberus | NRRL 3505T | EF652284.1 | EF652372.1 |
| A. protuberus | NRRL 58942 | JN853956.1 | JN854061.1 |
| A. protuberus | NRRL 58748 | JN853967.1 | JN854060.1 |
| A. amoenus | NRRL35600 | JN853952.1 | JN854033.1 |
| A. amoenus | NRRL 4838T | EF652304.1 | EF652392.1 |
| A. amoenus | NRRL 226 | JN853939.1 | JN854021.1 |
| A. amoenus | NRRL 236 | JN853940.1 | JN854028.1 |
| A. tabacinus | NRRL 4791T | EF652302.1 | EF652390.1 |
| A. tabacinus | NRRL A-23173 | JN853960.1 | JN854065.1 |
| A. tabacinus | NRRL 5031 | JN853947.1 | JN854036.1 |
| A. tabacinus | LEMI968 | KJ767002 | KJ766992 |
| A. versicolor | NRRL 13145 | JN853950.1 | JN854012.1 |
| A. versicolor | NRRL 13144 | JN853949.1 | JN854011.1 |
| A. versicolor | NRRL 13146 | JN853951.1 | JN854013.1 |
| A. fructus | NRRL 241 | JN853943.1 | JN854031.1 |
| A. niger | CBS 554.65T | FJ629288.1 | FN594540.1 |
| A. niger | LEMI975 | KJ777807 | KJ777813 |
| A. thermomutatus | LEMI918 | KJ767003 | KJ766993 |
| A. ochraceus | LEMI966 | KJ767724 | KJ767730 |
| A. clavatus | LEMI40 | KJ767723 | KJ767734 |
| A. terreus | LEMI941 | KJ777806 | KJ777812 |
| A. calidoustus | LEMI749 | KJ777803 | KJ790258 |
| A. fumigatus | LEMI864 | KJ777805 | KJ777810 |
T, type strain; NT, neotype strain.
Antifungal susceptibility tests.
Antifungal susceptibility testing was performed as outlined in the Clinical and Laboratory Standards Institute (CLSI) M38-A2 protocol (20). The isolates were cultured on PDA and incubated at 25°C for 7 days to prepare the fungal inocula. Briefly, 100-μl culture preparations in RPMI 1640 (Vitrocell, Campinas, São Paulo, Brazil) were inoculated into the flat-bottom wells of 96-well microtiter plates containing 100 μl of the drug dilutions. The final inoculum concentration ranged from 0.4 × 104 to 5 × 104 CFU/ml. The drugs tested were provided by the manufacturers as pure powders and included itraconazole (ITC) (Sigma, Janssen Pharmaceutica, Beerse, Antwerp, Belgium), voriconazole (VRC) (Sigma, Pfizer, Inc., New York, NY, USA), and posaconazole (PSC) (Schering-Plough, Inc., Kenilworth, IL, USA). The MIC values were determined visually as the lowest concentrations that resulted in complete growth inhibition. Tests were performed in duplicate, and when the results did not concur, the test was repeated and the mode of the MICs was considered (11, 21).
RESULTS
Screening of Aspergillus sections.
Based on morphological characterization, 133 isolates were classified into 9 different sections: Fumigati (n = 72), Flavi (n = 37), Nigri (n = 13), Nidulantes (n = 5), Terrei (n = 2), Circumdati (n = 1), Usti (n = 1), Flavipedes (n = 1), and Clavati (n = 1).
BLAST analysis of the ITS region sequences confirmed our morphological findings and the phenotypic identification of those sections.
Identification of species by polyphasic approach.
BLAST of calmodulin and β-tubulin gene sequences together with morphological and ITS characterization provided consistent identification at the species level of 82 out of the 133 isolates tested. All of the species from Fumigati, Circumdati, Usti, Terrei, Flavipedes, and Clavati sections were identified using the polyphasic approach (morphology, thermotolerance, and sequencing) without requiring further phylogeny analyses. However, those tools did not identify 35 isolates of the Flavi section, 11 isolates from the Nigri section, and 5 isolates from the Nidulantes section.
Thermotolerance testing was useful in the discrimination of closely related species within the sections Circumdati (A. ochraceus versus A. westerdijkiae), Usti (A. ustus versus A. calidoustus), and Fumigati (A. fumigatus versus A. thermomutatus). Another interesting finding was that only the A. fumigatus strains grew at 50°C. The 51 isolates from the Flavi, Nigri, and Nidulantes sections that were not identified at the species level using the strategies outlined above underwent further phylogenetic analysis.
Phylogenetic analysis of the Flavi, Nigri, and Nidulantes sections.
The results of the partition homogeneity test on sections Flavi, Nigri, and Nidulantes showed that the sequence data sets for the two selected loci (BT2 and CAL) were congruent and could therefore be combined (P = 0.1667, 0.1667, and 0.333, respectively).
The combined gene fragments generated by BT2 and CAL sequencing from Flavi, Nigri, and Nidulantes sections were 990 bp, 1,047 bp, and 1,372 bp, respectively. Unambiguous sequences of type and neotype strains corresponding to potential species to be identified were inserted into the analysis (Table 1). The analysis also included previously identified strains to function as outgroups.
Section Flavi.
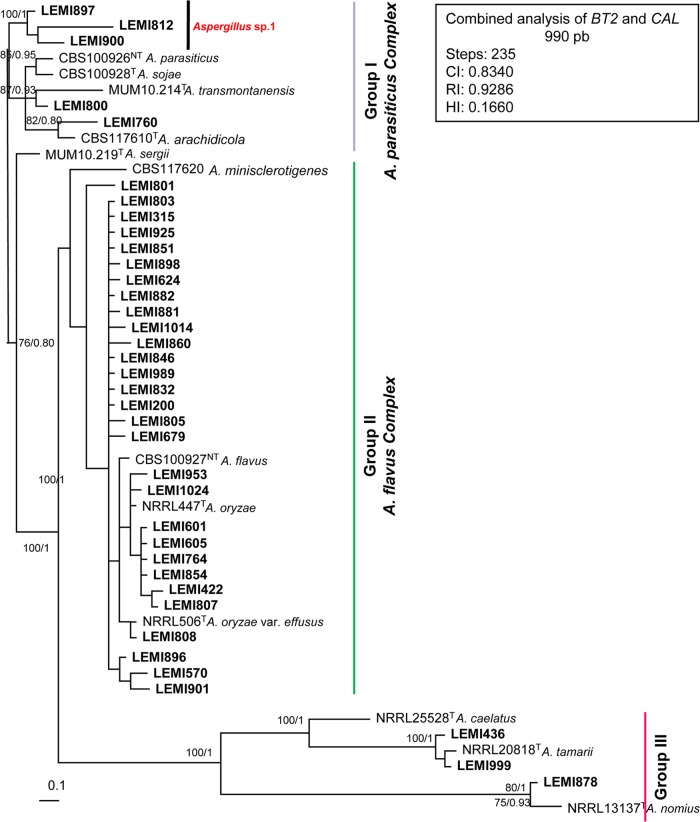
A most-parsimonious tree of 235 steps in length with a CI of 0.8340, HI of 0.1660, and RI of 0.9286 was produced from a heuristic search using the combined data set of 990 characters from two loci (Fig. 1), including 811 constant, 130 variable parsimony-informative, and 49 variable and parsimony-uninformative characters. After the analysis, we noted the presence of 3 main groups with well-supported clades. (i) Group I, denominated A. parasiticus complex, with 85% of bootstrap (bs) and Bayesian posterior probability (bpp) of 0.95 included 5 clinical isolates, type and neotype strains of A. parasiticus, A. sojae, A. transmontanensis, and A. arachidicola. One isolate (LEMI760) allocated as A. arachidicola and another isolate (LEMI800) as A. transmontanensis. Three isolates grouped apart from the reference strains and were named Aspergillus sp. 1. (ii) Group II, named A. flavus complex (100% bs and 1 bpp), contained 29 clinical isolates and type and neotype strains of A. flavus, A. oryzae, and A. oryzae var. effusus. The type strain sequences of A. minisclerotigenes and A. sergii were phylogenetically close to the A. flavus complex but were not grouped with any isolate. (iii) Group III (100% bs and 1 bpp) was subdivided into two subgroups. One subgroup included two clinical isolates and a type strain of A. tamarii. The other group included one clinical isolate and a type strain of A. nomius. Notably, A. tamarii and A. nomius were previously identified using morphological features and BLAST analysis.
FIG 1.
Aspergillus section Flavi. Most-parsimonious tree obtained from a heuristic search based on parsimony analysis of the data produced from the BT2 and CAL genes combined. The numbers at the nodes of the branches are bootstrap values/Bayesian posterior probability. Clinical isolates are designated in bold. T, type strain; NT, neotype strain. Bar, 0.1 nucleotide changes between taxa.
Section Nigri.
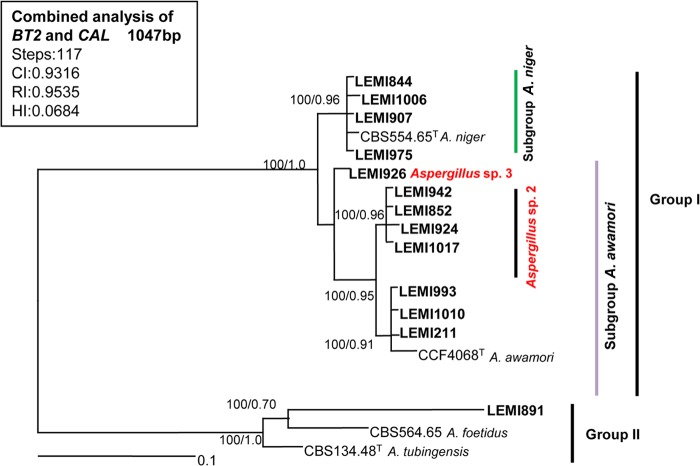
A most-parsimonious tree of 117 steps in length with a CI of 0.9316, HI of 0.0684, and RI of 0.9535 was produced from a heuristic search using the combined data set of 1,047 characters from two loci (Fig. 2), including: 943 constant, 65 variable parsimony informative, and 39 variable and parsimony uninformative. After analysis, we noted the presence of 2 main groups with well-supported clades. Group I (100% bs and 1 bpp) was subdivided into other 2 subgroups, (i) the subgroup A. niger (100% bs and 0.96 bpp), which contained 4 clinical isolates and one type strain of A. niger, and (ii) the subgroup A. awamori (100% bs and 0.95 bpp), which contained 7 clinical isolates and one type strain of A. awamori. This subgroup was also subdivided into 2 groups. One of them contained 4 clinical isolates and no reference sequences (Aspergillus sp. 2), and the other contained 3 clinical isolates and one type strain of A. awamori. The isolate LEMI926 was related to subgroup A. awamori but was not identified. We referred to this isolate as Aspergillus sp. 3. Group 2, also with 100% bs and 1 bpp, included one clinical isolate, one type strain of A. foetidus, and one separate branch with one type strain sequence of A. tubingensis.
FIG 2.
Aspergillus section Nigri. Most-parsimonious tree obtained from a heuristic search based on parsimony analysis of the data produced from the BT2 and CAL genes combined. The numbers at the nodes of the branches are bootstrap values/Bayesian posterior probability. Clinical isolates are highlighted in bold type. T, type strain; NT, neotype strain. Bar, 0.1 nucleotide changes between taxa.
Section Nidulantes.
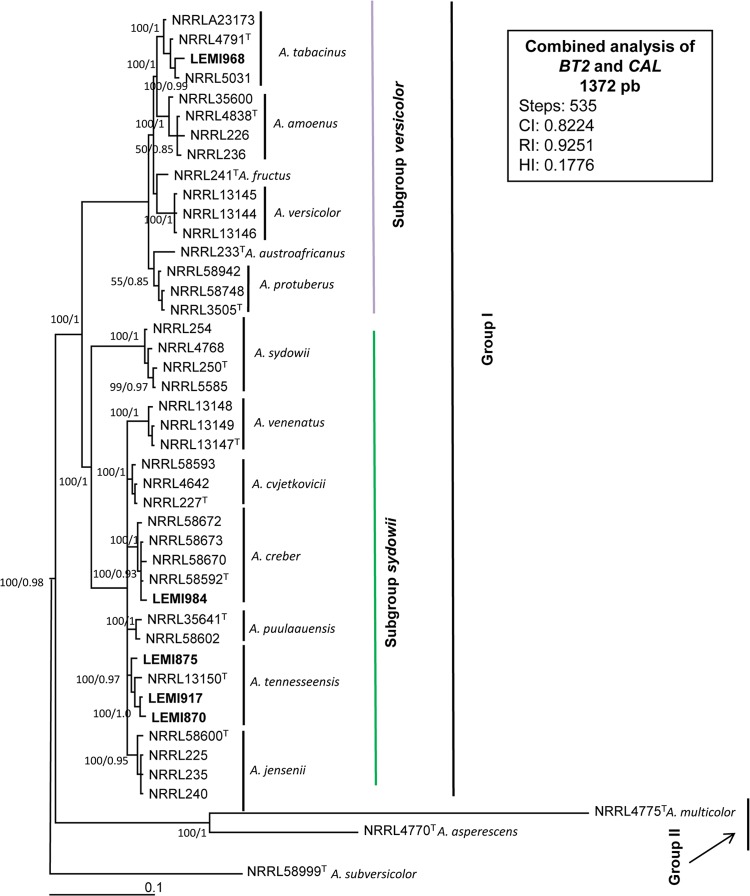
A most-parsimonious tree of 535 steps long with a CI of 0.8224, HI of 0.1776, and RI of 0.9251 was produced from a heuristic search using the combined data set of 1,372 characters from two loci (Fig. 3), including 1,009 constant, 188 variable parsimony informative, and 175 variable and parsimony uninformative. After the analysis, we noted the presence of 2 main groups. Group I was a well-supported clade (100% bs and 1 bpp) subdivided into two other subgroups, versicolor and sydowii. The subgroup versicolor (100% bs and 1 bpp) contained one clinical isolate and one type strain from each of the following species: A. tabacinus, A. fructus, A. versicolor, A. austroafricanus, and A. protuberus. The LEMI968 isolate grouped with the A. tabacinus type strain. The subgroup sydowii (100% bs and 1 bpp) contained 4 clinical isolates and 1 type strain of A. sydowii, A. venenatus, A. cvjetkovicii, A. creber, A. puulaauensis, A. tennesseensis, and A. jensenii. The clinical isolate LEMI984 clustered with the reference sequences of A. creber with 100% bs and 1 bpp. The other 3 clinical isolates (LEMI875, LEMI917, and LEMI870) grouped with reference sequences of A. tennesseensis. Group II, with 100% bp and 0.97 bpp, consisted of A. asperescens and A. multicolor sequences. The reference sequences of A. subversicolor were used as outgroups.
FIG 3.
Aspergillus section Nidulantes. Most-parsimonious tree obtained from a heuristic search based on parsimony analysis of the data produced from the BT2 and CAL genes combined. The numbers at the nodes of the branches are bootstrap values/Bayesian posterior probability. Clinical isolates are highlighted in bold type. T, type strain; NT, neotype strain. Bar, 0.1 nucleotide changes between taxa.
Final species identification and susceptibility profiles to triazoles.
According to data generated by conventional molecular tools and phylogenetic analysis, A. fumigatus was found to be the most prevalent species, with 71 (53%) isolates, followed by A. flavus with 29 (22%) isolates and A. niger with only 4 (3%) isolates. The following less frequent or rare species were also found: A. clavatus (n = 1), A. flavipes (n = 1), and A. terreus (n = 2). Among the cryptic species found in Brazilian clinical samples, the most frequent was Aspergillus sp. 2 undescribed species (4 isolates, section Flavi), followed by A. awamori (section Nigri), Aspergillus sp. 1 undescribed species (section Flavi), and A. tennesseensis (section Nidulantes), with 3 isolates each. Other species found were related to Flavi (2 A. tamarii strains, 1 A. arachidicola strain, 1 A. transmontanensis strain, and 1 A. nomius strain), Fumigati (1 A. thermomutatus strain), Circumdati (1 A. ochraceus strain), Nidulantes (1 A. tabacinus strain and 1 A. creber strain), Nigri (1 A. foetidus strain and 1 Aspergillus sp. 3 strain), and Usti (1 A. calidoustus strain). We observed that 8 isolates from our study (32% of cryptic species) were not identified by any of our 3 applied methodologies and possibly represent new Aspergillus species (Table 2).
TABLE 2.
Cryptic and rare Aspergillus species distributions, in vitro susceptibility profiles, and sources of isolates
| Section (no. of isolates) | Species identification (no. of isolates) | Source (no. of isolates) | MIC data (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|---|---|
| Itraconazole |
Voriconazole |
Posaconazole |
||||||
| Range | GMa | Range | GM | Range | GM | |||
| Clavati (1) | A. clavatus (1) | Respiratory tract biopsy specimen (1) | 1.0 | 1.0 | 1.0 | 0.5 | 0.5 | |
| Circumdati (1) | A. ochraceus (1) | Respiratory secretion (1) | 4.0 | 1.0 | 1.0 | 0.5 | 0.5 | |
| Flavi (8) | A. arachidicola (1) | Respiratory tract biopsy specimen (1) | 0.5 | 1.0 | 1.0 | 0.125 | 0.125 | |
| Aspergillus sp. 1 (3) | Respiratory secretion (2), respiratory tract biopsy specimen (1) | 0.5–1.0 | 0.63 | 0.5–1.0 | 0.63 | 0.125–0.25 | 0.16 | |
| A. tamarii (2) | Tissue biopsy specimen (1), respiratory tract biopsy specimen (1) | 0.25–0.5 | 0.35 | 0.125–0.5 | 0.25 | 0.03–0.125 | 0.06 | |
| A. transmontanensis (1) | Respiratory tract biopsy specimen (1) | 0.25 | 1.0 | 0.125 | ||||
| A. nomius (1) | Respiratory secretion (1) | 0.5 | 1.0 | 0.25 | ||||
| Flavipedes (1) | A. flavipes (1) | Tissue biopsy specimen (1) | 0.125 | 0.125 | 0.25 | |||
| Fumigati (1) | A. thermomutatus (1) | Tissue biopsy specimen (1) | 2.0 | 16.0 | 0.5 | |||
| Nidulantes (5) | A. tennesseensis (3) | Respiratory tract secretion (2), skin biopsy specimen (1) | 0.5–1.0 | 0.63 | 0.5–1.0 | 0.63 | 0.25 | 0.25 |
| A. tabacinus (1) | Respiratory secretion (1) | 1.0 | 1.0 | 0.25 | ||||
| A. creber (1) | Respiratory secretion (1) | 0.5 | 0.25 | 0.5 | ||||
| Nigri (9) | A. awamori (3) | Respiratory secretion (2), skin biopsy specimen (1) | 0.25–2.0 | 0.5 | 0.25–0.5 | 0.4 | 0.125–0.25 | 0.16 |
| Aspergillus sp. 2 (4) | Respiratory secretion (3), ear secretion (1) | 0.5–1.0 | 0.84 | 0.25–1.0 | 0.6 | 0.25–0.5 | 0.3 | |
| Aspergillus sp. 3 (1) | Respiratory secretion (1) | 1.0 | 1.0 | 0.25 | ||||
| A. foetidus (1) | Respiratory secretion (1) | 2.0 | 0.25 | 0.25 | ||||
| Terrei (2) | A. terreus (2) | Tissue biopsy specimen (1), ear secretion (1) | 0.25–0.5 | 0.35 | 0.25–1.0 | 0.5 | 0.25 | 0.25 |
| Usti (1) | A. calidoustus (1) | Tissue biopsy specimen (1) | >32 | 4.0 | 4.0 | |||
GM, geometric mean.
Of note, only three cryptic species isolates (A. calidoustus, A. thermomutatus, and A. ochraceus) presented high MIC values against at least one of the triazoles tested (Table 2). The A. calidoustus isolate presented high MIC values against all triazoles tested (MICs of 4 μg/ml against voriconazole and posaconazole and >32 μg/ml against itraconazole). The isolate of A. thermomutatus showed high MIC values against itraconazole and voriconazole, 2 and 16 μg/ml, respectively. The A. ochraceus isolate presented a high MIC value (4 μg/ml) only against itraconazole. The other isolates of Aspergillus cryptic species showed MIC values ranging from 0.03 and 2 μg/ml for all triazoles studied.
DISCUSSION
Advances in genomic and molecular tools have provided improved conditions for the classification of all microorganisms, including fungal species. Following the application of new molecular tools in taxonomic studies of the Aspergillus genus, a large number of new species have been described in the past few decades (6). This new information has driven the attention of the medical community to an increasing number of invasive fungal infections caused by rare and cryptic species of Aspergillus that in the recent past had been misidentified and most likely underestimated (6).
A variety of factors have all contributed to an increase in the number of people at risk of developing fungal infections, including by agents that had never been described as human pathogens. These factors include an aging population, large numbers of patients with degenerative and neoplastic diseases, patients who have had solid organ and hematopoietic stem cell transplantations, and patients under immunosuppressive therapy (22, 23).
The most common species implicated in IA is Aspergillus fumigatus. Other Aspergillus species, including A. flavus, A. niger, A. terreus, and A. ustus, have also been reported as pathogens. Aspergillus flavus has been described as the second most common Aspergillus species in several medical centers from Europe and the United States, whereas A. terreus is particularly frequent in Austria (6th Trends in Medical Mycology Workshop, w 10.1) (24, 25). In countries such as Saudi Arabia, Sudan, and Taiwan, with semi-arid, arid dry, and tropical weather conditions, A. flavus appears to be the main etiological agent of invasive aspergillosis (26, 27).
In the study samples described here, A. flavus was the second most common species, responsible for 22% of all clinical isolates. The prevalence of A. flavus in our collection is substantially higher than the rates demonstrated in recent multicenter studies from the United States and Spain (9 to 10%) (28, 29). Indeed, this observed prevalence is also considerably higher than the 10% rates of A. flavus found in a worldwide collection represented by 771 Aspergillus clinical samples obtained from 62 medical centers (30).
In addition to the different species we found within the section Flavi, we characterized a large spectrum of cryptic species related to sections Circumdati, Fumigati, Nigri, Usti, and Nidulantes. Overall, cryptic species of Aspergillus represented 19% of our collection. This number agrees closely with the rate recently found by Alastruey-Izquierdo et al. (29) by testing 325 isolates in filamentous fungi. The most frequently identified cryptic species being reported in both Europe and the United States are Aspergillus alliaceus (section Flavi), Aspergillus tubingensis (section Nigri), Aspergillus calidoustus (section Usti), and Aspergillus lentulus (section Fumigati) (28, 29, 31). In Brazil, unlike in Europe and the United States, A. tamarii and A. awamori were the two cryptic species most commonly found within the sections Flavi and Nigri, respectively.
Of note, some cryptic species described in our study had never been reported in human hosts such as A. arachidicola, A. tabacinus, A. tennesseensis, A. creber, and A. transmontanensis. As illustrated in Table 2, all of these strains were related to episodes of colonization and/or infection of the respiratory tract. In addition, we also found 5 other cryptic species already described in the literature as agents of human infections (A. ochraceus, A. tamarii, A. nomius, A. thermomutatus, and A. foetidus [3, 11, 32]).
In terms of antifungal susceptibility, it is notable that almost 90% of all cryptic species exhibited susceptibility to the 3 triazoles tested. The exceptions were strains representative of A. calidoustus, A. ochraceus, and A. thermomutatus that were less susceptible in vitro against at least one of the three triazoles tested. The reduced in vitro susceptibility to triazoles of A. thermomutatus and A. calidoustus has also been demonstrated in other studies (10, 33).
We conclude that there is a great diversity of species belonging to the Aspergillus genus causing human colonization and/or infections in Brazil, with a higher occurrence of Aspergillus section Flavi compared to U.S. and European medical centers. We emphasized the importance of accurate identification and in vitro susceptibility tests of clinical Aspergillus species. This will allow the generation of consistent data about potential peculiarities of infections caused by rare and cryptic species with regard to reservoirs, natural history, and clinical response to antifungal drugs.
ACKNOWLEDGMENTS
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (grant 2012/01134-8). C.E.N. received a Master's degree fellowship from FAPESP (2012/01548-7). S.S.G. received a postdoctoral fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil (PNPD 23038.007393/2011-11). H.X. received a doctoral fellowship from FAPESP (2012/19103-1).
We declare no conflicts of interest.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Segal BH, Steinbach WJ, Stevens DA, van Burik JA, Wingard JR, Patterson TF, Infectious Diseases Society of America 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46:327–360. 10.1086/525258 [DOI] [PubMed] [Google Scholar]
- 2.Barnes PD, Marr KA. 2006. Aspergillosis: spectrum of disease, diagnosis, and treatment. Infect. Dis. Clin. North Am. 20:545–561, vi [DOI] [PubMed] [Google Scholar]
- 3.Buzina W. 2013. Aspergillus—classification and antifungal susceptibilities. Curr. Pharm. Des. 19:3615–3628. 10.2174/1381612811319200005 [DOI] [PubMed] [Google Scholar]
- 4.De Hoog GS, Guarro J, Figueras MJ. 2011. Atlas of clinical fungi. CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands: http://www.clinicalfungi.org [Google Scholar]
- 5.Balajee SA, Marr KA. 2006. Phenotypic and genotypic identification of human-pathogenic aspergilli. Future Microbiol. 1:435–445. 10.2217/17460913.1.4.435 [DOI] [PubMed] [Google Scholar]
- 6.Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. 2012. Current section and species complex concepts in Aspergillus: recommendations for routine daily practice. Ann. N. Y. Acad. Sci. 1273:18–24. 10.1111/j.1749-6632.2012.06822.x [DOI] [PubMed] [Google Scholar]
- 7.Samson RA, Varga J, Witiak SM, Geiser DM. 2007. The species concept in Aspergillus: recommendations of an international panel. Stud. Mycol. 59:71–73. 10.3114/sim.2007.59.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonçalves SS, Cano JF, Stchigel AM, Melo AS, Godoy-Martinez PC, Correa B, Guarro J. 2012. Molecular phylogeny and phenotypic variability of clinical and environmental strains of Aspergillus flavus. Fungal Biol. 116:1146–1155. 10.1016/j.funbio.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 9.Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226. 10.3852/mycologia.100.2.205 [DOI] [PubMed] [Google Scholar]
- 10.Van Der Linden JW, Warris A, Verweij PE. 2011. Aspergillus species intrinsically resistant to antifungal agents. Med. Mycol. 49(Suppl 1):S82–S89. 10.3109/13693786.2010.499916 [DOI] [PubMed] [Google Scholar]
- 11.Gonçalves SS, Stchigel AM, Cano J, Guarro J, Colombo AL. 2013. In vitro antifungal susceptibility of clinically relevant species belonging to Aspergillus section Flavi. Antimicrob. Agents Chemother. 57:1944–1947. 10.1128/AAC.01902-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samson RA, Hoekstra ES, Frisvad JC. (ed). 2004. Introduction to food and airborne fungi, 7th ed. Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands [Google Scholar]
- 13.Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swofford DL. 2002. PAUP. Phylogenetic analysis using parsimony (and other methods), version 4.0b 10 (Alvitec). Sinauer Associates, Sunderland, MA [Google Scholar]
- 15.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 16.Baum DA, Shaw KL. 1995. Genealogical perspectives on the species problem, p 289–303 In Hoch PC, Stephenson AG. (ed), Experimental and molecular approaches to plant biosystematics. Missouri Botanical Garden, St Louis, MO [Google Scholar]
- 17.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- 18.Jurjevic Z, Peterson SW, Horn BW. 2012. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 3:59–79. 10.5598/imafungus.2012.03.01.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dettman JR, Jacobson DJ, Taylor JW. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703–2720. 10.1554/03-073 [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed. CLSI document M38-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 21.Pfaller M, Boyken L, Hollis R, Kroeger J, Messer S, Tendolkar S, Diekema D. 2011. Comparison of the broth microdilution methods of the European Committee on Antimicrobial Susceptibility Testing and the Clinical and Laboratory Standards Institute for testing itraconazole, posaconazole, and voriconazole against Aspergillus isolates. J. Clin. Microbiol. 49:1110–1112. 10.1128/JCM.02432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayr A, Lass-Florl C. 2011. Epidemiology and antifungal resistance in invasive aspergillosis according to primary disease: review of the literature. Eur. J. Med. Res. 16:153–157. 10.1186/2047-783X-16-4-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meersseman W, Lagrou K, Maertens J, Van Wijngaerden E. 2007. Invasive aspergillosis in the intensive care unit. Clin. Infect. Dis. 45:205–216. 10.1086/518852 [DOI] [PubMed] [Google Scholar]
- 24.Mortensen KL, Mellado E, Lass-Flör C, Rodriguez-Tudela JL, Johansen HK, Arendrup MC. 2010. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrob. Agents Chemother. 54:4545–4549. 10.1128/AAC.00692-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lass-Flör C, Griff K, Mayr A, Petzer A, Gastl G, Bonatti H, Freund M, Kropshofer G, Dierich MP, Nachbaur D. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br. J. Haematol. 131:201–207. 10.1111/j.1365-2141.2005.05763.x [DOI] [PubMed] [Google Scholar]
- 26.Kameswaran M, al-Wadei A, Khurana P, Okafor BC. 1992. Rhinocerebral aspergillosis. J. Laryngol Otol. 106:981–985. 10.1017/S0022215100121528 [DOI] [PubMed] [Google Scholar]
- 27.Hsueh PR, Lau YJ, Chuang YC, Wan JH, Huang WK, Shyr JM, Yan JJ, Yu KW, Wu JJ, Ko WC, Yang YC, Liu YC, Teng LJ, Liu CY, Luh KT. 2005. Antifungal susceptibilities of clinical isolates of Candida species, Cryptococcus neoformans, and Aspergillus species from Taiwan: surveillance of multicenter antimicrobial resistance in Taiwan program data from 2003. Antimicrob. Agents Chemother. 49:512–517. 10.1128/AAC.49.2.512-517.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balajee SA, Kano R, Baddley JW, Moser SA, Marr KA, Alexander BD, Andes D, Kontoyiannis DP, Perrone G, Peterson S, Brandt ME, Pappas PG, Chiller T. 2009. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. J. Clin. Microbiol. 47:3138–3141. 10.1128/JCM.01070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alastruey-Izquierdo A, Mellado E, Pelaez T, Pemán J, Zapico S, Alvarez M, Rodríguez-Tudela JL, Cuenca-Estrella M; Study Group FILPOP. 2013. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob. Agents Chemother. 57:3380–3387. 10.1128/AAC.00383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller MA, Messer SA, Boyken L, Rice C, Tendolkar S, Hollis RJ, Diekema DJ. 2008. In vitro survey of triazole cross-resistance among more than 700 clinical isolates of Aspergillus species. J. Clin. Microbiol. 46:2568–2572. 10.1128/JCM.00535-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balajee SA, Houbraken J, Verweij PE, Hong SB, Yaghuchi T, Varga J, Samson RA. 2007. Aspergillus species identification in the clinical setting. Stud Mycol. 59:39–46. 10.3114/sim.2007.59.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2009. Species identification and antifungal susceptibility patterns of species belonging to Aspergillus section Nigri. Antimicrob. Agents Chemother. 53:4514–4517. 10.1128/AAC.00585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varga J, Houbraken J, Van Der Lee HA, Verweij PE, Samson RA. 2008. Aspergillus calidoustus sp. nov., causative agent of human infections previously assigned to Aspergillus ustus. Eukaryot. Cell 7:630–638. 10.1128/EC.00425-07 [DOI] [PMC free article] [PubMed] [Google Scholar]