LETTER
Carbapenem resistance in Acinetobacter baumannii is typically mediated by the acquisition of class D β-lactamases belonging to gene clusters of blaOXA-23-like, blaOXA-40-like, and blaOXA-58-like (1). In the Enterobacteriaceae, carbapenem-hydrolyzing class D β-lactamases are encoded by a dissimilar set of genes, the blaOXA-48-like genes (2). The presence of blaOXA-23 in Enterobacteriaceae is exceptional and has been described once in Proteus mirabilis, where it was chromosomally encoded (3).
As a result of national surveillance efforts to characterize non-carbapenem-susceptible Enterobacteriaceae isolates, we detected a case of blaOXA-23 in Escherichia coli, which to the best of our knowledge is the first description of plasmid-borne blaOXA-23 in E. coli.
The isolate was recovered from the urine sample of an elderly woman. However, the isolate was not likely to be a cause of infection, as urine microscopy did not show an increased number of white blood cells. The isolate exhibited resistance to β-lactams, fluoroquinolones, and chloramphenicol. Sensitivity to tigecycline, colistin, and aminoglycosides was observed (Table 1).
TABLE 1.
MICs of antibiotics for the Escherichia coli isolate harboring blaOXA-23
| Antibiotic | MIC (mg/liter) |
|---|---|
| Aztreonam | 32 |
| Piperacillin-tazobactam | >256 |
| Cefoxitin | 128 |
| Cefotaxime | 32 |
| Ceftazidime | 16 |
| Imipenem | 3 |
| Meropenem | 4 |
| Ertapenem | 6 |
| Tigecycline | 0.125 |
| Tetracycline | 86 |
| Levofloxacin | >32 |
| Ciprofloxacin | >32 |
| Amikacin | 0.125 |
| Gentamicin | 2 |
| Chloramphenicol | 16 |
| Colistin | 0.125 |
Comprehensive PCR screening and sequencing for β-lactamase genes was performed (4, 5). The isolate carried blaOXA-23, which had 100% identity with A. baumannii blaOXA-23. It was also positive for TEM-1, OXA-1, and CMY-2 but was negative for other carbapenemases and extended spectrum β-lactamases. The isolate had a multilocus sequence type of ST471. (http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html).
Two large plasmids of approximately 50 and 100 kb were detected (6), with Southern blot hybridization localizing blaOXA-23 to the 50-kb plasmid.
Solid-medium conjugation assays were performed to assess the transferability of blaOXA-23 from the clinical isolate to the azide-resistant recipient E. coli J53 and to a spontaneous rifampin-resistant mutant of A. baumannii, ATCC 17978. However, we did not manage to isolate any transconjugants, suggesting that blaOXA-23 was harbored on a non-self-conjugative plasmid.
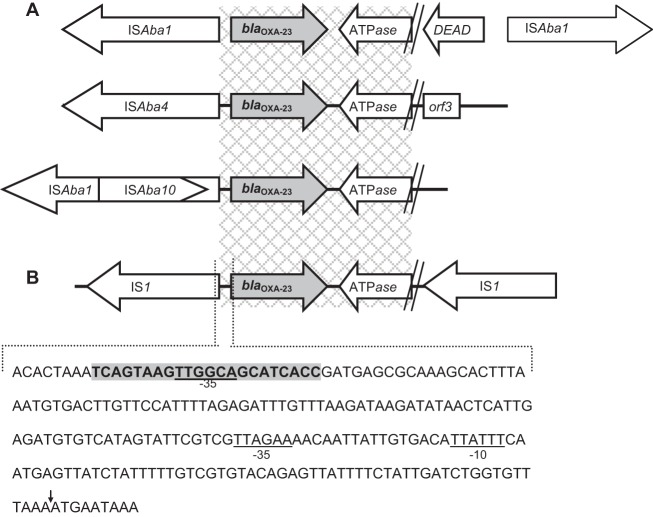
The immediate nucleotide sequences flanking blaOXA-23 were determined by inverse PCR and the primer walking approach (7). E. coli blaOXA-23 is flanked by two copies of insertion sequence IS1 (Fig. 1). In contrast, in A. baumannii, blaOXA-23 is found as part of a transposon (Tn2006, Tn2007, Tn2008) associated with insertion sequence ISAba1 or ISAba4 (8, 9) (Fig. 1).
FIG 1.
Schematic representation of the genetic organization of blaOXA-23. (A) In Acinetobacter baumannii, ISAba insertion sequence elements are typically found immediately upstream of blaOXA-23 (8, 9). (B) In the clinical Escherichia coli isolate, blaOXA-23 is flanked by two copies of Enterobacteriaceae insertion sequence IS1. The terminal left inverted repeat (IRL) of IS1 is indicated by the boldface nucleotides with gray shading. The putative −35 promoter of the IRL is underlined and may form hybrid promoters with existing −10 promoters, enabling transcription (11). The ISAba1-associated −35 and −10 promoters are underlined (10). The ATG start codon of OXA-23 is indicated by the vertical arrow (↓). Gray checked regions represent 100% identity between E. coli and A. baumannii. ATPase, gene encoding putative AAA ATPase; DEAD, gene encoding the putative DEAD (Asp-Glu-Ala-Asp) helicase.
One hundred eighty-three bases upstream, the E. coli blaOXA-23 start codon was completely identical to ISAba1 (GenBank accession number GQ861438.1). Within this region, −35 and −10 ISAba1-associated promoters were identified and likely to be functional (10) (Fig. 1). The 764-bp stretch after the blaOXA-23 stop codon comprised a partially truncated putative AAA ATPase identical to that in A. baumannii Tn2006, Tn2007, and Tn2008 (Fig. 1).
We hypothesized that E. coli blaOXA-23 may be carried within a transposon, not unlike those identified in A. baumannii but interrupted by IS1. However, attempts to PCR map the region using both ISAba1 and ISAba4 as reference sequences did not yield amplicons, suggesting that it was in a genetic configuration different from that found in A. baumannii.
In summary, we describe the novel detection of an E. coli isolate carrying an Acinetobacter blaOXA-23 gene associated with Enterobacteriaceae IS1 on a 50-kb non-self-conjugative plasmid. The discovery highlights the potential for the tremendous spread of carbapenemases.
Nucleotide sequence accession number.
The blaOXA-23 sequence from the clinical E. coli isolate has been deposited into GenBank under accession number KJ716226.
ACKNOWLEDGMENTS
This work was supported by a National University Health System Bridging Fund grant (SBRO13/NS04G) provided to Jeanette W. P. Teo.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 16 July 2014
REFERENCES
- 1.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 3.Bonnet R, Marchandin H, Chanal C, Sirot D, Labia R, De Champs C, Jumas-Bilak E, Sirot J. 2002. Chromosome-encoded class D beta-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004–2006. 10.1128/AAC.46.6.2004-2006.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac. Surveil. Response J. 3:19–24. 10.5365/wpsar.year.2011.2.4.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balm MN, Ngan G, Jureen R, Lin RT, Teo JW. 2013. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect. Dis. 13:58–62. 10.1186/1471-2334-13-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- 7.Jong AY, T'ang A, Liu DP, Huang SH. 2002. Inverse PCR. Genomic DNA cloning. Methods Mol. Biol. 192:301–317. 10.1385/1-59259-177-9:301 [DOI] [PubMed] [Google Scholar]
- 8.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40. 10.3201/eid1601.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim CK, Lee H, Jeong SH, Yong D, Lee K. 2011. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the bla(OXA-23) gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:361–363. 10.1128/AAC.01672-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal H, Jacobson RK, Garny S, Bamford CM, Elisha BG. 2007. Extended −10 promoter in ISAba-1 upstream of blaOXA-23 from Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3040–3041. 10.1128/AAC.00594-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prentki P, Teter B, Chandler M, Galas DJ. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 5:383–393 [DOI] [PubMed] [Google Scholar]