Abstract
The detection of pathogens associated with gastrointestinal disease may be important in certain patient populations, such as immunocompromised hosts, the critically ill, or individuals with prolonged disease that is refractory to treatment. In this study, we evaluated two commercially available multiplex panels (the FilmArray gastrointestinal [GI] panel [BioFire Diagnostics, Salt Lake City, UT] and the Luminex xTag gastrointestinal pathogen panel [GPP] [Luminex Corporation, Toronto, Canada]) using Cary-Blair stool samples (n = 500) submitted to our laboratory for routine GI testing (e.g., culture, antigen testing, microscopy, and individual real-time PCR). At the time of this study, the prototype (non-FDA-cleared) FilmArray GI panel targeted 23 pathogens (14 bacterial, 5 viral, and 4 parasitic), and testing of 200 μl of Cary-Blair stool was recommended. In contrast, the Luminex GPP assay was FDA cleared for the detection of 11 pathogens (7 bacterial, 2 viral, and 2 parasitic), but had the capacity to identify 4 additional pathogens using a research-use-only protocol. Importantly, the Luminex assay was FDA cleared for 100 μl raw stool; however, 100 μl Cary-Blair stool was tested by the Luminex assay in this study. Among 230 prospectively collected samples, routine testing was positive for one or more GI pathogens in 19 (8.3%) samples, compared to 76 (33.0%) by the FilmArray and 69 (30.3%) by the Luminex assay. Clostridium difficile (12.6 to 13.9% prevalence) and norovirus genogroup I (GI)/GII (5.7 to 13.9% prevalence) were two of the pathogens most commonly detected by both assays among prospective samples. Sapovirus was also commonly detected (5.7% positive rate) by the FilmArray assay. Among 270 additional previously characterized samples, both multiplex panels demonstrated high sensitivity (>90%) for the majority of targets, with the exception of several pathogens, notably Aeromonas sp. (23.8%) by FilmArray and Yersinia enterocolitica (48.1%) by the Luminex assay. Interestingly, the FilmArray and Luminex panels identified mixed infections in 21.1% and 13.0% of positive prospective samples, respectively, compared to only 8.3% by routine methods.
INTRODUCTION
Infectious diarrhea affects millions of people each year and can cause significant morbidity and mortality (1, 2). In the United States, certain risk groups have been associated with deaths due to diarrhea and are more likely to be tested (3–5). Rapid and accurate detection of gastrointestinal (GI) pathogens is vitally important so that appropriate therapy can be initiated and proper infection control and epidemiologic measures can be taken to help curb the spread of disease (6–8). Conventional identification techniques, such as culture, microscopy, and antigen detection as well as individual real-time PCR assays, are often laborious and time-consuming, and they typically test for a limited number of pathogens (9). Furthermore, health care providers are required to select the appropriate tests despite the fact that diseases caused by many GI pathogens are clinically indistinguishable (4). Due to the limitations of conventional methods, there has been interest in the development of multiplex molecular assays for the detection and identification of infectious diseases. In this study, we evaluated and compared two commercial multiplex panels for the detection of GI pathogens from clinical stool samples, the FilmArray GI panel (BioFire, Inc., Salt Lake City, UT) and the Luminex xTag GI pathogen panel (GPP) (Luminex Corporation, Toronto, Canada). At the time of this study, the prototype (non-FDA-cleared) FilmArray GI panel included 23 targets (14 bacterial, 5 viral, and 4 parasitic) within a single pouch. The Luminex xTag GPP assay is FDA cleared and tests for 11 GI pathogens (7 bacterial, 2 viral, and 2 parasitic). Four additional pathogens (2 bacterial, 1 viral, and 1 parasitic) are also available on the Luminex panel under research-use-only (RUO) status, and these targets were included in this evaluation. We set out to compare the performance characteristics of these multiplex tests to those of routine assays commonly used in the diagnosis of GI disease.
MATERIALS AND METHODS
Study design.
Stool specimens (n = 500) in Cary-Blair medium were submitted to our laboratory for routine testing (e.g., culture, microscopy, antigen testing, and/or individual real-time PCR) and were analyzed in a blinded fashion by the FilmArray GI panel and the Luminex xTag GPP (Table 1). Prospective stool samples (n = 230) were received at ambient temperature, stored at 4°C, and tested by the multiplex tests within 5 days. Characterized stool samples (n = 270) with known results for analytes represented on the multiplex panels were stored at either −20°C (samples collected at the Mayo Clinic) or 4°C (samples collected at the Minnesota Department of Health) prior to analysis by the multiplex assays.
TABLE 1.
Routine methods for detection of gastrointestinal pathogens and targets represented on two commercial multiplex assays
| Targeta | Method(s) used for routine testing and discordant analysis | Target included on multiplex panelb |
|
|---|---|---|---|
| FilmArray | Luminex | ||
| Aeromonas | Culture | IUO | |
| Campylobacter | Culture, real-time PCR | ✓ | ✓ |
| Clostridium difficile toxin A/B tcdC gene | Real-time PCR | ✓ | ✓ |
| Plesiomonas shigelloides | Culture | ✓ | |
| Salmonella | Culture, real-time PCR | ✓ | ✓ |
| Yersinia enterocolitica | Culture, real-time PCR | ✓ | RUO |
| Vibrio spp. | Culture | ✓ | RUO |
| EAEC | NDc | ✓ | |
| EPEC | ND | ✓ | |
| ETEC | ND | ✓ | ✓ |
| STEC | Real-time PCR | ✓ | ✓ |
| E. coli 0157 | Culture, real-time PCR | ✓ | ✓ |
| EIECd/Shigella | Culture, real-time PCR | ✓ | ✓ |
| Cryptosporidium | Antigen, microscopy | ✓ | ✓ |
| Cyclospora cayetanensis | Microscopy | ✓ | |
| Entamoeba histolytica | Microscopy | ✓ | RUO |
| Giardia lamblia | Antigen | ✓ | ✓ |
| Adenovirus 40/41e | Real-time PCR | ✓ | RUO |
| Norovirus GI/GIIe | ND | ✓ | ✓ |
| Rotavirus Ae | Antigen | ✓ | ✓ |
| Sapoviruse | ND | ✓ | |
| Astroviruse | ND | ✓ | |
EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli (lt/st); STEC, Shiga-like toxin-producing E. coli (stx1/stx2); EIEC, enteroinvasive E. coli.
IUO, investigational use only; RUO, research use only.
ND, not done.
The Luminex panel does not specifically target EIEC; however, the Luminex Shigella assay does cross-react with EIEC.
These targets were also tested at the Minnesota Department of Health by individual real-time PCR tests.
Reference standard.
A reference standard positive result was defined as (i) an organism identified by culture or microscopy or (ii) a “consensus positive,” in which an organism was detected by at least two tests (e.g., multiplex PCR and antigen testing). The reference standard for a negative result was defined as a negative result by at least two of three assays. Whenever possible, samples showing a discordant result(s) were tested by an additional method. In some cases, we were unable to arbitrate discordant test results due to (i) the lack of an additional test method or (ii) insufficient remaining specimen volume. These exceptions are noted in the text, and samples with discordant results that were not arbitrated by additional testing were not included in the calculation of performance characteristics.
Routine methods.
Based on the order placed by the health care provider, one or more of the following in-house, routine tests were performed, (i) bacterial culture (Salmonella, Shigella, Campylobacter, Yersinia, Plesiomonas shigelloides, Aeromonas, or Vibrio species) using standard methods; (ii) real-time PCR for Shiga toxin (stx1/stx2)-producing Escherichia coli, Campylobacter jejuni/coli, Yersinia enterocolitica, Salmonella sp., and Shigella sp. as previously described (10); (iii) Clostridium difficile toxin A/B (tcdC) lab-developed real-time PCR (11); (iv) adenovirus real-time PCR (12); (v) viral culture using standard methods and inoculation of medical research council 5 (MRC-5) and rhesus monkey kidney (RMK) cells (Diagnostic Hybrids, Athens, OH); (vi) ova and parasite (O&P) examination that included a concentrated wet preparation as well as permanent stains; (vii) a modified safranin stain for Cyclospora (13); (viii) Giardia and/or Cryptosporidium ProSpecT antigen enzyme immunoassay (EIA) (Remel/Thermo Fisher Scientific, Lenexa, KS) according to the manufacturer's instructions; and (ix) rotavirus Premier Rotaclone antigen EIA (Meridian Bioscience, Cincinnati, OH) as recommended by the manufacturer (Table 1).
Multiplex analysis.
Testing on the FilmArray platform was performed according to the manufacturer's instructions using 200 μl of Cary-Blair stool, which was the volume of sample recommended by the manufacturer. Testing by the Luminex assay was performed on the MagPix platform (Luminex) with minor modifications to the manufacturer's FDA-cleared protocol. In brief, 100 μl of Cary-Blair stool (instead of 100 μl raw stool, as recommended by the manufacturer) and 10 μl of xTAG MS2 internal control (Luminex) were placed in a Bertin SK28 tube containing silica beads and 1 ml of lysis buffer (Luminex). The tube was vortexed for 5 min. The sample was placed at room temperature for 10 to 15 min, and then centrifuged at 14,000 rpm for 2 min. Subsequently, 200 μl of the supernatant was extracted on a MagNA Pure LC (Roche Diagnostics, Indianapolis, IN), instead of the EasyMag (bioMérieux) as recommended by the manufacturer. Next, 10 μl of nucleic acid extract was combined with 15 μl PCR mastermix (Luminex) in a 96-well plate. Amplification was performed on the Veriti cycler (Life Technologies, Grand Island, NY) using the following cycling conditions: 20 min at 53°C for 1 cycle; 15 min at 95°C for 1 cycle; 30 s at 95°C, 30 s at 58°C, and 30 s at 72°C for 38 cycles; 2 min at 72°C for 1 cycle; and a 4°C hold. Of the amplified material, 5 μl was then transferred into the well of a 96-well plate containing 20 μl of fluorescently labeled beads (Luminex) and allowed to hybridize at 60°C for 3 min, followed by 45°C for 45 min in an Eppendorf thermocycler (Eppendorf, Hauppauge, NY). Following the hybridization step, each well was analyzed on the MagPix instrument using the supplied RUO software version 1.2 (Luminex). Samples tested by the Luminex assay were processed in batches of 25 to 30.
Discordant analysis.
Samples showing discordant results between FilmArray, Luminex, and/or routine methods were further analyzed by an additional assay, where available (Table 1). Samples showing discordant results for norovirus genogroup I (GI)/GII, adenovirus F 40/41, or rotavirus A, as well as additional detections by the FilmArray GI panel for sapovirus and astrovirus, were tested by real-time PCR at the Minnesota Department of Health (MDH) as previously described (14–18). In some cases, we were unable to complete discordant analysis due to inadequate remaining sample volume or the lack of a confirmatory test.
Statistics and data analysis.
Percent sensitivity, specificity, and 95% confidence intervals were calculated using GraphPad QuickCalcs. The 95% confidence intervals were calculated using the modified Wald method.
RESULTS
Prospective study.
Testing by the FilmArray was completed using 200 μl of Cary-Blair stool (as recommended by the manufacturer), while analysis by the Luminex panel was performed using 100 μl of Cary-Blair stool (instead of 100 μl raw stool as recommended in the FDA-cleared product insert). Among 230 prospective stool samples, routine testing was positive for one or more GI pathogens in 19 (8.3%) samples, compared to 76 (33.0%) by the FilmArray and 69 (30.0%) by the Luminex assay (Fig. 1A and 1B). Following discordant analysis, the number of samples with a confirmed positive result(s) by the FilmArray and Luminex assays was 65 (28.3%) and 46 (20.0%), respectively. Upon exclusion of false-positive results, Clostridium difficile and norovirus were two of the most commonly detected pathogens among prospective samples by both FilmArray (C. difficile, 24/230 [10.4%]; norovirus GI/GII, 11/230 [4.8%]) and Luminex (C. difficile, 23/230) [10.0%]; norovirus GI/GI, 12/230 [5.2%]). Sapovirus was also detected in 13/230 (5.6%) samples tested by the FilmArray panel and these results were confirmed by a second molecular assay performed at the Minnesota Department of Health. The percent specificity of all targets on the multiplex panels was high (>96%), with the exception of the Luminex norovirus assay, which demonstrated a specificity of 90.8% (198/218) (Table 2).
FIG 1.
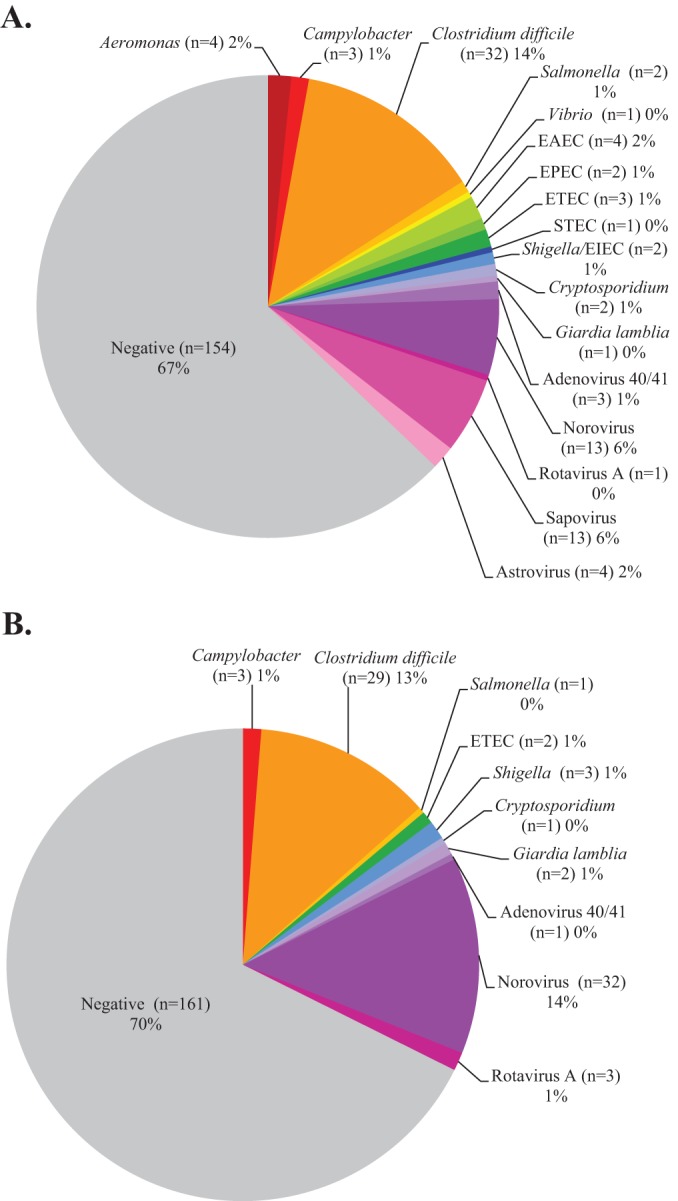
Distribution of pathogens detected by FilmArray (A) and Luminex (B) multiplex assays using prospective clinical specimens (n = 230).
TABLE 2.
Comparison of two commercial multiplex panels for routine tests using prospective stool samples (n = 230)a
| Targetb | FilmArrayc |
Luminex |
FilmArrayd |
Luminexe |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TN | TP | FP | FN | TN | TP | FP | FN | Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | |
| Aeromonas spp. | 84 | 1 | 0 | 0 | 100 (16.7, 100) | 100 (94.8, 100) | ||||||
| Campylobacter spp. | 227 | 3 | 0 | 0 | 227 | 3 | 0 | 0 | 100 (38.3, 100) | 100 (98, 100) | 100 (38.3, 100) | 100 (98, 100) |
| Clostridium difficile toxin A/B | 198 | 24 | 7 | 0 | 200 | 23 | 6 | 1 | 100 (83.7, 100) | 96.6 (93.0, 98.5) | 95.8 (78.1, 99.9) | 97.2 (93.8, 98.8) |
| Plesiomonas shigelloides | 88 | 0 | 0 | 0 | ND | 100 (95, 100) | ||||||
| Salmonella spp. | 228 | 1 | 1 | 0 | 229 | 1 | 0 | 0 | 100 (16.7, 100) | 99.6 (97.3, 99.9) | 100 (16.7, 100) | 100 (98, 100) |
| Yersinia enterocolitica | 230 | 0 | 0 | 0 | 230 | 0 | 0 | 0 | ND | 100 (98, 100) | ND | 100 (98, 100) |
| Vibrio spp. | 5 | 0 | 0 | 0 | ND | 100 (51.1, 100) | ||||||
| Vibrio cholerae | 230 | 0 | 0 | 0 | 230 | 0 | 0 | 0 | ND | 100 (98, 100) | ND | 100 (98, 100) |
| EAEC | 0 | 0 | 0 | 0 | ND | ND | ||||||
| EPEC | 0 | 0 | 0 | 0 | ND | ND | ||||||
| ETEC | 227 | 2 | 0 | 0 | 227 | 2 | 0 | 0 | 100 (38.3, 100) | 100 (98, 100) | 100 (29, 100) | 100 (98, 100) |
| STEC | 229 | 0 | 1 | 0 | 230 | 0 | 0 | 0 | ND | 99.6 (97.3, 100) | ND | 100 (98, 100) |
| E. coli 0157 | 230 | 0 | 0 | 0 | 230 | 0 | 0 | 0 | ND | 100 (98, 100) | ND | 100 (98, 100) |
| Shigella/EIEC | 228 | 2 | 0 | 0 | 227 | 2 | 1 | 0 | 100 (29, 100) | 100 (98, 100) | 100 (29, 100) | 99.6 (97.3, 99.9) |
| Cryptosporidium spp. | 228 | 1 | 0 | 0 | 229 | 1 | 0 | 0 | 100 (16.7, 100) | 100 (98, 100) | 100 (16.7, 100) | 100 (98, 100) |
| Cyclospora cayetanensis | 15 | 0 | 0 | 0 | ND | 100 (76.1, 100) | ||||||
| Entamoeba histolytica | 230 | 0 | 0 | 0 | 230 | 0 | 0 | 0 | ND | 100 (98, 100) | ND | 100 (98, 100) |
| Giardia lamblia | 229 | 1 | 0 | 0 | 228 | 1 | 0 | 0 | 100 (16.7, 100) | 100 (98, 100) | 100 (16.7, 100) | 100 (98, 100) |
| Adenovirus 40/41 | 227 | 1 | 1 | 0 | 229 | 1 | 0 | 0 | 100 (16.7, 100) | 99.6 (97.3, 100) | 100 (16.7, 100) | 100 (98, 100) |
| Norovirus GI/GII | 216 | 11 | 1 | 1 | 198 | 12 | 20 | 0 | 91.7 (62.5, 100) | 99.5 (97.2, 100) | 100 (71.8, 100) | 90.8 (86.2, 94.0) |
| Rotavirus A | 229 | 0 | 1 | 0 | 227 | 0 | 2 | 0 | ND | 99.6 (97.3, 100) | ND | 99.1 (96.7, 100) |
| Sapovirus | 0 | 13 | 0 | 0 | 100 (73.4, 100) | ND | ||||||
| Astrovirus | 0 | 4 | 0 | 0 | 100 | ND | ||||||
The reference standard for a positive result was defined as an organism detected by (i) bacterial culture or microscopy or (ii) ≥2 methods. Empty cells, not present on the panel.
EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli (lt/st); STEC, Shiga-like toxin-producing E. coli (stx1/stx2; E. coli 0157); EIEC, enteroinvasive E. coli.
TN, true negative; TP, true positive; FP, false positive; FN, false negative.
FilmArray yielded additional detections for Aeromonas (n = 3), Vibrio spp. (n = 1), EAEC (n = 4), EPEC (n = 2), Cryptosporidium (n = 1), C. difficile (n = 1), adenovirus (n = 1), norovirus (n = 1), and rotavirus (n = 1) that were not confirmed due to the lack of a confirmatory test or insufficient remaining sample volume. These specimens were not included in the calculation of performance characteristics. CI, confidence interval; ND, not done.
Luminex yielded additional detections for Giardia (n = 1) and rotavirus (n = 1) that were not confirmed due to the lack of a confirmatory method or insufficient remaining sample volume. These specimens were not included in the calculation of performance characteristics.
Characterized samples.
Due to the relatively low number of prospective samples testing positive for analytes represented on the multiplex panels, previously characterized stool samples (n = 270) with known results (27 negative and 243 positive) were also analyzed by both multiplex assays. Among the positive samples, the majority of targets on both the FilmArray and Luminex panels showed percent sensitivities of >90%; however, there were several targets that demonstrated lower sensitivity (Table 3). The FilmArray Aeromonas sp. assay was positive in only 5 of 21 (23.8%) characterized samples. In addition, the FilmArray Entamoeba histolytica test was negative in a single sample that was determined to be positive by both routine microscopy and the Luminex assay. Interestingly, a number of pathogens were detected by the FilmArray that were not identified during initial, routine testing (Table 3). Notably, enteropathogenic E. coli (EPEC) and enteroaggregative E. coli (EAEC) were detected by FilmArray in 27 and 11 samples, respectively; however, these results were not confirmed due to an inadequate volume of remaining sample or the lack of a confirmatory test. In comparison, the Luminex assay demonstrated a sensitivity of <90% for several targets, including Campylobacter (82.1%; 23/28), Salmonella (83.3%; 20/24), Y. enterocolitica (48.1%; 13/27), and adenovirus 40/41 (80%; 8/10) (Table 3). The percent specificity of all targets for both multiplex assays was generally high (>97%) among the characterized sample set, with the exception of the Luminex norovirus assay, which showed a specificity of 86% (196/228) (Fig. 2). However, when a new lot of Luminex reagents was used to retest these 32 samples, 31 (96.9%) were negative, indicating that the false reactivity may have been due to a lot-specific reagent issue, although we were unable to rule out the possibility of amplicon or specimen contamination.
TABLE 3.
Performance of two gastrointestinal multiplex panels using characterized stool samples (n = 270)a
| Targetb | FilmArrayc |
Luminex |
FilmArrayd |
Luminex |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TN | TP | FP | FN | TN | TP | FP | FN | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | |
| Aeromonas spp. | 248 | 5 | 0 | 16 | 23.8 (10.2, 47.1) | 100 (98.2, 100) | ||||||
| Campylobacter spp. | 240 | 28 | 1 | 1 | 241 | 23 | 0 | 6 | 96.6 (81.4, 99.9) | 99.6 (97.4, 100) | 79.3 (61.3, 90.5) | 100 (98.1, 100) |
| Clostridium difficile | 229 | 33 | 5 | 3 | 229 | 33 | 4 | 3 | 91.7 (77.4, 97.9) | 97.9 (95.0, 99.2) | 91.7 (77.4, 97.9) | 98.3 (95.5, 99.5) |
| Plesiomonas shigelloidese | 265 | 2 | 0 | 0 | 100 (29, 100) | 100 (98.3, 100) | ||||||
| Salmonella spp. | 246 | 24 | 0 | 0 | 246 | 20 | 0 | 4 | 100 (83.7, 100) | 100 (98.1, 100) | 83.3 (63.5, 93.9) | 100 (98.1, 100) |
| Yersinia enterocolitica | 242 | 27 | 1 | 0 | 243 | 13 | 0 | 14 | 100 (85.2, 100) | 99.6 (97.5, 100) | 48.1 (30.7, 66) | 100 (98.1, 100) |
| Vibrio spp. | 270 | 0 | 0 | 0 | ND | ND | ||||||
| Vibrio cholerae | 270 | 0 | 0 | 0 | 270 | 0 | 0 | 0 | ND | ND | ND | ND |
| EAEC | 256 | 0 | 0 | 0 | ND | ND | ||||||
| EPEC | 243 | 0 | 0 | 0 | ND | ND | ||||||
| ETEC | 267 | 0 | 0 | 0 | 270 | 0 | 0 | 0 | ND | ND | ND | ND |
| STEC | 240 | 28 | 2 | 0 | 241 | 27 | 1 | 1 | 100 (85.7, 100) | 99.2 (96.8, 100) | 96.4 (80.8, 99.9) | 99.6 (97.5, 100) |
| E. coli 0157f | 258 | 11 | 0 | 0 | 257 | 10 | 1 | 1 | 100 (70, 100) | 100 (98.2, 100) | 90.9 (60.1, 99.9) | 99.6 (97.6, 100) |
| Shigella/EIEC | 258 | 10 | 1 | 1 | 259 | 9 | 0 | 2 | 90.9 (60.1, 99.9) | 99.6 (97.6, 100) | 81.8 (51.1, 96) | ND |
| Cryptosporidium spp. | 264 | 5 | 0 | 0 | 265 | 5 | 0 | 0 | 100 (51.1, 100) | 100 (98.3, 100) | 100 (51.1, 100) | 100 (98.3, 100) |
| Cyclospora cayetanensis | 269 | 0 | 0 | 0 | ND | ND | ||||||
| Entamoeba histolytica | 269 | 0 | 0 | 1 | 268 | 1 | 0 | 0 | 0 (0, 83.3) | 100 (98.3, 100) | 100 (16.8, 100) | 100 (98.3, 100) |
| Giardia lamblia | 263 | 7 | 0 | 0 | 263 | 7 | 0 | 0 | 100 (60, 100) | 100 (98.3, 100) | 100 (60, 100) | 100 (98.3, 100) |
| Adenovirus 40/41 | 258 | 9 | 2 | 1 | 259 | 8 | 0 | 2 | 90 (57.4, 99.9) | 99.2 (97.1, 100) | 80 (47.9, 95.4) | 100 (98.2, 100) |
| Norovirus GI/GII | 225 | 41 | 0 | 3 | 194 | 41 | 32 | 3 | 93.2 (81.3, 98.6) | 100 (98.0, 100) | 93.2 (81.3, 98.6) | 85.8 (80.7, 89.8) |
| Rotavirus | 255 | 14 | 1 | 0 | 254 | 13 | 1 | 1 | 100 (74.9, 100) | 99.6 (97.6, 100) | 92.9 (66.5, 99.9) | 99.6 (97.6, 100) |
| Sapovirus | 237 | 28 | 0 | 3 | 90.3 (74.3, 97.4) | 100 (98.1, 100) | ||||||
| Astrovirus | 257 | 11 | 0 | 0 | 100 (70, 100) | 100 (98.2, 100) | ||||||
The reference standard for a positive result was defined as an organism detected by (i) bacterial culture or microscopy or (ii) 2 methods.
EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli (lt/st); STEC, Shiga-like toxin-producing E. coli (stx1/stx2; E. coli 0157); EIEC, enteroinvasive E. coli; empty cells, not present on the panel.
TP, true positive; TN, true negative; FP, false positive; FN, false negative.
CI, confidence interval; ND, not done.
FilmArray yielded additional detections for P. shigelloides (n = 3), EAEC (n = 14), EPEC (n = 27), ETEC (n = 3), E. coli O157 (n = 1), Aeromonas (n = 1), Cryptosporidium (n = 1), Cyclospora (n = 1), norovirus (n = 1), sapovirus (n = 2), and astrovirus (n = 2) that could not be confirmed due to lack of a confirmatory method or insufficient remaining sample volume. These samples were not included in the calculation of performance characteristics.
Luminex yielded additional detections for E. coli O157 (n = 1), E. histolytica (n = 1), C. difficile (n = 1), adenovirus (n = 1), and rotavirus (n = 1) that could not be confirmed due to lack of a confirmatory method or insufficient remaining sample volume. These samples were not included in the calculation of performance characteristics.
FIG 2.
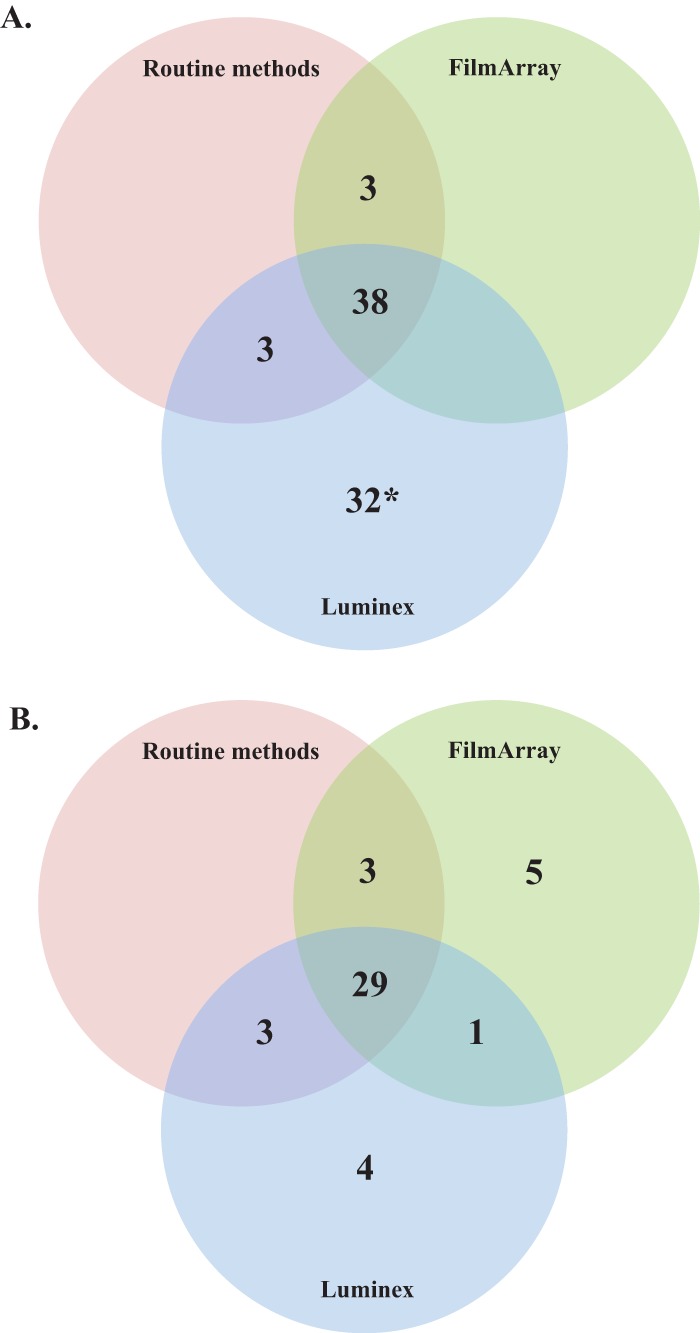
Distribution of positive samples by routine methods, FilmArray and Luminex, for norovirus (A) and Clostridium difficile (B) in characterized stool specimens. *, 31/32 samples were negative upon repeat testing with a new reagent lot.
Detection of mixed infections.
Among positive characterized and prospective samples, the FilmArray and Luminex assays showed overall rates of mixed infections of 27.0% (86/318) and 14.1% (44/312), respectively (Table 4). All mixed infections were confirmed by at least two methods, with the exception of EPEC and EAEC, for which no confirmatory tests were available. Depending on the panel used for testing, certain pathogens tended to be present at higher rates in mixed infections, with EPEC (n = 20), Y. enterocolitica (n = 19), and norovirus (n = 15) being most commonly represented in samples positive by FilmArray. In comparison, norovirus (n = 13), C. difficile (n = 12), and Shiga toxin-producing E. coli (STEC) (n = 11) were most commonly found in mixed infections detected by the Luminex assay (Fig. 3).
TABLE 4.
Comparison of features between two commercial multiplex platforms for the detection of gastrointestinal pathogens
| Feature | FilmArray | Luminex |
|---|---|---|
| Maximum no. of targets | 23 | 15 |
| Processing time per run (min) | 2 | 45 |
| Separate extraction required (min) | No | Yes (∼45) |
| Time/run (h) | ∼1 | ∼3.5 |
| Throughput (no. specimens/run) | 1 | 96 |
| Technology | Nested PCR plus melting curve | PCR plus xTag (fluorescent bead-based detection) |
| Regulatory status | FDA cleared (22 targets) | FDA cleared (11 targets); RUOa (4 additional targets) |
| Open or closed system | Closed | Open |
| Results | Qualitative | Qualitative |
| Footprint size | Small | Moderate |
| List price per instrument ($) | 39,500 | 37,000 |
| List price reagent cost per specimen ($) | 155 | 80–90 |
RUO, research use only.
FIG 3.
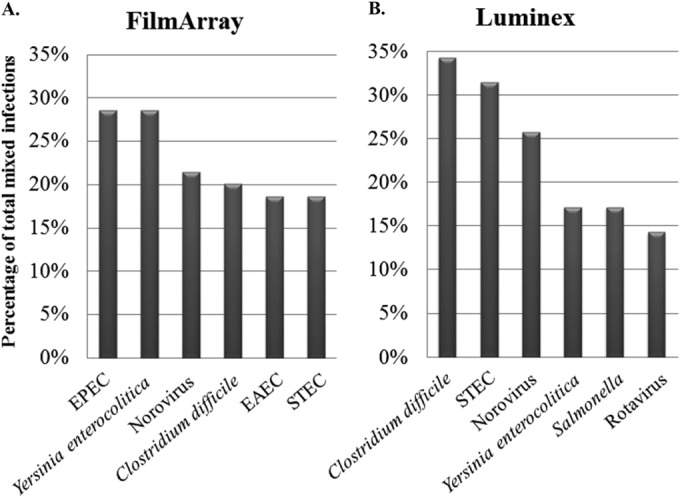
Pathogens most frequently observed in mixed infections as a percentage of all confirmed mixed infections detected by FilmArray (n = 70) (A) and Luminex (n = 35) (B). EPEC, enteropathogenic E. coli; STEC, Shiga-like toxin-producing E. coli (stx1/stx2); EAEC, enteroaggregative E. coli.
DISCUSSION
In this study, we compared the performances of two commercial multiplex panels to that of routine testing using clinical stool samples (n = 500). The data from this evaluation demonstrate that the majority of targets represented on these panels show high sensitivity and specificity (>90%); however, there are analytes on both panels that showed poorer performance, and these deserve further discussion. Among our characterized sample set, the FilmArray Aeromonas assay demonstrated 23.8% (5/21) sensitivity compared to routine bacterial culture. All of these samples had been stored at −20°C for >1 month, so we are unable to rule out the possibility of nucleic acid degradation in these samples. Samples showing discordant results (e.g., culture positive and FilmArray negative) for Aeromonas were recultured, but as expected from a sample that had been frozen and thawed, only 2/21 stool specimens regrew the organism. Interestingly, the Aeromonas target was not included in the initial FilmArray GI panel FDA clearance, due to discordance with culture methods during the clinical evaluations (Wade Stevenson, BioFire, personal communication). The Luminex assay showed low percent sensitivity for Y. enterocolitica, detecting 13 (48.1%) of 27 positive samples. Nucleic acid degradation is an unlikely explanation for these discrepant results, as the FilmArray assay was positive in 100% (27/27) of these samples. Other notable findings from our characterized samples were (i) the low specificity (86%; 196/228) of the Luminex norovirus assay and (ii) the high number of samples that were positive by FilmArray for EAEC (n = 11) or EPEC (n = 27), which are not part of our routine stool testing panel. In regard to the Luminex norovirus target, the low observed specificity may have been due to a lot-specific reagent issue, as retesting of these discordant samples using a new kit lot showed negative results in 31/32 samples that were falsely positive upon initial testing (Fig. 2A). However, the Luminex platform is an open system, and therefore we cannot rule out the possibility of amplicon contamination. Unfortunately, the additional positive results for EAEC and EPEC by FilmArray were not confirmed, due to insufficient volume of remaining sample or the lack of an alternative method for discordant analysis. Interestingly, data from the manufacturer's clinical trials showed good performance (>97% positive and negative agreement) for EAEC and EPEC among over 1,500 specimens tested (FilmArray GI, package insert).
Among prospective clinical specimens, C. difficile and norovirus were the pathogens most commonly detected by both panels in our patient population, with sapovirus also being detected in a substantial percentage of samples by FilmArray (Fig. 1A and B). The fact that we tested Cary-Blair stool samples may have affected the rate of C. difficile detection, as it is not possible to differentiate between formed and diarrheal stool when samples are submitted in Cary-Blair medium. Whether the positive results for C. difficile (Fig. 2B) represented bacteria associated with disease or colonization is an important area for future study. Interestingly, sapovirus was positive by the FilmArray in 13 (5.6%) of 230 prospective samples. Identification of viral pathogens (e.g., sapovirus and norovirus) in diarrheal stool has important implications for antibiotic stewardship, since these pathogens are not susceptible to common antibiotics used to treat gastroenteritis (19–21).
An important aspect of this study was the observation that a high percentage of stool samples were positive for ≥2 pathogens. Among positive samples, routine testing identified at least two pathogens in 19 (8.3%) samples, while the FilmArray and Luminex assays showed overall rates of mixed infections of 27.0% (86/318) and 14.1% (44/312), respectively. Certain organisms were found more commonly in mixed infections, with EAEC, Y. enterocolitica, and norovirus being detected most frequently by FilmArray, while C. difficile, STEC, and norovirus were most commonly detected in mixed infections by the Luminex assay (Fig. 3). These differences are likely influenced by the composition of each multiplex panel. For example, EAEC is not included on the Luminex assay. The association of pathogens with mixed infection is likely dependent on a variety of factors, including the geographic location and patient population (22, 23). For example, a study performed in India showed that Vibrio cholerae and rotavirus were highly associated with coinfections (24). Further investigations of the mechanisms of coinfection, the organisms associated with them, and the impact on clinical outcomes will be important areas of future research.
This study has several important limitations that should be discussed. First, different volumes of stool were tested by the FilmArray (200 μl) and Luminex (100 μl) assays. The use of 200 μl of Cary-Blair stool is recommended by BioFire for testing on the FilmArray, while the Luminex assay is FDA cleared for testing 100 μl of raw stool. Our laboratory receives stool only in Cary-Blair medium, so 100 μl of sample was tested by the Luminex assay. However, we emphasize that Cary-Blair stool is more dilute than raw stool and is not considered an FDA-cleared source for the Luminex assay. Therefore, it should be underscored that testing a higher volume (e.g., up to 400 μl) of this sample type may enhance the sensitivity of the Luminex assay. A second limitation is that we were unable to correlate the results of testing with clinical information and treatment decisions. A third limitation is that our prospective study identified a relatively low number of positive results for many of the targets and did not identify any samples testing positive for several analytes, including P. shigelloides, Vibrio spp., Cyclospora, and E. histolytica. Additional studies are needed to better characterize the performance characteristics of the multiplex assays for these targets. Finally, the multiplex assays detected a number of pathogens (e.g., EPEC and EAEC) that are not tested for by our routine stool panel and were not confirmed, because of inadequate amounts of remaining sample or the lack of a confirmatory test. Discerning whether these findings represent true- or false-positive results will be an important area of future research.
Our evaluation demonstrated that multiplex GI panels yield an increased percent positive rate compared to routine testing (33.3% and 30.3%, respectively, detected by FilmArray and Luminex versus 8.3% by routine methods). However, after exclusion of false-positive results, percent positivity by the FilmArray and Luminex decreased to 28.3% and 20.0%, respectively. Furthermore, the FilmArray GI and Luminex GPP panels detected mixed infections in 27.0% and 14.1% of positive samples, respectively, indicating that the presence of multiple pathogens in diarrheal stool samples may be underestimated by current routine tests.
In terms of workflow and laboratory implementation, both multiplex platforms offer certain advantages, but there are differences that deserve discussion (Table 4). The FilmArray is a closed system that offers a rapid result (∼1 h turnaround time) with minimal hands-on time (∼5 min) and requires minimal user training. However, the FilmArray analyzes 1 sample at a time and therefore is best suited for laboratories requiring a lower throughput. A single FilmArray system accommodates 7 to 8 samples in an 8-h shift. In contrast, the Luminex platform is an open system, requires ∼60 min of hands-on time, and has turnaround time of 5 to 6 h. Despite the longer turnaround time, the Luminex system processes up to 96 samples in an 8-h shift, making it more suitable for high-volume reference laboratories. Importantly, open molecular platforms pose an increased risk of amplicon contamination, and therefore laboratories using the Luminex system should use unidirectional workflow and follow strict good laboratory practice. Both systems also have several advantages compared to routine laboratory methods. Results are available in <6 h, whereas routine testing commonly requires up to several days if culture-based methods are used. This may have a significant impact on patient management. In addition, test utilization and antimicrobial stewardship may be affected by the use of multiplex testing platforms. In our prospective study, there was an average of 3 routine tests ordered per sample. The ability to cover for a broad spectrum of GI pathogens in a single test is an appealing advantage of multiplex technology. However, the impact of multiplex tests on the management and treatment of patients with GI illness is still unclear, as no standard therapy exists for some of the pathogens represented on these panels. It is possible that the detection of common viral causes of GI disease may help curb the use of antibiotic therapy. This will be an exciting area for future research as multiplex panels become a more common feature in diagnostic laboratories.
ACKNOWLEDGMENTS
We thank BioFire and Luminex for providing the kits and instruments used in this study.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1.Petri WA, Jr, Miller M, Binder HJ, Levine MM, Dillingham R, Guerrant RL. 2008. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Invest. 118:1277–1290. 10.1172/JCI34005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 3.Esposito DH, Holman RC, Haberling DL, Tate JE, Podewils LJ, Glass RI, Parashar U. 2011. Baseline estimates of diarrhea-associated mortality among United States children before rotavirus vaccine introduction. Pediatr. Infect. Dis. J. 30:942–947. 10.1097/INF.0b013e3182254d19 [DOI] [PubMed] [Google Scholar]
- 4.Hennessy TW, Marcus R, Deneen V, Reddy S, Vugia D, Townes J, Bardsley M, Swerdlow D, Angulo FJ. 2004. Survey of physician diagnostic practices for patients with acute diarrhea: clinical and public health implications. Clin. Infect. Dis. 38(Suppl 3):S203–S211. 10.1086/381588 [DOI] [PubMed] [Google Scholar]
- 5.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.09-1101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlenker C, Surawicz CM. 2009. Emerging infections of the gastrointestinal tract. Best Pract Res. Clin. Gastroenterol. 23:89–99. 10.1016/j.bpg.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 7.Malecki M, Schildgen V, Kamm M, Mattner F, Schildgen O. 2012. Rapid screening method for multiple gastroenteric pathogens also detects novel enterohemorrhagic Escherichia coli O104:H4. Am. J. Infect. Control 40:82–83. 10.1016/j.ajic.2011.07.019 [DOI] [PubMed] [Google Scholar]
- 8.Hatchette TF, Farina D. 2011. Infectious diarrhea: when to test and when to treat. CMAJ 183:339–344. 10.1503/cmaj.091495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar SA. 2013. Molecular revolution entering GI diagnostic testing. MLO Med. Lab. Obs. 45:28. [PubMed] [Google Scholar]
- 10.Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. 2010. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J. Clin. Microbiol. 48:2929–2933. 10.1128/JCM.00339-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karre T, Sloan L, Patel R, Mandrekar J, Rosenblatt J. 2011. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 49:725–727. 10.1128/JCM.01028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckwalter SP, Teo R, Espy MJ, Sloan LM, Smith TF, Pritt BS. 2012. Real-time qualitative PCR for 57 human adenovirus types from multiple specimen sources. J. Clin. Microbiol. 50:766–771. 10.1128/JCM.05629-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visvesvara GS, Moura H, Kovacs-Nace E, Wallace S, Eberhard ML. 1997. Uniform staining of Cyclospora oocysts in fecal smears by a modified safranin technique with microwave heating. J. Clin. Microbiol. 35:730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trujillo AA, McCaustland KA, Zheng DP, Hadley LA, Vaughn G, Adams SM, Ando T, Glass RI, Monroe SS. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405–1412. 10.1128/JCM.44.4.1405-1412.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu FT, White PA, Takeda N. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 78:1347–1353. 10.1002/jmv.20699 [DOI] [PubMed] [Google Scholar]
- 16.Grant L, Vinje J, Parashar U, Watt J, Reid R, Weatherholtz R, Santosham M, Gentsch J, O'Brien K. 2012. Epidemiologic and clinical features of other enteric viruses associated with acute gastroenteritis in American Indian infants. J. Pediatr. 161:110–115, e111. 10.1016/j.jpeds.2011.12.046 [DOI] [PubMed] [Google Scholar]
- 17.Freeman MM, Kerin T, Hull J, McCaustland K, Gentsch J. 2008. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J. Med. Virol. 80:1489–1496. 10.1002/jmv.21228 [DOI] [PubMed] [Google Scholar]
- 18.Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239. 10.1002/jmv.10382 [DOI] [PubMed] [Google Scholar]
- 19.Stefanoff P, Rogalska J, Czech M, Staszewska E, Rosinska M. 2013. Antibacterial prescriptions for acute gastrointestinal infections: uncovering the iceberg. Epidemiol. Infect. 141:859–867. 10.1017/S0950268812001173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotwani A, Chaudhury RR, Holloway K. 2012. Antibiotic-prescribing practices of primary care prescribers for acute diarrhea in New Delhi, India. Value Health 15:S116–S119. 10.1016/j.jval.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 21.Ekwochi U, Chinawa JM, Obi I, Obu HA, Agwu S. 2013. Use and/or misuse of antibiotics in management of diarrhea among children in Enugu, Southeast Nigeria. J. Trop. Pediatr. 59:314–316. 10.1093/tropej/fmt016 [DOI] [PubMed] [Google Scholar]
- 22.Navidad JF, Griswold DJ, Gradus MS, Bhattacharyya S. 2013. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J. Clin. Microbiol. 51:3018–3024. 10.1128/JCM.00896-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTAG(R) gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J. Microbiol. Biotechnol. 23:1041–1045. 10.4014/jmb.1212.12042 [DOI] [PubMed] [Google Scholar]
- 24.Lindsay B, Ramamurthy T, Sen Gupta S, Takeda Y, Rajendran K, Nair GB, Stine OC. 2011. Diarrheagenic pathogens in polymicrobial infections. Emerg. Infect. Dis. 17:606–611. 10.3201/eid1704.100939, [DOI] [PMC free article] [PubMed] [Google Scholar]


