LETTER
We have read the article by Zong et al. with considerable interest (1). They described the molecular phylogeny of Coxsackievirus A16 (CVA16) in Shenzhen, China, from 2005 to 2009. In their paper, they claimed that “Subtype B2 could be further divided into clusters B2a, B2b, and B2c with bootstrap support of 98%. The phylogenetic classification in our study was consistent with those of previous studies. A single difference compared with studies of other researchers was the definition of genotype B1.”
That article illustrated the history of molecular evolution across the world as follows. Since 1981, the mainstream genotype of CVA16 was B1 until it was replaced by B2 gradually after 1997. Over the past 10 years, cluster B2a and B2b CVA16 strains have been predominant in mainland China and neighboring countries and regions. However, this conclusion differs from that of other studies (2–4).
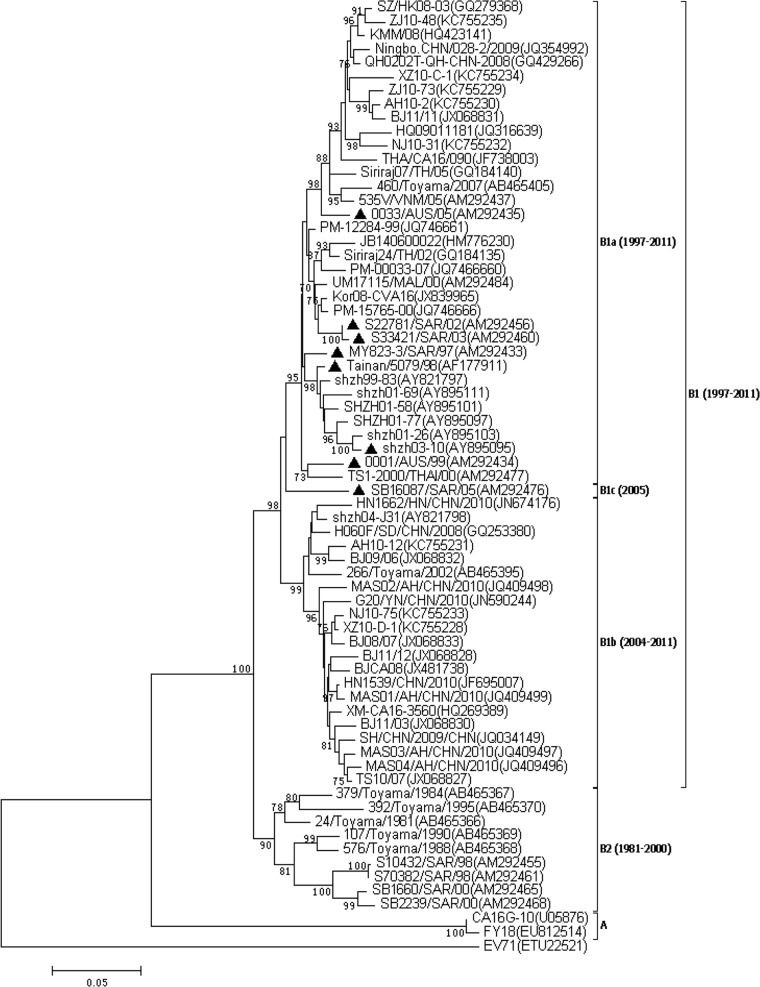
The study by Zong et al. was the only study on the molecular phylogeny of CVA16 to mention subgenotype B2a, B2b, or B2c. Most of the B2a, B2b, and B2c strains evaluated by Zong et al. were classified as B1a, b1b, and b1c (2, 3) by strain name and collection year, such as Tainan/5079/98 (AF177911), 0033/AUS/05 (AM292435), S22781/SAR/02 (AM292456), MY823-3/SAR/97 (AM292433), shzh03-10 (AY895095), and so on. These strains are marked with triangles in Fig. 1. In our opinion, this difference in results was caused by the misidentification of subtype B1 in the report of Zong et al.
FIG 1.
Phylogenetic dendrogram constructed by the neighbor-joining method on the basis of the Kimura two-parameter model with MEGA 6.05, based on the complete VP1 gene sequences of 68 CVA16 strains obtained from GenBank. The prototype EV-A71 strain (ETU22521) was used as an outgroup. Bootstrap values (1,000 replicates) are shown next to the branches. Strains marked by triangles are those with inconsistencies in studies and were classified as genotype B2 by Zong et al. (1).
For further investigation, all CVA16 reference sequences (a total of 187 strains; data not shown) were downloaded from the NCBI GenBank database. Sequences classified by researchers as having matching subgenotypes were marked, and then all of the CVA16 strains were used to construct phylogenetic dendrograms based on their complete VP1 gene regions by using MEGA 6.05. After phylogenetic analysis, the method of classification used by Zhang et al. and Chen et al. (2, 3) was found to be more scientific and credible than that of Zong et al. and other methods.
Discrepancies were attributed to a lack of criteria for genotype classification. To establish criteria for the genotype classification of CVA16 strains, the phylogenetic tree was optimized by cutting off the branches with low tree option values. Then 68 genotype-identified CVA16 and EV-A71 strains were reserved and used to construct another phylogenetic tree, as shown in Fig. 1.
It is obvious that the prevalent genotypes of CVA16 have changed twice since they were first identified in South Africa in 1951. From 1981 to 2000, a plurality of CVA16 strains belonged to genotype B2. Since 1997, genotype B1 has emerged and gradually replaced genotype B2 as the most commonly detected genotype in the world. From 2004 to 2011, CVA16 strain B1a and B1b groups became the predominant types of virus in mainland China and neighboring regions, including Taiwan, Japan, Vietnam, Thailand, Malaysia, and Australia. Different regions usually show the same prevalent genotypes of CVA16 in the same period. This has been reported and is widely accepted as true. Taken together, all Chinese CVA16 strains belong to either subgenotype B1a or B1b, which have been continuously circulating in mainland China since 1997 (2, 3, 4).
Current analysis indicates that the genotype identification of CVA16 and the corresponding conclusions drawn by Zong et al. may have been wrong. Mistakes are amplified if misidentified genotypes and incorrect conclusions are referred to and quoted by subsequent authors. In fact, these mistakes have already been quoted in an otherwise excellent review (5). Considering that the molecular phylogeny of CVA16 is associated with epidemiology and vaccine development research, the comprehensive recognition of genotype identifications by researchers could be extremely important.
We look forward to a reply from Zong et al. to confirm the genotype classification and improve our understanding of the molecular phylogeny of CVA16 across the world.
Footnotes
Ed. Note: The authors of the published article declined to respond.
REFERENCES
- 1.Zong WP, He YQ, Yu SY, Yang H, Xian HX, Liao YX, Hu GF. 2011. Molecular phylogeny of Coxsackievirus A16 in Shenzhen, China, from 2005 to 2009. J. Clin. Microbiol. 49:1659–1661. 10.1128/JCM.00010-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Wang DY, Yan DM, Zhu SL, Liu JF, Wang HY, Zhao SC, Yu DS, Nan LJ, An JJ, Chen L, An HQ, Xu AQ, Xu WB. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 Coxsackievirus A16-associated hand, foot, and mouth disease in China. J. Clin. Microbiol. 48:619–622. 10.1128/JCM.02338-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen XP, Tan XJ, Li J, Jin Y, Gong LM, Hong M, Shi YL, Zhu SL, Zhang BM, Zhang S, Zhang Y, Mao NY, Xu WB. 2013. Molecular epidemiology of Coxsackievirus A16: intratype and prevalent intertype recombination identified. PLoS One. 8(12):e82861. 10.1371/journal.pone.0082861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Xu WB. 2013. Molecular epidemiology of enteroviruses associated with hand, foot, and mouth disease in the mainland of China. Biomed. Environ. Sci. 26:875–876. 10.3967/bes2013.015 [DOI] [PubMed] [Google Scholar]
- 5.Mao QY, Wang YP, Yao X, Bian LL, Wu X, Xu M, Liang ZL. 2014. Coxsackievirus A16 epidemiology, diagnosis, and vaccine. Hum. Vaccin. Immunother. 10:360–367. 10.4161/hv.27087 [DOI] [PMC free article] [PubMed] [Google Scholar]