Abstract
Invasive aspergillosis is a difficult-to-diagnose infection with a high mortality rate that affects high-risk groups such as patients with neutropenia and hematologic malignancies. We performed a bivariate meta-analysis of diagnostic data for an Aspergillus sp. PCR assay with blood specimens from high-risk hematology patients. We included all studies involving human subjects that assessed the performance of any PCR assay for invasive aspergillosis in whole blood or serum and that used the European Organization for the treatment of Cancer/Mycoses Study Group criteria as a reference standard. Three investigators independently searched the literature for eligible studies and extracted the data. Out of a total of 37 studies, 25 met strict quality criteria and were included in our evidence synthesis. Twenty-five studies with 2,595 patients were analyzed. The pooled diagnostic performance of whole-blood and serum PCR assays was moderate, with a sensitivity and specificity of 84% (95% confidence interval [CI], 75 to 91%) and 76% (95% CI, 65 to 84%), respectively, suggesting that a positive or negative result is unable, on its own, to confirm or exclude a suspected infection. The performance of a PCR assay of serum was not significantly different from that of whole blood. Notably, at least two positive PCR test results were found to have a specificity of 95% and a sensitivity of 64% for invasive infection, achieving a high positive likelihood ratio of 12.8. Importantly, the European Aspergillus PCR Initiative (EAPCRI) recommendations improved the performance of the PCR even further when at least two positive specimens were used to define PCR positivity. In conclusion, two positive PCR results should be considered highly indicative of an active Aspergillus sp. infection. Use of the EAPCRI recommendations by clinical laboratories can further enhance PCR performance.
INTRODUCTION
Despite advances in treatment and supportive care, invasive aspergillosis (IA) is associated with significant morbidity and mortality rates, especially among patients with hematologic malignancies and hematopoietic stem cell transplant (HSCT) recipients (1, 2). Systemic antifungal prophylaxis is widely used in this patient population (3, 4), and patients with recurrent or persistent fever and prolonged neutropenia frequently require empirical coverage with antifungal agents (5). Timely and accurate diagnosis of an active infection is needed in order to initiate targeted antifungal therapy and avoid unnecessary antifungal treatment, which is often accompanied by a multitude of side effects and the cumulative risk of resistance.
There is a need for the development of newer diagnostic techniques that would ideally be able to identify IA rapidly, noninvasively, and at an early stage. To aid in this endeavor, the European Organization for the treatment of Cancer/Mycoses Study Group (EORTC/MSG) developed specific criteria for the diagnosis of IA in 2002 (6), which were later revised in 2008 (7), in an attempt to provide the elusive “gold standard” to which any newer diagnostic test should be compared. Dominant among the novel diagnostic methods is the PCR assay, which allows pathogen DNA detection and identification to the species level in a variety of clinical samples. A number of PCR assays are now available to detect Aspergillus spp. in various clinical specimens and have a high diagnostic yield in vitro, but their clinical performance is debated because of contradictory reports from clinical trials. Previous systematic reviews have assessed Aspergillus sp. PCR assay performance with promising findings (8) but did not succeed in convincing the guideline-issuing organizations to incorporate the test in their algorithms (6, 7). The main argument has been that differences in the PCR protocols used in different studies do not allow sufficient interlaboratory comparisons to be made and thus preclude widespread implementation (9). In an effort to determine the value of the Aspergillus sp. PCR assay in clinical diagnostics as a screening or a confirmatory tool and to evaluate different parameters that could contribute to contradictory reports regarding PCR performance from the existent literature, we performed a meta-analysis of clinical trials that evaluated the accuracy of the PCR assay for IA performed with serum and whole blood from high-risk patients.
MATERIALS AND METHODS
Literature search.
A systematic literature search of the MEDLINE (1951 to December 2013) and Cochrane Central Register of Controlled Trials (2010 to June 2013) databases was conducted to identify all of the published studies involving human subjects and evaluating the performance of a PCR assay for Aspergillus spp. using as a reference standard the EORTC/MSG criteria. We searched only full publications and not unpublished studies or conference abstracts. We used the search term “Aspergil* AND (PCR OR PCR).” The date of last access was 20 January 2014.
Three investigators (M.A., I.M.Z., and F.N.Z.) independently identified and scrutinized studies for potential inclusion. Studies published in languages other than English were excluded. Our meta-analysis is in line with PRISMA (preferred reporting items for systematic reviews and meta-analyses) recommendations (10).
Eligibility criteria and definitions.
We included all studies involving human subjects that assessed the performance of any PCR assay for IA in whole blood or serum and that used as a reference standard the EORTC/MSG criteria (either the 2002 [6] or the revised 2008 [7] criteria). Studies of series of patients with a positive-only (probable/definite) or negative-only (no infection) reference outcome were not considered because it is impossible to extrapolate all of the pertinent diagnostic estimates from these studies. We defined as “true positive” the cases that were positive by PCR and were classified as definite or probable by the EORTC/MSG criteria and as “true negative” the cases that were negative by PCR and were classified as possible or unlikely infection by the EORTC/MSG criteria. Possible IA cases were categorized as negative in accordance with the recommendations of the Cochrane group (11). For our primary analysis, we defined as PCR positive any case that had at least one positive PCR result. In multiarm studies that used the same patients more than once, we included the strata with the highest reported performance estimates. Also, in trials that examined PCR assay performance in both a high-risk group and a control group of patients at low risk for IA, we excluded the control arm to avoid a case-control design that could spuriously increase performance estimates.
Outcomes of interest.
The primary outcomes of interest were the summary sensitivity and specificity of an Aspergillus PCR assay of blood products compared to the reference standard. Our secondary goal was to evaluate whether the choice of sample, the type of PCR, the choice of primers, or any of the methodological aspects proposed by the European Aspergillus PCR Initiative (EAPCRI) (12, 13) for optimum PCR assay performance had a significant impact on the test's accuracy. Finally, we performed a sensitivity analysis to evaluate how PCR assay performance would be affected if two positive results were used to define a positive PCR outcome.
Data extraction and quality assessment.
Data from eligible studies were independently extracted by three reviewers (M.A., I.M.Z., and F.N.Z.). Discrepancies between authors were resolved by consensus. The methodological quality of each trial was evaluated independently by three of us (M.A., I.M.Z., and F.N.Z.) for potential sources of bias by using standard criteria. The QUADAS-2 score was used to test for potential sources of bias in each study (14). To have high-quality estimates, we excluded from further analysis the studies with high-bias QUADAS-2 elements. Specifically, we excluded studies that had a nonrandom population or a case-control design, and we also excluded studies that did not have a prespecified threshold of PCR positivity. We chose not to eliminate trials in which the investigators had knowledge of the EORTC/MSG classification of the patient before PCR interpretation because this knowledge cannot alter the interpretation of a positive or negative finding as long as the positivity threshold is prespecified. Finally, we excluded studies that had a >10% loss to follow-up.
Statistical analysis.
We calculated the independent sensitivity and specificity of each study by using a two-by-two contingency table. Each table consisted of true positives, true negatives, false positives, and false negatives according to the results of the Aspergillus sp. PCR assay (the index test) compared to the classification of the patient as definite/probable versus possible/unlikely aspergillosis on the basis of the EORTC/MSG criteria (the reference standard). We estimated the combined (pooled) sensitivity, specificity, likelihood ratios (LRs), and diagnostic accuracy (area under the curve) by using a bivariate mixed-effects binomial regression model that accounts for variability both within a study and between studies (15–17). We used empirical thresholds of >10 for LR+ and <0.1 for LR− to rate the test as of high value in the decision to rule in (i.e., probable/definite aspergillosis) or rule out infection (18). Publication bias was assessed by using the Deeks regression test for asymmetry, with P < 0.05 for the slope coefficient denoting significant asymmetry (19). We did not use heterogeneity tests (16, 20), as such tests can mislead systematic reviews of diagnostic test accuracy, are not recommended by the Cochrane diagnostic test accuracy group (16), and may be misleading in diagnostic accuracy studies, since computing separate I2 statistics for sensitivity and specificity will usually overestimate statistical heterogeneity. Effects were plotted as summary receiver operating characteristic (SROC) curves (15).
We calculated subgroup estimates to account for variation in laboratory PCR methodologies, provided that we had at least four pertinent studies in each group to draw the quadrature points and provided that we had data on the magnitude and difference of average sensitivity and specificity estimates between groups. We also evaluated whether the use of the old (2002) or the revised (2008) EORTC/MSG criteria as a reference standard affected PCR performance estimates. The significance of differences between average specificities or sensitivities was assessed by metaregression, and a P value was reported.
The bivariate meta-analysis was performed with Stata v11 (StataCorp, College Station, TX) by using the Midas set of commands (21, 22). The metaregression was performed with MetaAnalyst software (23).
RESULTS
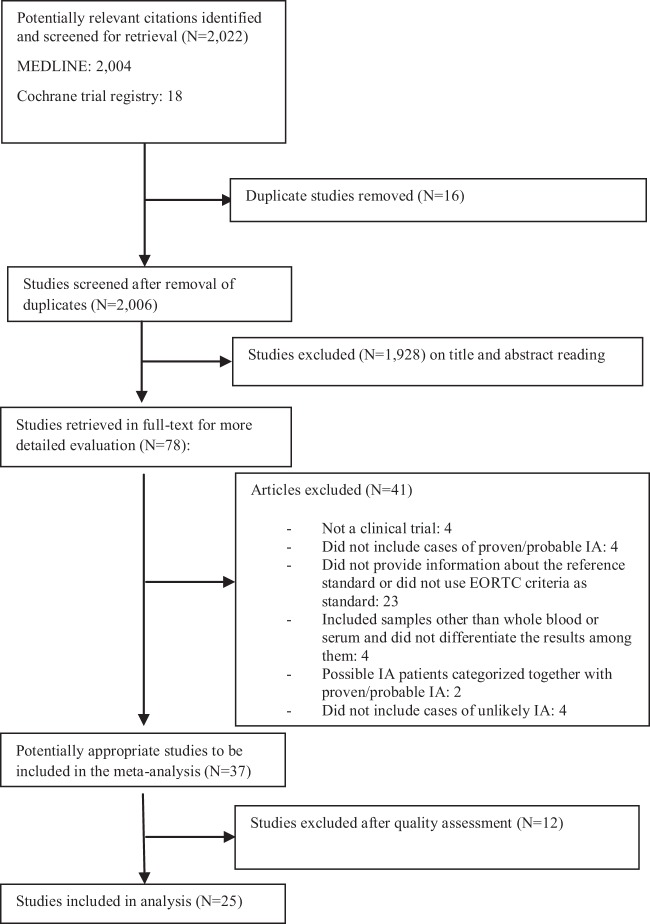
Two thousand six nonduplicate studies were identified by the initial search, of which 37 met the inclusion criteria (Fig. 1). Twenty-three studies provided data for PCR based on the 2002 EORTC/MSG criteria (24–46), and 14 did so by using the revised (2008) criteria (47–60) (Table 1).
FIG 1.
Flow diagram of the inclusion criteria used in this study.
TABLE 1.
Characteristics of studies included
| Reference | Study design | Population studied | PCR type | Specimen tested | Primer(s) used | No. of proven/probable IA episodes | No. of unlikely IA episodes |
|---|---|---|---|---|---|---|---|
| 25 | Retrospective | 218 patients with hematologic malignancies | Nested | Whole blood | 18S rDNA | 36 | 182 |
| 24 | Prospective | 54 patients with cancer and pulmonary infiltrates | Traditional | Whole blood | Mitochondrial DNA and alkaline protease gene | 11 | 36 |
| 36 | Prospective | 17 hematology patients | Nested | Whole blood | Mitochondrial DNA | 3 | 10 |
| 39 | Retrospective | 20 Patients with hematologic malignancies who had proven or probable IA | Real time | Serum | Mitochondrial DNA | 20 | 10 |
| 33 | Prospective | 8 patients at risk for IFI,a 45 negative controls | Real time | Whole blood | 18S rDNA | 2 | 4 |
| 45 | Retrospective | 106 patients at risk for IFIs | Real time | Serum | 5.8S rDNA | 41 | 35 |
| 41 | Prospective | 96 patients at risk for IA | Real time | Whole blood | 18S rRNA gene | 11 | 125 |
| 35 | Retrospective | 25 patients with hematologic malignancies | PCR-ELISA | Whole blood | 18S rDNA | 5 | 20 |
| 40 | Retrospective | 29 patients with at least one positive galactomannan test | Real time | Serum | Mitochondrial DNA | 7 | 11 |
| 32 | Prospective | 78 immunocompromised hemato-oncology patients | Real time | Whole blood | 18S rDNA | 1 | 106 |
| 34 | Retrospective | 77 hematologic malignancy patients or solid-organ transplant recipients at high risk for IA | Real time | Whole blood | 18S rDNA | 3 | 42 |
| 26 | Prospective | 29 adults and 36 children with febrile neutropenia undergoing intensive chemotherapy for hematologic malignancy or having received HSCT | Nested | Whole blood | 18S rRNA gene | 13 | 61 |
| 46 | Prospective | 91 febrile neutropenic pediatric patients | Traditional | Serum | 18S rDNA | 28 | 49 |
| 29 | Prospective | 203 patients at risk for IFI | Real time | Whole blood | 28S rRNA | 13 | 149 |
| 38 | Prospective | 201 patients with hematologic malignancies | PCR-ELISA | Serum | Mitochondrial DNA | 33 | 106 |
| 27 | Prospective | 194 patients with hematologic malignancies | PCR-ELISA | Whole blood | rRNA | 15 | 150 |
| 30 | Prospective | 62 pediatric patients at risk for IA | Real time | Whole blood | 18S rRNA gene | 8 | 54 |
| 42 | Retrospective | 25 patients with at least 1 GMb-positive serum sample | Real time | Serum | Mitochondrial DNA | 11 | 12 |
| 43 | Prospective | 124 patients with hematologic malignancies undergoing chemotherapy or HSCT | Real time with fluorescent probes | Serum | 28S rRNA | 15 | 121 |
| 37 | Prospective | 125 hematology patients | Real time | Whole blood | 18S rDNA | 7 | 84 |
| 31 | Prospective | 127 patients at risk for IA | Real time | Whole blood | 18S rRNA | 5 | 105 |
| 57 | Prospective | 172 patients who received high-dose chemotherapy | Traditional | Serum | 18S rRNA | 20 | 173 |
| 28 | Prospective | 91 patients in AmBiLoad trial | Nested | Whole blood | 18S rRNA | 59 | 1 |
| 47 | Prospective | 82 bone marrow transplant recipients | Real time | Whole blood | 18S rDNA | 10 | 67 |
| 48 | Prospective | 46 patients receiving either allogeneic stem cell transplant or myeloablative chemotherapy | Real time | Whole blood | Multicopy ribosomal operon region from ITS1d to 5.8S region | 3 | 24 |
| 59 | Retrospective | 44 patients with two sequential positive serum galactomannan results and an IA risk factor | Two different real-time assays | Serum | Assay 1, mitochondrial DNA; assay 2, 18S rRNA | 26 | 18 |
| 60 | Retrospective | 31 patients (10 with proven/probable IA vs 21 with no IA) | Two different real-time PCR assays | Serum | Assay 1, 28S rRNA; assay 2, 18S rRNA | 10 | 21 |
| 49 | Retrospective | 38 adult patients with a high clinical suspicion of IA | Real time | Serum and whole blood | ITS1 | 18 | 4 |
| 58 | Prospective | 62 pediatric patients at increased risk for IA | Nested | Serum | Not reported | 10 | 26 |
| 50 | Prospective | 63 patients with allogeneic stem cell transplant and myeloablative chemotherapy | Real time | Whole blood | ITS1-5.8S | 3 | 41 |
| 51 | Retrospective multicenter | 47 patients with proven/probable IA and 31 controls | Various real-time PCR assays | Serum and whole blood | Various | 47 | 31 |
| 52 | Prospective (two different centers) | 278 patients undergoing intensive chemotherapy or HSCT | Two different real-time PCR assays | Whole blood | 28S rRNA (nested) ITS (single run) | 15/30 | 120/83 |
| 54 | Prospective | 72 patients with hematologic malignancies with fever, 4 with normal temperatures, 10 healthy volunteers | Real time | Whole blood | 28S-ITS2 rRNA genes | 22 | 41 |
| 53 | Prospective | 51 patients with hematologic malignancies at risk for IA | Nested | Whole blood | 18S | 6 | 47 |
| 55 | Prospective | 103 adult hematology patients at high risk for IA | Real time | Whole blood | 28S rDNA | 22 | 59 |
| 44 | Prospective | 185 febrile neutropenic patients treated with chemotherapy for AMLc | Real time | Serum | Mitochondrial DNA | 11 | 174 |
| 56 | Prospective | 62 hematologic malignancy patients | Real time | Whole blood | 18S rDNA | 5 | 44 |
IFI, invasive fungal infection.
GM, galactomannan.
AML, acute myeloid leukemia.
ITS1, internal transcribed spacer 1.
Quality assessment.
Most of the studies were of high quality, as determined by the QUADAS-2 assessment tool (Table 2). After all of the low-quality studies were excluded, 25 studies remained and were further analyzed (25, 26, 28–37, 43–48, 50, 52–54, 56–58).
TABLE 2.
Quality assessment and final determination of inclusion versus exclusion based on QUADAS-2 toola
| Sample type used for PCR assay and reference | Patient selection |
Index test |
Reference standard |
Flow and timing |
Applicability |
Final determination | No. of deviations from EAPCRI recommendations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Random patient selection | Case-control design | Inappropriate exclusions | Test results without knowledge of standard | Prespecified threshold | Reference standard interpreted without knowledge of index test | Reference standard likely to correctly classify condition | Appropriate interval between index test and reference standard | Did all patients receive a reference standard? | Was the reference standard the same for all patients? | Were all patients included in the analysis? | Are there concerns that the patients included do not match the review question? | Are there concerns that the target condition defined by the reference standard does not match the review question? | Are there concerns that the index test differs from the review question? | |||
| Whole blood | ||||||||||||||||
| 24 | No | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 25 | Yes | No | No | Yes | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 26 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 3 |
| 27 | Unclear | No | Unclear | Unclear | Yes | Yes | Yes | Unclear | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 28 | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 29 | Yes | No | No | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 30 | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 4 |
| 31 | Yes | No | No | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 47 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 48 | Yes | No | No | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 0 |
| 49 | No | No | Unclear | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 51 | No | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 52 | Yes | No | No | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 0 |
| 52 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 54 | Yes | No | No | Yes | Yes | Yes | Yes | Unclear | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 53 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 50 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Low | Yes | Yes | Included | 0 |
| 55 | No | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 32 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 33 | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 4 |
| 34 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 35 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 36 | Yes | No | Unclear | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 37 | Yes | No | No | Unclear | No | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| 56 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 2 |
| Serum | ||||||||||||||||
| 38 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Low | Yes | Yes | Excluded | |
| 57 | Yes | No | No | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | Unclear | Low | Yes | Yes | Included | 2 |
| 58 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 3 |
| 39 | No | Yes | No | No | No | Yes | Yes | No | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 41 | Yes | No | No | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 40 | No | No | No | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Low | Yes | Yes | Excluded | |
| 42 | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 43 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 59 | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 60 | No | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 49 | No | Yes | No | No | Yes | No | Yes | Yes | Yes | Yes | No | Low | Yes | Yes | Excluded | |
| 51 | No | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Excluded | |
| 44 | Yes | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 1 |
| 45 | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Low | Yes | Yes | Included | 3 |
| 46 | Yes | No | No | Unclear | Yes | Yes | Yes | Unclear | Yes | Yes | Yes | Low | Yes | Yes | Included | 3 |
Boldface indicates characteristics that predict high risk of bias; italics indicate uncertain bias.
The 25 eligible studies (26 strata) included a total of 2,595 patients. The number of patients enrolled varied from 8 to 218 (median, 91). The target patient population was patients with hematologic malignancies and/or HSCT recipients in 17/25 studies, bone marrow transplant recipients in 1/25, and a mixed population of patients with hematologic malignancies and other immunocompromised individuals in 7/25. Six studies performed PCR assays of serum (23%), and the rest used whole-blood assays (77%). The setting was heterogeneous regarding quality laboratory criteria. Of note, only 3 studies (all 3 using a whole-blood PCR assay) were found to be fully compliant with all EAPCRI criteria, while 7 studies (5 using whole blood and 2 using serum) were compliant with all of the criteria but one, 10 (9 using whole blood and 1 using serum) deviated by 2 criteria, and 6 (4 using whole-blood and 2 using serum) deviated by 3 or more.
Pooled diagnostic estimates.
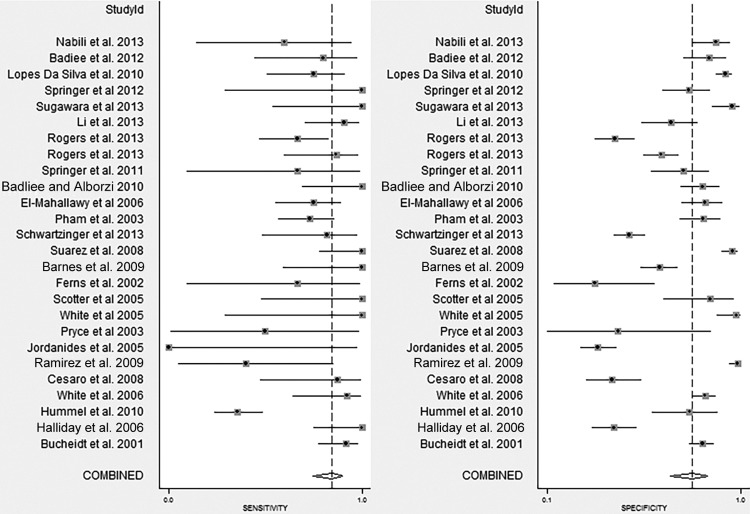
Across the 25 eligible studies, the individual sensitivity estimates ranged from 0 to 1.0, while the individual specificity estimates ranged from 0.29 to 0.98 (Fig. 2). In bivariate meta-analysis, the pooled sensitivity was 0.84 (0.75 to 0.91) and the corresponding pooled specificity was 0.76 (0.65 to 0.84) (Table 2; Fig. 3). There was no evidence of significant funnel plot asymmetry (Deeks' bias = −0.71, P = 0.9), indicating that small study effects were not present. The combined effects show that the Aspergillus sp. PCR assay has a positive LR of 3.5 and a negative LR of 0.21, suggesting that its diagnostic performance is moderate (61). Specifically, in settings where the expected pretest probability of IA is low (≤5%), PCR will yield a positive predictive value (PPV) of 0.08 (range, 0.06 to 0.11) and a negative predictive value (NPV) of 0.99 (range, 0.97 to 1.00), while in settings where IA is strongly suspected (pretest probability, ≥10%) PCR is expected to yield a PPV of 0.76 (range, 0.71 to 0.81) and an NPV of 0.71 (range, 0.66 to 0.77), respectively.
FIG 2.
Forest plot of independent sensitivity and specificity estimates. The studies cited correspond to references 25, 26, 28 to 37, 43 to 48, 50, 52 to 54, and 56 to 58.
FIG 3.
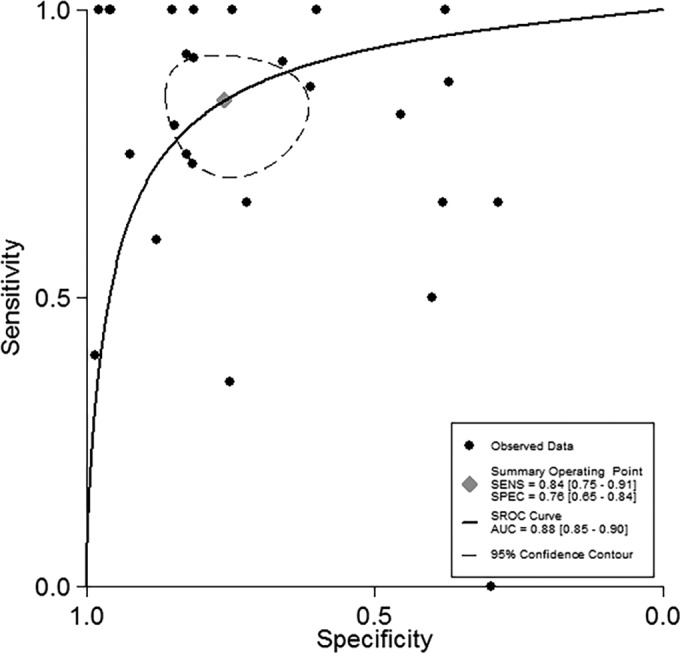
SROC curve of PCR performance.
Subgroup analysis.
We performed a subgroup analysis to evaluate changes in sensitivity and specificity caused by a series of different parameters in each study. We also assessed whether the effect of these covariates on the average sensitivity estimates were significant. Specifically, across studies where the updated EORTC/MSG 2008 criteria were implemented, the diagnostic estimates were 0.83 (range, 0.72 to 0.90) for sensitivity and 0.79 (range, 0.67 to 0.87) for specificity, respectively. These estimates did not differ from those of studies that used the EORTC/MSG 2002 criteria as the reference index. Interestingly, studies that used PCR assays of serum yielded higher specificity (85% versus 73%) and lower sensitivity (78% versus 86%) estimates than those that used whole-blood assays, but these differences did not reach statistical significance. It is of note that the average specificity of serum was significantly higher than that of whole blood without bead beating (P = 0.04).
Regarding the assay methodology used, no methodological parameter significantly affected PCR performance (Table 3). Further, when the studies that deviated by no more than one of the EAPCRI criteria were compared to studies that deviated by two or more, no significant changes in sensitivity (87% versus 82%, respectively) or specificity (77% versus 75%, respectively) were noted. Of note, we could not assess the effect of white blood cell and red blood cell lysis steps, as these were undertaken by almost all of studies in which whole blood was used. We also were unable to evaluate the effect of the use of mitochondrial versus ribosomal primers because only two studies used the former (36, 44), while the difference in PCR performance among different ribosomal primers (18S rRNA versus others) was not significant.
TABLE 3.
Results of subgroup analysisc
| Group or subgroup (no.) of studies with ≥1 positive result | Sensitivity (95% CI)d | Specificity (95% CI) | LR+/LR− |
|---|---|---|---|
| All studies (26) | 0.84 (0.75–0.91) | 0.76 (0.65–0.84) | 3.5/0.21 |
| Criteria used | |||
| 2008 (10) | 0.83 (0.72–0.90) | 0.79 (0.67–0.87) | 3.9/0.22 |
| 2002 (16) | 0.85 (0.69–0.93) | 0.74 (0.57–0.86) | 3.2/0.21 |
| Study design | |||
| Prospective (20) | 0.86 (0.77–0.92) | 0.76(0.63–0.85) | 3.5/0.18 |
| Retrospective (6) | 0.76 (0.49–0.91) | 0.77 (0.55–0.90) | 3.2/0.31 |
| Specimen type | |||
| Serum (6) | 0.78 (0.69–0.85) | 0.85 (0.70–0.93) | 5.1/0.25 |
| Whole blood (10) | 0.86 (0.73–0.93] | 0.73 (0.59–0.83) | 3.1/0.19 |
| Elution vol (μl) of: | |||
| ≤100 (13) | 0.86 (0.76–0.93) | 0.72 (0.59–0.82) | 3.1/0.19 |
| >100 (8) | 0.82 (0.54–0.95) | 0.75 (0.46–0.91) | 3.3/0.24 |
| Sample vol | |||
| Largea (18) | 0.83 (0.70–0.91) | 0.79 (0.68–0.87) | 4.0/0.21 |
| Smallb (7) | 0.89 (0.77–0.95) | 0.64 (0.38–0.83) | 2.4/0.17 |
| Internal control | |||
| Yes (19) | 0.84 (0.73–0.91) | 0.76 (0.63–0.85) | 3.4/0.21 |
| No (7) | 0.79 (0.67–0.87) | 0.77 (0.56–0.90) | 3.4/0.27 |
| Primer type | |||
| 18S (16) | 0.84 (0.69–0.93) | 0.80 (0.67–0.89) | 4.3/0.19 |
| Other ribosomal (7) | 0.85 (0.70–0.93) | 0.74 (0.57–0.86) | 3.3/0.20 |
| Bead beating (for whole blood only) | |||
| Yes (8) | 0.78 (0.62–0.89) | 0.82(0.64–0.92) | 4.3/0.27 |
| No (12) | 0.90 (0.68–0.98) | 0.64 (0.47–0.78) | 2.5/0.15 |
| Compliance with criteria | |||
| 0–1 deviation (10) | 0.87 (0.74–0.94) | 0.77 (0.60–0.88) | 3.8/0.17 |
| >1 deviation (16) | 0.82 (0.68–0.90) | 0.75 (0.60–0.86) | 3.3/0.24 |
Sample volumes used: ≥3 ml of whole blood, ≥0.5 ml of serum.
Sample volumes used: <3 ml of whole blood, <0.5 ml of serum.
No statistically significant differences were found in any comparison of subgroups.
CI, confidence interval.
Sensitivity analysis.
Thirteen of our 26 studies (12 using whole blood and 1 using serum) included data on the diagnostic performance of the PCR assay when at least two PCR results were considered positive. By analyzing the performance estimates of these studies using differential criteria for PCR positivity (one versus two or more positive PCR results), we found that PCR specificity was greatly increased (73% versus 95%, respectively) with the latter method, giving a high positive LR of 12.8, whereas sensitivity decreased (85% versus 64%, respectively). Moreover, two or more positive assays for low-risk individuals (≤5% pretest probability) give a PPV and an NPV of 0.23 and 0.99, respectively, while among high-risk patients (≥10% pretest probability), the PPV increases to 0.90 while the NPV drops to 0.62.
Interestingly, when we compared the effect of the EAPCRI recommendations on this sensitivity analysis (Table 4), we found that bead beating significantly improved the specificity of the test (96% versus 86%, metaregression P = 0.006), with a nonsignificant drop in sensitivity (50% versus73%). Further, we found that compliant studies (no more than one deviation from the EAPCRI criteria) showed higher specificity (98% versus 85%, metaregression P = 0.003) with an additional nonsignificant increase in sensitivity (67% versus 61%).
TABLE 4.
Results of sensitivity analysis
| Group or subgroup (no.) of studies with ≥2 positive results | Sensitivity (95% CI)f | Specificity (95% CI) | LR+/LR− |
|---|---|---|---|
| All studies (13) | 0.64 (0.38–0.84) | 0.95 (0.88–0.98) | 12.8/0.38 |
| Criteria used | |||
| 2008 (5) | 0.64 (0.28–0.89) | 0.96 (0.91–0.98) | 15.0/0.38 |
| 2002 (8) | 0.63 (0.27–0.89) | 0.94 (0.80–0.98) | 10.7/0.39 |
| Study design | |||
| Prospective (11) | 0.71 (0.45–0.88) | 0.95(0.87–0.98) | 13.8/0.31 |
| Retrospective (2) | NAa | NA | NA |
| Specimen type | |||
| Serum (1) | NA | NA | NA |
| Whole blood (12) | 0.62 (0.34–0.84) | 0.93 (0.86–0.97) | 9.5/0.41 |
| Elution vol (μl) of: | |||
| ≤100 (5) | 0.74 (0.31–0.95) | 0.95 (0.90–0.97) | 14.1/0.28 |
| >100 (6) | 0.68 (0.31–0.91) | 0.95 (0.71–0.99) | 14.2/0.34 |
| Sample vol | |||
| Largeb (8) | 0.45 (0.18–0.75) | 0.97 (0.89–0.99) | 15.2/0.57 |
| Smallc (5) | 0.83 (0.57–0.95) | 0.89 (0.76–0.95) | 7.6/0.19 |
| Internal control | |||
| Yes (11) | 0.66 (0.34–0.88) | 0.95 (0.89–0.98) | 14.5/0.36 |
| No (2) | NA | NA | NA |
| Primer type | |||
| 18S (8) | 0.64 (0.25–0.91) | 0.92 (0.80–0.97) | 8.0/0.39 |
| Other ribosomal (5) | 0.64 (0.34–0.86) | 0.98 (0.88–1.0) | 27.4/0.37 |
| Bead beating (for whole blood only) | |||
| Yes (6)d | 0.50(0.25–0.76) | 0.96(0.92–0.98) | 13.7/0.51 |
| No (6) | 0.73 (0.16–0.97) | 0.86 (0.71–0.94) | 5.1/0.31 |
| Compliance with criteria | |||
| 0–1 deviation (7)e | 0.67 (0.34–0.89) | 0.98 (0.93–0.99) | 28.1/0.33 |
| >1 deviation (6) | 0.61 (0.23–0.89) | 0.85(0.71–0.93) | 4.2/0.46 |
NA, not applicable (too few studies to pool).
Sample volumes used: <3 ml of whole blood, <0.5 ml of serum.
Sample volumes used: ≥3 ml of whole blood, ≥0.5 ml of serum.
Average specificity significantly improves for bead beating (P = 0.006).
Average specificity significantly improves for the more compliant studies (P = 0.003). All other subgroup comparisons not significant.
CI, confidence interval.
DISCUSSION
Despite the fact that multiple Aspergillus sp. PCR assays of blood specimens are now available, a consensual conclusion about the role that this test can play in the diagnosis of IA has yet to be reached. Shortly after its introduction in the 1990s, it was thought that this method could, in fact, revolutionize the diagnosis and management of this severe disease (62). However, the multitude of clinical studies performed since then failed to prove beyond doubt whether the test can rule an active infection in or out, thus often leaving clinicians baffled when trying to interpret a positive or negative result. Our findings indicate that whole-blood and serum Aspergillus sp. PCR assays have moderate diagnostic performance, which suggests that a positive or negative result is unable, on its own, to confirm or exclude a suspected infection in high-risk patients. We should note, however, that the pooled PCR performance estimates in our study were not inferior to the performance estimates of serum galactomannan (sensitivity, 71%; specificity, 89%) (63) or beta-glucan (sensitivity, 76.8%; specificity, 85.3%) (64) tests obtained by previous meta-analyses.
Our choice to define as PCR positive all episodes that had at least one PCR-positive specimen, coupled with the fact that most of the studies tested more than one specimen per episode, may have overestimated the average sensitivity and underestimated the specificity of the method. To see whether this is true, we performed a sensitivity analysis of studies that had extractable data about at least two versus one PCR-positive specimen per patient. Indeed, we found that PCR specificity was increased dramatically by this approach to 95%, which leads to a PPV of 90% for high-risk individuals. Therefore, the presence of at least two positive whole-blood PCR specimens in a high-risk patient should be considered very indicative, if not confirmatory, of IA. On the other hand, the sensitivity of this approach was lower at 64%, which suggests that it would be more valuable as a confirmatory tool than as a screening tool. Although our results are in accordance with previous reports (8), we observed a significant variation of results reported by individual studies involved in our analysis, which led us to search for reasons behind these inconsistencies.
One of the potential explanations is the choice of the specimen, namely, whole blood or serum, with which the PCR assay is performed. Two previous clinical studies comparing Aspergillus sp. PCR assays performed with serum and whole blood from the same high-risk patients failed to show significant differences in accuracy (49, 51), with one study suggesting a nonsignificant trend toward increased sensitivity and reduced specificity of the whole-blood PCR assay (51). In accordance with these studies, our results suggest that PCR assays performed with serum had nonsignificantly lower sensitivity and higher specificity than those performed with whole blood. Taking into consideration the facts that a serum PCR assay is faster and easier to perform and that serum can also be used for other biomarker diagnostic tests, such as galactomannan, serum has the potential to be the preferred specimen for PCR testing.
Another potential moderator of effect among different trials could be the use of different versions of the EORTC/MSG criteria for the diagnosis of IA. Compared to the old version, the revised criteria (7) kept the terminology but expanded the definition of probable while reducing the scope of possible IA. The adoption of the new classification may have an unclear impact on outcomes in future trials. Indeed, a recent retrospective evaluation stated that the majority of “possible” IA cases can be downgraded to “unclassifiable” but also probable cases showed 75% reductions (65). While this impact may or may not hold true in clinical practice, our data indicated that PCR performance was unaffected.
Finally, the use of different methodological parameters in DNA extraction and amplification is considered one of the most important sources of inconsistencies between different trials (66). To circumvent this problem and standardize Aspergillus sp. PCR assays among different laboratories, the EAPCRI has recently issued a series of recommendations on how to perform Aspergillus PCR assays with whole blood and serum on the basis of the results of two multicenter studies with spiked samples (12, 13). These include the use of a larger sample volume, smaller elution volumes, an internal control, and thorough cell lysis with bead beating and enzymatic white and red blood cell lysis steps for whole blood. Notably, in the clinical setting we studied, none of these methodological characteristics significantly altered the sensitivity or specificity estimates in our primary analysis.
The interpretation of this finding is challenging, mainly because we do not know in what form Aspergillus spp. circulate in the blood. Of note, many studies suggest that most of the circulating Aspergillus DNA is present not in the form of conidia but rather as free DNA (67, 68). This provides an explanation for the fact that blood cultures have such a low sensitivity for aspergillosis (69), as well as our finding that whole-blood testing is not superior to serum testing. This would also explain why sequential cell lysis methods could not improve PCR assay performance for patients with IA, in contrast to what would be expected for samples spiked with Aspergillus sp. conidia (13). In addition, it is unclear whether any form of Aspergillus DNA can be found in the circulation at all times during an active infection. As suggested by a recent in vitro study, release of DNA is intermittent and happens only during certain stages of fungal growth (68). If, in fact, circulating fungal DNA is released intermittently only during mycelial breakdown, it is plausible that its levels in the blood would have such a wide fluctuation that it could make differences in sample size or elution volume relatively insignificant.
Nevertheless, it is of note that compliance with the EAPCRI recommendations significantly increased the specificity of PCR and was also accompanied by an increase in sensitivity when at least two positive results were used to define PCR positivity. An explanation for this seemingly contradictory finding would be that other factors that we were unable to assess (such as PCR testing algorithms and the use of prophylactic antifungal agents) could have served as potential confounders in the assessment of the effect of the EAPCRI recommendations on our primary analysis, and those factors should be evaluated in future trials. Compliance with these simple methodological guidelines should, in fact, be encouraged since it would allow interlaboratory comparisons and has the potential to improve PCR performance in clinical practice.
Several systematic reviews of Aspergillus sp. PCR assays of bronchoalveolar lavage fluid have been previously performed, with very promising findings (70). However, despite the large number of new studies on the issue, to our knowledge, there is only one meta-analysis of whole-blood and serum PCR performance to date. That study, by Mengoli et al. (8), reported that PCR assay performance is moderate but specificity can be increased when two PCR results are used to define a positive PCR finding. That study was published before the EAPCRI recommendations were issued and included a smaller number of trials, so the authors could not reach any conclusions regarding the performance of different PCR assay protocols. It is of note that our study confirms the finding that a single positive PCR assay result has moderate diagnostic accuracy, whereas a strategy that uses at least two positive PCR assay results per suspicious episode to define positivity is able to achieve superior specificity. Moreover, by investigating the effects of different protocols on PCR assay performance, our study shows that a serum PCR assay is not inferior to a whole-blood PCR assay and that compliance with the EAPCRI recommendations has the potential to improve PCR assay performance in clinical practice.
Important limitations of our study are the facts that (i) only a subset of the studies included had extractable data regarding PCR assay performance for at least two positive specimens and (ii) the vast majority of these studies used whole blood and not serum as the test specimen. Therefore, the results of this sensitivity analysis should be interpreted with caution. Also, all of the studies included in our analysis enrolled individuals who had a high risk of invasive fungal infection, as this is the patient population for which the Aspergillus sp. PCR test will be the most valuable. Therefore, our performance estimates may not be accurate for the general population.
In summary, for high-risk individuals, two whole-blood or serum specimens PCR assay positive for IA should be considered very indicative, if not confirmatory, of a clinically suspected infection. Given the superior specificity of this approach, it has the potential to be used along with other circulating biomarker detection assays, such as galactomannan, as a criterion to define a probable infection. Compliance with the EAPCRI Aspergillus sp. PCR assay recommendations can further increase the specificity of this approach and will allow better interlaboratory comparisons. However, given the complexity of IA diagnosis, it is unlikely that a single noninvasive test will be able to be used alone in clinical decision making. Consequently, the focus of future clinical trials should include the development of decision algorithms that would take into account multiple parameters to guide the management of high-risk groups. In this context, serum and whole-blood PCR assays have a concrete potential to improve our ability to detect and diagnose IA.
ACKNOWLEDGMENTS
A.M.C. is an active member of the Aspergillus Technology Consortium (AsTeC) group.
A.M.C. and E.M. have received financial support from T2 Biosystems for research unrelated to this report. The rest of us have no potential conflicts of interest to report.
Footnotes
Published ahead of print 13 August 2014
REFERENCES
- 1.Ramos ER, Jiang Y, Hachem R, Kassis C, Kontoyiannis DP, Raad I. 2011. Outcome analysis of invasive aspergillosis in hematologic malignancy and hematopoietic stem cell transplant patients: the role of novel antimold azoles. Oncologist 16:1049–1060. 10.1634/theoncologist.2010-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omer AK, Ziakas PD, Anagnostou T, Coughlin E, Kourkoumpetis T, McAfee SL, Dey BR, Attar E, Chen YB, Spitzer TR, Mylonakis E, Ballen KK. 2013. Risk factors for invasive fungal disease after allogeneic hematopoietic stem cell transplantation: a single center experience. Biol. Blood Marrow Transplant. 19:1190–1196. 10.1016/j.bbmt.2013.05.018 [DOI] [PubMed] [Google Scholar]
- 3.Ziakas PD, Kourbeti IS, Voulgarelis M, Mylonakis E. 2010. Effectiveness of systemic antifungal prophylaxis in patients with neutropenia after chemotherapy: a meta-analysis of randomized controlled trials. Clin. Ther. 32:2316–2336. 10.1016/j.clinthera.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 4.Ziakas PD, Kourbeti IS, Mylonakis E. 2014. Systemic antifungal prophylaxis after hematopoietic stem cell transplantation: a meta-analysis. Clin. Ther. 36:292–306 e291. 10.1016/j.clinthera.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 5.Morrissey CO, Chen SC, Sorrell TC, Milliken S, Bardy PG, Bradstock KF, Szer J, Halliday CL, Gilroy NM, Moore J, Schwarer AP, Guy S, Bajel A, Tramontana AR, Spelman T, Slavin MA, Australasian Leukaemia Lymphoma Group, Australia and New Zealand Mycology Interest Group 2013. Galactomannan and PCR versus culture and histology for directing use of antifungal treatment for invasive aspergillosis in high-risk haematology patients: a randomised controlled trial. Lancet Infect. Dis. 13:519–528. 10.1016/S1473-3099(13)70076-8 [DOI] [PubMed] [Google Scholar]
- 6.Ascioglu S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, Denning DW, Donnelly JP, Edwards JE, Erjavec Z, Fiere D, Lortholary O, Maertens J, Meis JF, Patterson TF, Ritter J, Selleslag D, Shah PM, Stevens DA, Walsh TJ, Invasive Fungal Infections Cooperative Group of the European Organization for Research and Treatment of Cancerr, Mycoses Study Group of the National Institute of Allergy and Infectious Diseases 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7–14. 10.1086/323335 [DOI] [PubMed] [Google Scholar]
- 7.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821. 10.1086/588660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. 2009. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9:89–96. 10.1016/S1473-3099(09)70019-2 [DOI] [PubMed] [Google Scholar]
- 9.Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. 2014. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin. Microbiol. Rev. 27:490–526. 10.1128/CMR.00091-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leeflang MM, Debets-Ossenkopp YJ, Visser CE, Scholten RJ, Hooft L, Bijlmer HA, Reitsma JB, Bossuyt PM, Vandenbroucke-Grauls CM. 2008. Galactomannan detection for invasive aspergillosis in immunocompromized [sic] patients. Cochrane Database Syst. Rev. 10.1002/14651858.CD007394 [DOI] [PubMed] [Google Scholar]
- 12.White PL, Bretagne S, Klingspor L, Melchers WJ, McCulloch E, Schulz B, Finnstrom N, Mengoli C, Barnes RA, Donnelly JP, Loeffler J, European Aspergillus PCR Initiative 2010. Aspergillus PCR: one step closer to standardization. J. Clin. Microbiol. 48:1231–1240. 10.1128/JCM.01767-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White PL, Mengoli C, Bretagne S, Cuenca-Estrella M, Finnstrom N, Klingspor L, Melchers WJ, McCulloch E, Barnes RA, Donnelly JP, Loeffler J, European Aspergillus PCR Initiative 2011. Evaluation of Aspergillus PCR protocols for testing serum specimens. J. Clin. Microbiol. 49:3842–3848. 10.1128/JCM.05316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, QUADAS-2 Group 2011. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155:529–536. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 15.Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. 2005. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol. 58:982–990. 10.1016/j.jclinepi.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 16.Mant J, Doust J, Roalfe A, Barton P, Cowie MR, Glasziou P, Mant D, McManus RJ, Holder R, Deeks J, Fletcher K, Qume M, Sohanpal S, Sanders S, Hobbs FD. 2009. Systematic review and individual patient data meta-analysis of diagnosis of heart failure, with modelling of implications of different diagnostic strategies in primary care. Health Technol. Assess. 13:1–207, iii. 10.3310/hta13320 [DOI] [PubMed] [Google Scholar]
- 17.Harbord RM, Whiting P, Sterne JA, Egger M, Deeks JJ, Shang A, Bachmann LM. 2008. An empirical comparison of methods for meta-analysis of diagnostic accuracy showed hierarchical models are necessary. J. Clin. Epidemiol. 61:1095–1103. 10.1016/j.jclinepi.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 18.Grimes DA, Schulz KF. 2005. Refining clinical diagnosis with likelihood ratios. Lancet 365:1500–1505. 10.1016/S0140-6736(05)66422-7 [DOI] [PubMed] [Google Scholar]
- 19.Deeks JJ, Macaskill P, Irwig L. 2005. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 58:882–893. 10.1016/j.jclinepi.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 20.Prodromou ML, Ziakas PD, Poulou LS, Karsaliakos P, Thanos L, Mylonakis E. 2014. FDG PET is a robust tool for the diagnosis of spondylodiscitis: a meta-analysis of diagnostic data. Clin. Nucl. Med. 39:330–335. 10.1097/RLU.0000000000000336 [DOI] [PubMed] [Google Scholar]
- 21.Ziakas PD, Poulou LS, Voulgarelis M, Thanos L. 2012. The Gordian knot of interim 18-fluorodeoxyglucose positron emission tomography for Hodgkin lymphoma: a meta-analysis and commentary on published studies. Leuk. Lymphoma 53:2166–2174. 10.3109/10428194.2012.685730 [DOI] [PubMed] [Google Scholar]
- 22.Dwamena BA. 2009. Evidence-based radiology: step 3—diagnostic systematic review and meta-analysis (critical appraisal). Semin. Roentgenol. 44:170–179. 10.1053/j.ro.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 23.Wallace BC, Schmid CH, Lau J, Trikalinos TA. 2009. Meta-Analyst: software for meta-analysis of binary, continuous and diagnostic data. BMC Med. Res. Methodol. 9:80. 10.1186/1471-2288-9-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raad I, Hanna H, Sumoza D, Albitar M. 2002. Polymerase chain reaction on blood for the diagnosis of invasive pulmonary aspergillosis in cancer patients. Cancer 94:1032–1036. 10.1002/cncr.10349 [DOI] [PubMed] [Google Scholar]
- 25.Buchheidt D, Baust C, Skladny H, Ritter J, Suedhoff T, Baldus M, Seifarth W, Leib-Moesch C, Hehlmann R. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428–435. 10.1086/321887 [DOI] [PubMed] [Google Scholar]
- 26.Halliday C, Hoile R, Sorrell T, James G, Yadav S, Shaw P, Bleakley M, Bradstock K, Chen S. 2006. Role of prospective screening of blood for invasive aspergillosis by polymerase chain reaction in febrile neutropenic recipients of haematopoietic stem cell transplants and patients with acute leukaemia. Br. J. Haematol. 132:478–486. 10.1111/j.1365-2141.2005.05887.x [DOI] [PubMed] [Google Scholar]
- 27.Badiee P, Kordbacheh P, Alborzi A, Ramzi M, Shakiba E. 2008. Molecular detection of invasive aspergillosis in hematologic malignancies. Infection 36:580–584. 10.1007/s15010-008-7385-8 [DOI] [PubMed] [Google Scholar]
- 28.Hummel M, Spiess B, Cornely OA, Dittmer M, Morz H, Buchheidt D. 2010. Aspergillus PCR testing: results from a prospective PCR study within the AmBiLoad trial. Eur. J. Haematol. 85:164–169. 10.1111/j.1600-0609.2010.01452.x [DOI] [PubMed] [Google Scholar]
- 29.White PL, Linton CJ, Perry MD, Johnson EM, Barnes RA. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42:479–486. 10.1086/499949 [DOI] [PubMed] [Google Scholar]
- 30.Cesaro S, Stenghele C, Calore E, Franchin E, Cerbaro I, Cusinato R, Tridello G, Manganelli R, Carli M, Palu G. 2008. Assessment of the LightCycler PCR assay for diagnosis of invasive aspergillosis in paediatric patients with onco-haematological diseases. Mycoses 51:497–504. 10.1111/j.1439-0507.2008.01512.x [DOI] [PubMed] [Google Scholar]
- 31.Ramírez M, Castro C, Palomares JC, Torres MJ, Aller AI, Ruiz M, Aznar J, Martin-Mazuelos E. 2009. Molecular detection and identification of Aspergillus spp. from clinical samples using real-time PCR. Mycoses 52:129–134. 10.1111/j.1439-0507.2008.01548.x [DOI] [PubMed] [Google Scholar]
- 32.Jordanides NE, Allan EK, McLintock LA, Copland M, Devaney M, Stewart K, Parker AN, Johnson PR, Holyoake TL, Jones BL. 2005. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplant. 35:389–395. 10.1038/sj.bmt.1704768 [DOI] [PubMed] [Google Scholar]
- 33.Pryce TM, Kay ID, Palladino S, Heath CH. 2003. Real-time automated polymerase chain reaction (PCR) to detect Candida albicans and Aspergillus fumigatus DNA in whole blood from high-risk patients. Diagn. Microbiol. Infect. Dis. 47:487–496. 10.1016/S0732-8893(03)00139-1 [DOI] [PubMed] [Google Scholar]
- 34.White PL, Archer AE, Barnes RA. 2005. Comparison of non-culture-based methods for detection of systemic fungal infections, with an emphasis on invasive Candida infections. J. Clin. Microbiol. 43:2181–2187. 10.1128/JCM.43.5.2181-2187.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scotter JM, Campbell P, Anderson TP, Murdoch DR, Chambers ST, Patton WN. 2005. Comparison of PCR-ELISA and galactomannan detection for the diagnosis of invasive aspergillosis. Pathology 37:246–253. 10.1080/00313020500099148 [DOI] [PubMed] [Google Scholar]
- 36.Ferns RB, Fletcher H, Bradley S, Mackinnon S, Hunt C, Tedder RS. 2002. The prospective evaluation of a nested polymerase chain reaction assay for the early detection of Aspergillus infection in patients with leukaemia or undergoing allograft treatment. Br. J. Haematol. 119:720–725. 10.1046/j.1365-2141.2002.03862.x [DOI] [PubMed] [Google Scholar]
- 37.Barnes RA, White PL, Bygrave C, Evans N, Healy B, Kell J. 2009. Clinical impact of enhanced diagnosis of invasive fungal disease in high-risk haematology and stem cell transplant patients. J. Clin. Pathol. 62:64–69. 10.1136/jcp.2008.058354 [DOI] [PubMed] [Google Scholar]
- 38.Florent M, Katsahian S, Vekhoff A, Levy V, Rio B, Marie JP, Bouvet A, Cornet M. 2006. Prospective evaluation of a polymerase chain reaction-ELISA targeted to Aspergillus fumigatus and Aspergillus flavus for the early diagnosis of invasive aspergillosis in patients with hematological malignancies. J. Infect. Dis. 193:741–747. 10.1086/500466 [DOI] [PubMed] [Google Scholar]
- 39.Costa C, Costa JM, Desterke C, Botterel F, Cordonnier C, Bretagne S. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224–2227. 10.1128/JCM.40.6.2224-2227.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millon L, Piarroux R, Deconinck E, Bulabois CE, Grenouillet F, Rohrlich P, Costa JM, Bretagne S. 2005. Use of real-time PCR to process the first galactomannan-positive serum sample in diagnosing invasive aspergillosis. J. Clin. Microbiol. 43:5097–5101. 10.1128/JCM.43.10.5097-5101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawazu M, Kanda Y, Nannya Y, Aoki K, Kurokawa M, Chiba S, Motokura T, Hirai H, Ogawa S. 2004. Prospective comparison of the diagnostic potential of real-time PCR, double-sandwich enzyme-linked immunosorbent assay for galactomannan, and a (1→3)-beta-d-glucan test in weekly screening for invasive aspergillosis in patients with hematological disorders. J. Clin. Microbiol. 42:2733–2741. 10.1128/JCM.42.6.2733-2741.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Botterel F, Farrugia C, Ichai P, Costa JM, Saliba F, Bretagne S. 2008. Real-time PCR on the first galactomannan-positive serum sample for diagnosing invasive aspergillosis in liver transplant recipients. Transpl. Infect. Dis. 10:333–338. 10.1111/j.1399-3062.2008.00323.x [DOI] [PubMed] [Google Scholar]
- 43.Suarez F, Lortholary O, Buland S, Rubio MT, Ghez D, Mahe V, Quesne G, Poiree S, Buzyn A, Varet B, Berche P, Bougnoux ME. 2008. Detection of circulating Aspergillus fumigatus DNA by real-time PCR assay of large serum volumes improves early diagnosis of invasive aspergillosis in high-risk adult patients under hematologic surveillance. J. Clin. Microbiol. 46:3772–3777. 10.1128/JCM.01086-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzinger M, Sagaon-Teyssier L, Cabaret O, Bretagne S, Cordonnier C, Investigators P. 2013. Performance of serum biomarkers for the early detection of invasive aspergillosis in febrile, neutropenic patients: a multi-state model. PLoS One 8(6):e65776. 10.1371/journal.pone.0065776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham AS, Tarrand JJ, May GS, Lee MS, Kontoyiannis DP, Han XY. 2003. Diagnosis of invasive mold infection by real-time quantitative PCR. Am. J. Clin. Pathol. 119:38–44. 10.1309/RQ05PP9NEG6DADXR [DOI] [PubMed] [Google Scholar]
- 46.El-Mahallawy HA, Shaker HH, Ali Helmy H, Mostafa T, Razak Abo-Sedah A. 2006. Evaluation of pan-fungal PCR assay and Aspergillus antigen detection in the diagnosis of invasive fungal infections in high risk paediatric cancer patients. Med. Mycol. 44:733–739. 10.1080/13693780600939955 [DOI] [PubMed] [Google Scholar]
- 47.Badiee P, Alborzi A. 2010. Detection of Aspergillus species in bone marrow transplant patients. J. Infect. Dev. Ctries. 4:511–516. 10.3855/jidc.807 [DOI] [PubMed] [Google Scholar]
- 48.Springer J, Loeffler J, Heinz W, Schlossnagel H, Lehmann M, Morton O, Rogers TR, Schmitt C, Frosch M, Einsele H, Kurzai O. 2011. Pathogen-specific DNA enrichment does not increase sensitivity of PCR for diagnosis of invasive aspergillosis in neutropenic patients. J. Clin. Microbiol. 49:1267–1273. 10.1128/JCM.01679-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernal-Martínez L, Gago S, Buitrago MJ, Gomez-Lopez A, Rodriguez-Tudela JL, Cuenca-Estrella M. 2011. Analysis of performance of a PCR-based assay to detect DNA of Aspergillus fumigatus in whole blood and serum: a comparative study with clinical samples. J. Clin. Microbiol. 49:3596–3599. 10.1128/JCM.00647-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Springer J, Schlossnagel H, Heinz W, Doedt T, Soeller R, Einsele H, Loeffler J. 2012. A novel extraction method combining plasma with a whole-blood fraction shows excellent sensitivity and reproducibility for patients at high risk for invasive aspergillosis. J. Clin. Microbiol. 50:2585–2591. 10.1128/JCM.00523-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer J, Morton CO, Perry M, Heinz WJ, Paholcsek M, Alzheimer M, Rogers TR, Barnes RA, Einsele H, Loeffler J, White PL. 2013. Multicenter comparison of serum and whole-blood specimens for detection of Aspergillus DNA in high-risk hematological patients. J. Clin. Microbiol. 51:1445–1450. 10.1128/JCM.03322-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers TR, Morton CO, Springer J, Conneally E, Heinz W, Kenny C, Frost S, Einsele H, Loeffler J. 2013. Combined real-time PCR and galactomannan surveillance improves diagnosis of invasive aspergillosis in high risk patients with haematological malignancies. Br. J. Haematol. 161:517–524. 10.1111/bjh.12285 [DOI] [PubMed] [Google Scholar]
- 53.Sugawara Y, Nakase K, Nakamura A, Ohishi K, Sugimoto Y, Fujieda A, Monma F, Suzuki K, Masuya M, Matsushima Y, Wada H, Nobori T, Katayama N. 2013. Clinical utility of a panfungal polymerase chain reaction assay for invasive fungal diseases in patients with haematologic disorders. Eur. J. Haematol. 90:331–339. 10.1111/ejh.12078 [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Gao L, Ding Y, Xu Y, Zhou M, Huang W, Jing Y, Li H, Wang L, Yu L. 2013. Establishment and application of real-time quantitative PCR for diagnosing invasive aspergillosis via the blood in hematological patients: targeting a specific sequence of Aspergillus 28S-ITS2. BMC Infect. Dis. 13:255. 10.1186/1471-2334-13-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White PL, Parr C, Thornton C, Barnes RA. 2013. Evaluation of real-time PCR, galactomannan enzyme-linked immunosorbent assay (ELISA), and a novel lateral-flow device for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 51:1510–1516. 10.1128/JCM.03189-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nabili M, Shokohi T, Janbabaie G, Hashemi-Soteh MB, Ali-Moghaddam K, Aghili SR. 2013. Detection of invasive aspergillosis in bone marrow transplant recipients using real-time PCR. J. Glob. Infect. Dis. 5:68–75. 10.4103/0974-777X.112296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopes da Silva R, Ribeiro P, Abreu N, Ferreira T, Fernandes T, Monteiro A, Costa F, Caldas J, Silva M, Carande L, Ferreira G, Conduto A, Cruz E, Sousa MH, Rodrigues AS, Costa I, Veiga J, de Sousa AB. 2010. Early diagnosis of invasive aspergillosis in neutropenic patients. Comparison between serum galactomannan and polymerase chain reaction. Clin. Med. Insights Oncol. 4:81–88. 10.4137/CMO.S5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badiee P, Alborzi A, Karimi M, Pourabbas B, Haddadi P, Mardaneh J, Moieni M. 2012. Diagnostic potential of nested PCR, galactomannan EIA, and beta-d-glucan for invasive aspergillosis in pediatric patients. J. Infect. Dev. Ctries. 6:352–357. 10.3855/jidc.2110 [DOI] [PubMed] [Google Scholar]
- 59.Millon L, Grenouillet F, Legrand F, Loewert S, Bellanger AP, Gbaguidi-Haore H, Scherer E, Henon T, Rohrlich P, Deconinck E. 2011. Ribosomal and mitochondrial DNA target for real-time PCR diagnosis of invasive aspergillosis. J. Clin. Microbiol. 49:1058–1063. 10.1128/JCM.01904-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White PL, Perry MD, Moody A, Follett SA, Morgan G, Barnes RA. 2011. Evaluation of analytical and preliminary clinical performance of Myconostica MycAssay Aspergillus when testing serum specimens for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 49:2169–2174. 10.1128/JCM.00101-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deeks JJ, Altman DG. 2004. Diagnostic tests 4: likelihood ratios. BMJ 329:168–169. 10.1136/bmj.329.7458.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. 1996. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J. Clin. Microbiol. 34:2464–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfeiffer CD, Fine JP, Safdar N. 2006. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta-analysis. Clin. Infect. Dis. 42:1417–1427. 10.1086/503427 [DOI] [PubMed] [Google Scholar]
- 64.Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. 2011. Beta-d-glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin. Infect. Dis. 52:750–770. 10.1093/cid/ciq206 [DOI] [PubMed] [Google Scholar]
- 65.Tsitsikas DA, Morin A, Araf S, Murtagh B, Johnson G, Vinnicombe S, Ellis S, Suaris T, Wilks M, Doffman S, Agrawal SG. 2012. Impact of the revised (2008) EORTC/MSG definitions for invasive fungal disease on the rates of diagnosis of invasive aspergillosis. Med. Mycol. 50:538–542. 10.3109/13693786.2011.630040 [DOI] [PubMed] [Google Scholar]
- 66.Blennow O, Mattsson J. 2011. Standardization of Aspergillus PCR is needed. Bone Marrow Transplant. 46:1018. 10.1038/bmt.2010.220 [DOI] [PubMed] [Google Scholar]
- 67.Loeffler J, Kloepfer K, Hebart H, Najvar L, Graybill JR, Kirkpatrick WR, Patterson TF, Dietz K, Bialek R, Einsele H. 2002. Polymerase chain reaction detection of Aspergillus DNA in experimental models of invasive aspergillosis. J. Infect. Dis. 185:1203–1206. 10.1086/339824 [DOI] [PubMed] [Google Scholar]
- 68.Mennink-Kersten MA, Ruegebrink D, Wasei N, Melchers WJ, Verweij PE. 2006. In vitro release by Aspergillus fumigatus of galactofuranose antigens, 1,3-beta-d-glucan, and DNA, surrogate markers used for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 44:1711–1718. 10.1128/JCM.44.5.1711-1718.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simoneau E, Kelly M, Labbe AC, Roy J, Laverdiere M. 2005. What is the clinical significance of positive blood cultures with Aspergillus sp. in hematopoietic stem cell transplant recipients? A 23 year experience. Bone Marrow Transplant. 35:303–306. 10.1038/sj.bmt.1704793 [DOI] [PubMed] [Google Scholar]
- 70.Sun W, Wang K, Gao W, Su X, Qian Q, Lu X, Song Y, Guo Y, Shi Y. 2011. Evaluation of PCR on bronchoalveolar lavage fluid for diagnosis of invasive aspergillosis: a bivariate metaanalysis and systematic review. PLoS One 6(12):e28467. 10.1371/journal.pone.0028467 [DOI] [PMC free article] [PubMed] [Google Scholar]