Abstract
The identification of mycobacteria outside biocontainment facilities requires that the organisms first be rendered inactive. Exposure to 70% ethanol (EtOH) either before or after mechanical disruption was evaluated in order to establish a safe, effective, and rapid inactivation protocol that is compatible with identification of Mycobacterium and Nocardia species using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). A combination of 5 min of bead beating in 70% EtOH followed by a 10-min room temperature incubation period was found to be rapidly bactericidal and provided high-quality spectra compared to spectra obtained directly from growth on solid media. The age of the culture, the stability of the refrigerated or frozen lysates, and freeze-thaw cycles did not adversely impact the quality of the spectra or the identification obtained.
INTRODUCTION
Different approaches have been developed to hasten the identification of clinically significant mycobacteria compared to traditional biochemical phenotyping. These include DNA probe hybridization- and nucleic acid amplification-based assays with or without sequencing (1–4) and high-performance liquid chromatography (HPLC) to evaluate the mycolic acid composition (1, 2). More recently, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been shown to be a reliable and rapid means for the identification of bacterial species, including mycobacteria through the recognition of specific proteins (5), PCR amplification profiles (6), or protein spectral profiles (7, 8). Advanced databases for use with the latter approach are currently being expanded. Regardless of which option is used, it is essential that cultures are rendered nonviable prior to processing outside appropriate biocontainment facilities for obvious safety reasons. Previous reports have identified a variety of ways in which mycobacteria, including Mycobacterium tuberculosis can be successfully inactivated prior to manipulation, including heat treatment (9), exposure to lethal irradiation with or without photosensitizing chemicals (10–12), alcohol exposure (13), or a combination of treatments (14–16). While most were successful, temperatures and/or exposure times varied in order to achieve uniform cell death. Treatment with disinfecting agents such as 2% glutaraldehyde was not considered an option as it can result in extensive cross-linking of the nucleic acids, proteins, and other cellular components that are necessary for the MALDI-TOF MS identification process. Within the past year, two publications described the successful adaptation of a mycobacterial inactivation/extraction protocol for use with MALDI-TOF MS-based identification (17, 18). Herein, we report a detailed description of an even more convenient process that was first presented in part in 2013 (19).
MATERIALS AND METHODS
All studies involving mycobacteria were performed in a biological safety level 3 (BSL-3) laboratory. For various aspects of this study, 28 strains encompassing 13 Mycobacterium species, including five strains of M. tuberculosis with varied isoniazid (INH) and rifampin (RIF) susceptibility were selected (Table 1). In addition, five species of Nocardia (N. cyriacigeorgica, 9 strains; N. farcinica, 4 strains; N. kruczakiae, 2 strains; N. nova, 7 strains; and N. otitidiscaviarum, 1 strain) were included. For the inactivation studies, mycobacteria were harvested from solid media: Middlebrook 7H11 (Remel, Lenexa, KS, USA) or Lowenstein-Jensen (LJ) medium or LJ medium with ferric ammonium sulfate (Remel). Nocardia strains were cultured on sheep blood agar until visible growth was detected. To provide an estimate of organism load for mycobacteria, a calibrated loopful (1 or 10 μl) taken from visible colonies was inoculated into a glass tube containing 2 ml Middlebrook 7H9 broth (Remel) and 1.5 to 2.0 cm sterile Ottawa sand (part NC0190353; Fisher Scientific, Waltham, MA, USA). The tube was vortexed for 15 to 30 s to break up clumps of cells and allowed to equilibrate for 5 min at room temperature to permit sand particles to settle out of suspension. The absorbance at 600 nm (A660 nm) was then determined for each tube after which serial 1:10 dilutions were performed in 7H9 broth, and 0.5 ml of the 10−5, 10−6, and 10−7 dilutions were uniformly spread over the surface of 7H11 plates in duplicate. Plates were incubated with 5% CO2 at 37°C, and colonies were counted after visible growth was obtained. This procedure was performed in duplicate using both sizes of calibrated loops for eight strains from six species of mycobacteria. The only modification to the protocol for testing Nocardia involved use of a sterile cytobrush (Puritan, Guilford, ME, USA) to harvest growth in place of inoculating loops. Estimates of CFU/ml were not determined for Nocardia species, but a visibly turbid suspension equal to or greater than a 0.5 McFarland standard was used.
TABLE 1.
Growth results of mycobacterial test strains following inactivation strategies including beat beating either before or after a 10-min exposure to 500 μl of 70% EtOH or exposure to EtOH alone without bead beating for 15 or 30 mina
| Mycobacterium species | Strain | Growth results of test strains after: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 5-min bead beating before ethanol incubation and 10 min inactivation of inoculum loop per strain (μl) of: |
5-min bead beating after ethanol incubation for 10 min of inoculum loop per strain (μl) of: |
No bead beating and 15-min inactivation of inoculum loop per strain (μl) of: |
No bead beating and 30-min inactivation of inoculum loop per strain (μl) of: |
||||||
| 1 | 10 | 1 | 10 | 1 | 10 | 1 | 10 | ||
| M. abscessus | 19977b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. abscessus | 31313c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. abscessus | 23003b | NG | NG | NG | NG | NG | G | NG | NG |
| M. avium | 25291b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. avium | 1155c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. chelonae | 19235b | NG | NG | G | NG | NG | NG | NG | G |
| M. chelonae | 35749b | NG | NG | NG | NG | NG | NG | NG | G |
| M. fortuitum | 6841b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. fortuitum | 4031c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. fortuitum | MF89d | NG | NG | NG | NG | NG | NG | NG | G |
| M. genavense | NLA001200214c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. genavense | NLA001300504c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. gordonae | 35756b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. haemophilum | NLA000701523c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. intracellulare | 13950b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. intracellulare | 789c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. intracellulare | 1235c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. kansasii | 864c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. kansasii | 12478b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. kansasii | 92-733d | NG | NG | NG | NG | NG | NG | NG | NG |
| M. senegalense | 2301c | NG | NG | NG | NG | NG | NG | NG | NG |
| M. scrofulaceum | 1998b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. smegmatis | 11759b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. tuberculosise | S-07d | NG | NG | NG | NG | NG | NG | NG | NG |
| M. tuberculosisf | S-09d | NG | NG | NG | NG | NG | NG | NG | NG |
| M. tuberculosisg | S-38d | NG | NG | NG | NG | NG | NG | NG | NG |
| M. tuberculosish | 35822b | NG | NG | NG | NG | NG | NG | NG | NG |
| M. tuberculosise | B2c | NG | NG | NG | NG | NG | NG | NG | NG |
Treated strains were transferred to LJ slants and MP bottles, incubated at 37°C, and observed for growth (G) or no growth (NG) after 42 days of incubation.
ATCC isolate.
Reference collection isolate.
Clinical isolate.
Isoniazid and rifampin sensitive.
Isoniazid and rifampin resistant.
Low isoniazid and rifampin resistant.
Isoniazid resistant and rifampin sensitive.
To evaluate organism inactivation, suspensions of mycobacterial cultures were made at known concentrations based on the A660 nm data obtained from the quantitation studies. In each case, the highest absorbance recorded for a 1- or 10-μl loopful of each organism was used to approximate the maximum number of CFU. A 2-ml sample of each suspension was centrifuged to create a pellet, and the supernatant was removed. Pellets were resuspended in 500 μl of 70% ethanol (EtOH) and transferred into a sterile screw-cap 2-ml microcentrifuge tube containing 0.2 ml of sterile 0.5-mm glass beads (Ozyme France, Montigny-le-Bretonneux, France). Suspensions were mechanically disrupted at 2,500 oscillations/min using a Mini-BeadBeater 16 (BioSpec Products, Bartlesville, OK, USA) for 5 min either before or after a 10-min exposure to 70% EtOH at room temperature. Suspensions were then transferred into empty vials and centrifuged. After removal of the EtOH supernatant, the pellet was resuspended in 1 ml of sterile water. A 500-μl aliquot of each suspension was inoculated onto solid (LJ slants) and into liquid (BacT/Alert MP bottle, bioMérieux) media and incubated at 37°C for 42 days. In addition, samples of each organism exposed to 70% EtOH alone for 15 and 30 min without bead disruption were included for comparison. An aliquot of the sterile water suspension prepared for each test strain prior to inactivation was cultured and served as a growth control. For spectra evaluation, samples were centrifuged after the inactivation step (mechanical disruption followed by a 10-min incubation in 70% EtOH), and the resulting pellet was reconstituted with 70% formic acid followed by the addition of acetonitrile (10 μl each). The samples were again centrifuged, and the supernatant was used for Vitek MS RUO analysis after the addition of the matrix. Spectra were selected with a minimum of 80 peaks acquired with Launchpad v2.8 software (bioMérieux, Inc.) and analyzed using the SARAMIS v4.10 database (bioMérieux, Inc.).
The age of the culture and the lysate stability were determined for seven different Mycobacterium species: M. avium, M. fortuitum, M. gordonae, M. intracellulare, M. kansasii, M. scrofulaceum, and M. tuberculosis. Growth from the same LJ slant for each organism was processed daily for 7 days. Spectra were generated and compared for the presence and intensity of major peaks. For stability, processed lysates were stored at 2 to 8°C and tested daily for 7 consecutive days. In addition, portions of the lysates were stored frozen (−70°C), thawed, and tested daily for 7 days and again after 14 and 21 days of storage. Lysates were also subjected to freeze-thaw cycles to determine if this would impact the generation of acceptable spectra.
RESULTS
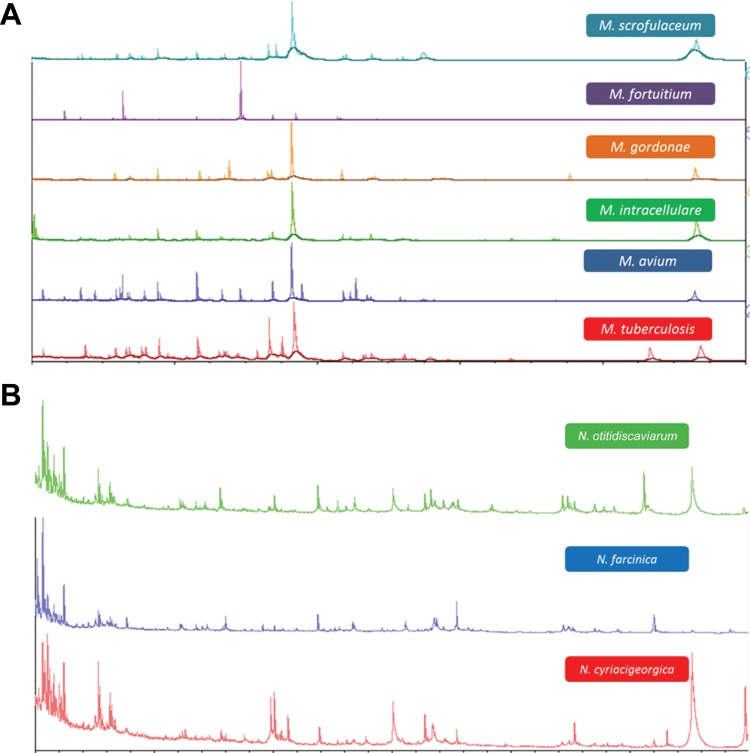
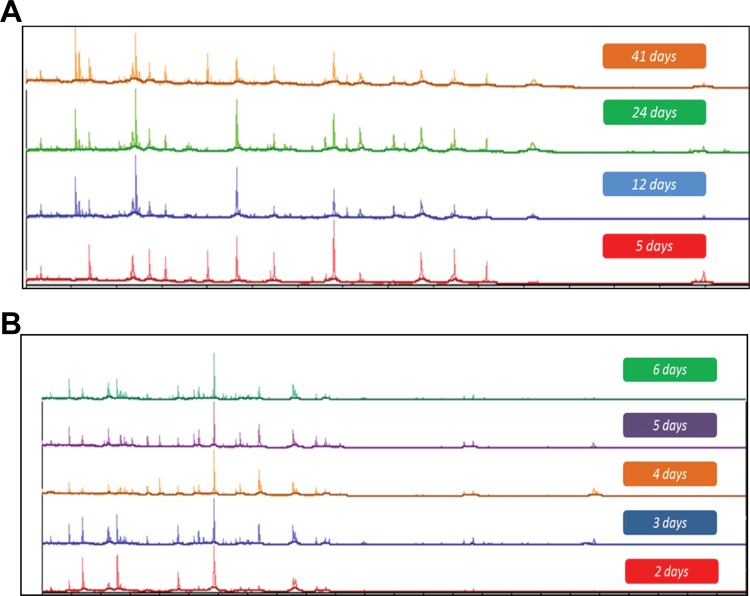
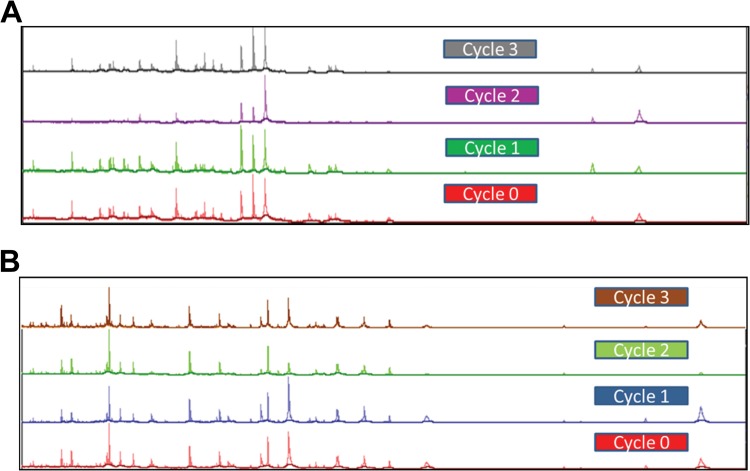
The cell density of inocula prepared using 1- and 10-μl loops varied considerably by mycobacterial species with M. tuberculosis strains generating the lowest CFU values followed by M. kansasii (Table 2). There was a 2- to 3-log difference in cell density between M. tuberculosis strains and those of M. intracellulare and the rapid growing mycobacteria (Table 2). Uniform inactivation was observed using inocula prepared from either 1- or 10-μl loops for all organisms when bead beating preceded a 10-min room temperature EtOH exposure, including all strains of M. tuberculosis regardless of INH and RIF susceptibility (Table 1). Similarly, all mycobacterial strains tested were inactivated when bead beating followed 10 min of EtOH exposure but only with the 1-μl loop inoculum. With bead beating after EtOH exposure, growth was observed with the 10-μl loop inoculum for one strain of M. chelonae and two strains of M. kansasii. The EtOH treatment alone for 15 or 30 min produced uniform killing of all mycobacterial strains tested but only with the 1-μl loopful of growth (Table 1). When the 10-μl inoculum was used, several failures were noted with either the 15- or 30-min EtOH inactivation period. These included one strain each of M. abscessus, M. gordonae, M. scrofulaceum, and M. smegmatis, two strains of M. kansasii for the 15-min preparation and both strains of M. chelonae, two strains of M. fortuitum, and one strain each of M. gordonae, M. intracellulare, and M. smegmatis for the 30-min inactivation period (Table 1). All growth controls (unprocessed starting inocula of each strain made to the same absorbance but resuspended in sterile water instead of 70% EtOH) were recovered from both liquid and solid media (not shown). Bead beating prior to the 10-min EtOH exposure resulted in total inactivation of all Nocardia strains tested (not shown). Spectra generated from Mycobacterium and Nocardia species inactivated using bead beating and the 10-min EtOH incubation or the extended EtOH only incubation generated single-choice identifications when paired with the Vitek MS RUO database. Examples of several spectra obtained via this process are shown in Fig. 1A and B. The diversity of mycobacteria processed in this study provided the opportunity to explore both the reproducibility of spectra obtained from lysates prepared from the same culture over time and the stability of lysates stored under different conditions. Lysates prepared daily for 7 days from the same LJ slant for seven different species of mycobacteria were analyzed. These results showed that the same numbers of peaks and expected identifications were obtained for all of the organisms tested, indicating that the same culture can be used for analysis over time rather than preparing fresh subcultures prior to testing (Fig. 2A and B). Similarly, lysates refrigerated for 7 days or frozen for up to 21 days all yielded single identifications and produced similar numbers of peaks in the spectra compared to those for initial testing (data not shown). Further, lysates from the same aliquots were subjected to at least three freeze-thaw cycles, and again similar peak distributions were obtained and resulted in the expected identification (Fig. 3A and B).
TABLE 2.
Estimation of mycobacterial density in CFU contained within 1- and 10-μl calibrated loopsa
| Mycobacterium species | Strain | Average population (CFU) per loopful of: |
|
|---|---|---|---|
| 1 μl | 10 μl | ||
| M. abscessus | 19977 | 1 × 109 | 2 × 109 |
| M. fortuitum | 6841 | 6 × 108 | 9 × 108 |
| M. intracellulare | 13950 | 1 × 108 | 9 × 109 |
| M. kansasii | 864 | 7 × 107 | 5 × 108 |
| M. senegalense | 2301 | 5 × 108 | 2 × 109 |
| M. tuberculosis | S-07 | 5 × 106 | 3 × 107 |
| S-09 | 6 × 106 | 4 × 107 | |
| S-38 | 4 × 106 | 1 × 107 | |
Estimates were generated using duplicate serial dilutions of a primary inoculum spread over the surface of 7H11 agar plates and enumerated after the appearance of visible colonies.
FIG 1.
Spectra generated from Mycobacterium (A) and Nocardia (B) species grown on LJ medium and Trypticase soy blood agar, respectively, Samples of growth were inactivated using mechanical disruption followed by a 10-min incubation in 70% EtOH. The samples were centrifuged, and the resulting pellet was reconstituted with 70% formic acid followed by the addition of acetonitrile (10 μl each). Samples were again centrifuged, and the supernatants were used for Vitek MS analysis after the addition of the matrix.
FIG 2.
Vitek MS spectra of M. kansasii 12478 (A) and M. fortuitum 6841 (B) grown on LJ medium. The samples were inactivated, and extraction was performed on different days from the same slants.
FIG 3.
Vitek MS spectra obtained from M. tuberculosis ATCC 25177 (A) and M. kansasii ATCC 12478 (B). The organisms were grown to log phase, and inactivation and extraction were performed. Aliquots from the protein extract were stored at −70°C and then subjected to three freeze-thaw cycles at a minimum of 24 h apart. The samples were analyzed after each thaw. The numbers of peaks obtained for M. tuberculosis were 116, 112, 87, and 108 at 0 (fresh sample prior to freeze), 1, 2, and 3 cycles, respectively. The numbers of peaks for M. kansasii were 120, 128, 108, and 121 at 0, 1, 2, and 3 cycles, respectively.
DISCUSSION
In this study, we demonstrate that a 15-min inactivation protocol using a 5-min bead-beating step in 70% EtOH followed by an additional 10-min room temperature 70% EtOH exposure produced uniform killing of a wide variety of Mycobacterium and Nocardia species and strains tested at two cell density levels. That bead beating before and not after EtOH exposure was more effective for higher cell density inocula is not surprising, given the hydrophobicity of mycobacterial cell walls and lipids in general, making cells prone to clumping with, hypothetically, less surface exposure to the antibacterial effects of EtOH (20). Longer periods of exposure to 70% EtOH (15 and 30 min) were effectively bactericidal to all organisms tested but only when used with the lower cell density preparation. Importantly, all of the inactivation protocols examined in this study were bactericidal for all M. tuberculosis strains tested, which is somewhat comforting from a biosafety standpoint, whereas 7 of 16 failures were noted with strains of M. kansasii and M. chelonae. That observation aside and given the sporadic failures of certain inactivation protocols, we selected an initial 5-min bead beating in 70% EtOH followed by an additional 10-min inactivation period for all future studies involving the identification of mycobacteria using MALDI-TOF mass spectrometry. Further, cultures can be used over an extended period of time rather than preparing fresh cultures for MALDI-TOF identification. The information gained by freezing lysates for various periods of time prior to processing opens avenues to support different workflow strategies within clinical laboratories as well as the ability to refer a suspension to another laboratory for testing, negating the need to send viable and potentially infectious organisms through courier services. For example, mycobacteria processed on Friday could be store refrigerated or frozen and analyzed the following week without loss of quality spectra and accurate identification. Expanded databases and improved algorithms will continue to enhance the performance of mycobacterial identification using the MALDI-TOF technology but a simple, rapid, and reliable inactivation process is essential for achieving the reproducibility of identification within a safe environment.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1.Lee AS, Jelfs P, Sintchenko V, Gilbert GL. 2009. Identification of non-tuberculous mycobacteria: utility of the GenoType Mycobacterium CM/AS assay compared with HPLC and 16S rRNA gene sequencing. J. Med. Microbiol. 58:900–904. 10.1099/jmm.0.007484-0 [DOI] [PubMed] [Google Scholar]
- 2.Toney NC, Toney SR, Butler WR. 2010. Utility of high-performance liquid chromatography analysis of mycolic acids and partial 16S rRNA gene sequence for routine identification of Mycobacterium spp. in a national reference laboratory. Diagn. Microbiol. Infect. Dis. 67:143–152. 10.1016/j.diagmicrobio.2010.02.011 [DOI] [PubMed] [Google Scholar]
- 3.Kim J-U, Cha C-H, An H-K. 2012. Multiplex real-time PCR assay and melting curve analysis for identifying Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J. Clin. Microbiol. 50:483–487. 10.1128/JCM.06155-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazzeri E, Santoro F, Oggioni MR, Iannelli F, Pozzi G. 2012. Novel primer-probe sets for detection and identification of mycobacteria by PCR-microarray assay. J. Clin. Microbiol. 50:3777–3779. 10.1128/JCM.02300-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soo P-C, Kung C-J, Horng Y-T, Chang K-C, Lee J-J, Peng W-P. 2012. Detonation nonodiamonds for rapid detection of clinical isolates of Mycobacterium tuberculosis complex in broth culture media. Anal. Chem. 84:7972–7978. 10.1021/ac301767z [DOI] [PubMed] [Google Scholar]
- 6.Lefmann M, Honisch C, Böcker S, Storm N, von Wintzingerode F, Schlötelburg C, Moter A, van den Boom D, Göbel UB. 2004. Novel mass spectrometry-based tool for genotypic identification of mycobacteria. J. Clin. Microbiol. 42:339–346. 10.1128/JCM.42.1.339-346.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hettick JM, Kashon ML, Slaven JE, Ma Y, Simpson JP, Siegel PD, Mazurek GN, Weissman DN. 2006. Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistics analysis. Proteomics 6:6416–6425. 10.1002/pmic.200600335 [DOI] [PubMed] [Google Scholar]
- 8.Pignone M, Greth KM, Cooper J, Emerson D, Tang J. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J. Clin. Microbiol. 44:1963–1970. 10.1128/JCM.01959-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doig C, Deagar AL, Watt B, Forbes KJ. 2002. The efficacy of the heat killing of Mycobacterium tuberculosis. J. Clin. Pathol. 55:778-779. 10.1136/jcp.55.10.778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David HL, Jones WD, Jr, Newman CM. 1971. Ultraviolet light inactivation and photoreactivation in the mycobacteria. Infect. Immun. 4:318–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feese E, Ghiladi RA. 2009. Highly efficient in vitro photodynamic inactivation of Mycobacterium smegmatis. J. Antimicrob. Chemother. 64:782–785. 10.1093/jac/dkp278 [DOI] [PubMed] [Google Scholar]
- 12.Murdoch LE, Maclean M, Endarko E, MacGregor SJ, Anderson JG. 2012. Bactericidal effects of 405 nm light exposure demonstrated by inactivation of Escherichia, Salmonella, Shigella, Listeria, and Mycobacterium species in liquid suspensions and on exposed surfaces. ScientificWorldJournal 2012:137805–137813. 10.1100/2012/137805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbir H, Abdel-Muhsin A-H, Babiker A. 2008. A one-step DNA PCR-based method for the detection of Mycobacterium tuberculosis complex grown on Lowenstein-Jensen media. Am. J. Trop. Med. Hyg. 78:316–317 [PubMed] [Google Scholar]
- 14.Bemer-Melchior P, Drugeon HB. 1999. Inactivation of Mycobacterium tuberculosis for DNA typing analysis. J. Clin. Microbiol. 37:2350–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chedore P, Th'ng C, Nolan DH, Churchwell GM, Sieffert DE, Hale YM, Jamieson F. 2002. Method for inactivating and fixing unstained smear preparations of Mycobacterium tuberculosis for improved laboratory safety. J. Clin. Microbiol. 40:4077–4080. 10.1128/JCM.40.11.4077-4080.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djelouagji Z, Drancourt M. 2006. Inactivation of cultured Mycobacterium tuberculosis organisms prior to DNA extraction. J. Clin. Microbiol. 44:1594–1595. 10.1128/JCM.44.4.1594-1595.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machen A, Kobayashi M, Connelly MR, Wang YF. 2013. Comparison of heat inactivation and cell disruption protocols for identification of mycobacteria from solid culture media by use of VITEK matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51:4226–4229. 10.1128/JCM.02612-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mather CA, Rivera SF, Butler-Wu SM. 2014. Comparison of the Bruker Biotyper and VITEK MS matrix assisted laser desorption ionization time-of-flight mass spectrometry systems for the identification of mycobacteria using simplified protein extraction protocols. J. Clin. Microbiol. 52:130–138. 10.1128/JCM.01996-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deol P, Girard V, Hyman J, Miller E, Dussoulier R, Mailler S, Schrenzel J, Beni A-M, Ninet Bescher B, Walsh J, Gates A, Arsac M, Chatellier S, Dunne W, van Nuenen M. 2013. Identification of mycobacteria by VITEK MS matrix assisted laser desorption–time of flight mass spectrometry. In Proceedings of MSACL 2013. MSACL, Inc., Del Ray, CA [Google Scholar]
- 20.Kremer L, Besra GS. 2005. A waxy tale, by Mycobacterium tuberculosis, p 287–305 In Cole ST, Eisenach KD, McMurray DN, Jacobs WR., Jr (ed), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC [Google Scholar]