Abstract
Staphylococcus pseudintermedius is the most common microorganism isolated from canine pyoderma and postoperative wound infections. The prevalence of methicillin-resistant S. pseudintermedius (MRSP) has increased, and recently, isolates that are resistant not only to methicillin but also to other classes of antibiotic drugs, including aminoglycosides, have become common. A total of 422 S. pseudintermedius isolates collected from 413 dogs were analyzed for amikacin and methicillin resistance using broth microdilution and disk diffusion testing. Methicillin-resistant isolates were significantly (P < 0.0001) more likely to be resistant to amikacin (37%, 31/84) than were methicillin-susceptible isolates (7%, 22/338). Additionally, resistance to non-β-lactam antibiotics was significantly associated with resistance to amikacin irrespective of methicillin resistance. Among the 422 isolates, 32 that tested positive for amikacin resistance by broth microdilution or disk diffusion testing were investigated further for the presence of aminoglycoside-modifying enzyme genes using multiplex PCR. Of these isolates, 66% (21/32) were methicillin resistant. In contrast to previous studies of Staphylococcus aureus, the most prevalent gene detected was aph(3′)-IIIa found in 75% (24/32) of isolates followed by aac(6′)/aph(2″) and ant(4′)-Ia in 12% (4/32) and 3% (1/32), respectively. Understanding the differences in antimicrobial resistance gene carriage between different species of Staphylococcus may improve antimicrobial drug selection for clinical therapy and provide insights into how resistance develops in S. pseudintermedius.
INTRODUCTION
Staphylococcus pseudintermedius is the most common bacterial agent isolated from canine pyoderma and wound infections (1, 2). Treatment of staphylococcal infections typically involves therapy with β-lactam antibiotics such as penicillin and cephalosporins. Resistance to this class of antimicrobial drug has increased in recent years, associated with the rise in methicillin resistance in S. pseudintermedius (3). In addition to being resistant to β-lactam antimicrobials, methicillin-resistant S. pseudintermedius (MRSP) strains are becoming increasingly resistant to other antimicrobials (4). A multicenter study from Europe and North America showed that MRSP isolates are commonly resistant to virtually all antimicrobial drug classes approved for use in dogs with 90% of MRSP isolates being resistant to ciprofloxacin, clindamycin, erythromycin, kanamycin, streptomycin, and trimethoprim and 57% being resistant to chloramphenicol (2, 5). The decreased susceptibility of MRSP to other antimicrobials has left relatively few options for therapy. The data for North America are limited, but many North American MRSP isolates were previously susceptible to chloramphenicol, rifampin, and amikacin. We have isolated MRSP that are resistant to chloramphenicol and have begun to identify isolates resistant to amikacin at our hospital (Table 1).
TABLE 1.
Amikacin-resistant and chloramphenicol-resistant Staphylococcus pseudintermedius canine isolates collected from the Texas A&M Veterinary Medical Teaching Hospital, 2010-2012
| Canine isolates | Results for yr: |
||
|---|---|---|---|
| 2010 | 2011 | 2012 | |
| Total S. pseudintermedius | 186 | 292 | 212 |
| Amikacin resistanta | 5 (2.7) | 30 (10.3) | 37 (17.5) |
| Chloramphenicol resistant (no. [%])b | 8 (4.3) | 48 (16.4) | 45 (21.2) |
Isolates with an amikacin MIC of ≥64 μg/ml or a zone of inhibition of ≤14 mm were considered resistant.
Isolates with a chloramphenicol MIC of ≥16 μg/ml or a zone of inhibition of ≤32 mm were considered resistant.
Aminoglycosides like amikacin inhibit bacterial protein synthesis by binding to the 30S ribosomal subunit (6). The most widespread mechanism of aminoglycoside resistance is drug inactivation by cellular aminoglycoside-modifying enzymes (AMEs) encoded on the chromosome or a plasmid or carried on a transposable element (7, 8). They can be divided into three classes: aminoglycoside acetyltransferases (AACs), aminoglycoside phosphotransferases (APHs), and aminoglycoside nucleotidyltransferases (ANTs) (9). In S. aureus, the aac(6′)/aph(2″) gene is the most frequently encountered aminoglycoside resistance gene followed by aph(3′)-IIIa and ant(4′)-Ia (9, 10).
The purpose of this study was to determine which genes encoding amikacin-modifying enzymes are present in amikacin-resistant S. pseudintermedius isolates collected from canine patients using a previously described multiplex PCR assay (9) and to see if an association exists between amikacin resistance, methicillin resistance, and resistance to non-β-lactam antibiotics. Although amikacin resistance has been noted in S. pseudintermedius and studies of aminoglycoside resistance in S. aureus have been published, no studies have assessed aminoglycoside resistance gene carriage in S. pseudintermedius (7, 9–11). Staphylococcus pseudintermedius is commonly associated with canine pyoderma and postoperative wound infections (1, 2). Amikacin resistance in S. pseudintermedius has significant repercussions for the treatment of canine MRSP infections, particularly as resistance to other classes of antimicrobial drugs, like fluoroquinolones, limits the antimicrobial choices for treatment. Understanding which aminoglycoside resistance mechanisms are present in S. pseudintermedius is crucial to the development of strategies to prevent resistance to this last line of therapy.
MATERIALS AND METHODS
Bacterial isolates.
A total of 422 canine Staphylococcus pseudintermedius isolates collected from the Texas A&M Veterinary Medical Teaching Hospital (VMTH) between 2010 and 2012 were available for study. All isolates were cultured from patient specimens by technicians in the VMTH clinical microbiology laboratory according to the standard operating procedures of the laboratory. Antimicrobial susceptibilities were determined according to the Clinical Laboratory Standards Institute (CLSI) performance standards (12) using a commercially available system (TREK Sensititre; TREK Diagnostics, Cleveland, OH, USA). In this study, isolates with intermediate susceptibility based on the CLSI interpretive criteria were considered to be resistant to the antimicrobial drug tested (12).
To identify methicillin resistance, every isolate was evaluated by three tests: the oxacillin broth microdilution test, the oxacillin disk diffusion test, and a PCR for detection of mecA. The oxacillin breakpoints used to confirm methicillin resistance were a MIC of ≥0.5 μg/ml and a disk diffusion zone of inhibition of ≤17 mm. These breakpoints were adopted by the clinical microbiology laboratory following recognition that the CLSI standards published in 2008 failed to identify some MRSP isolates (13, 14). Any isolate in which at least two of the three tests indicated methicillin resistance was deemed MRSP. PCRs were performed using previously described primers (15) and methods (16), with S. aureus ATCC 43300 and S. aureus ATCC 21923 (ATCC, Manassas, VA, USA) used as positive and negative controls, respectively. The disk diffusion and broth microdilution tests were performed in accordance with the CLSI performance standards (12).
Amikacin susceptibility of staphylococcal isolates was not routinely tested in the clinical microbiology laboratory prior to 2010. As such, only isolates collected between 22 October 2010 and 31 December 2012 were included in this study. From this inclusion period, two data sets were generated. Data set 1 included 422 isolates that were tested for amikacin resistance. This data set was used to determine the prevalence of amikacin resistance and the association of amikacin resistance with resistance to other antimicrobial drugs. A second data set included only the 32 amikacin-resistant isolates from data set 1 that were available for further testing. The isolates in the second data set were analyzed by PCRs for the presence of resistance genes that encode aminoglycoside-modifying enzymes.
Amikacin disk diffusion.
Isolates not tested for amikacin resistance at the time of the initial culture (e.g., antimicrobial susceptibility testing originally performed on isolates from urine) were screened for amikacin resistance by disk diffusion. The disk diffusion tests were performed according to the CLSI performance standards (12). Isolates with a zone of inhibition of ≤14 mm were considered resistant to amikacin.
DNA isolation and purification.
Of the 422 initial isolates, 32 of the total 53 amikacin-resistant isolates were available for genetic testing. All isolates were stored at −80°C at the time of collection and revived by inoculating them onto Trypticase soy agar supplemented with 5% sheep blood (blood agar plates) (BD Diagnostic Systems, Franklin Lakes, NJ, USA) and incubated at 37°C for 24 h. A single colony was used to inoculate 10 ml of L broth (LB), which contained 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl2 and incubated at 37°C overnight. The LB components were tryptone and yeast extract (BD Diagnostic Systems) and NaCl2 (Mallinckrodt Chemicals, St. Louis, MO, USA). DNA was purified from the broth culture using a DNeasy blood and tissue kit (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions for Gram-positive bacteria. DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA).
PCR.
Previously published primers for aac(6′)/aph(2′) (5′-GAAGTACGCAGAAGAGA-3′ and 5′-ACATGGCAAGCTCTAGGA-3′), aph(3′)-IIIa (5′-AAATACCGCTGCGTA-3′ and 5′-CATACTCTTCCGAGCAA-3′), ant(4′)-Ia (5′-AATCGGTAGAAGCCCAA-3′ and 5′-GCACCTGCCATTGCTA-3′), and mecA (5′-CCTAGTAAAGCTCCGGAA-3′ and 5′-CTAGTCCATTCGGTCCA-3′) were used to generate PCR products of 491, 242, 135, and 314 bp, respectively (Sigma-Aldrich, St. Louis, MO, USA) (9). PCR amplification was carried out as previously described using 50-μl mixtures containing 0.2 μM forward and reverse primers, 1× Taq buffer, 3 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate, and 1 U of Ex Taq DNA polymerase and inoculation with 2 μl of purified chromosomal template DNA. All PCR reagents were supplied by the TaKaRa Bio Company (Otsu, Shiga, Japan). The PCR assays were performed using an Applied Biosystems 2720 thermal cycler (Life Technologies, Grand Island, NY, USA) with an initial 5-min denaturation step at 95°C, followed by 30 cycles consisting of a 2-min denaturation step at 95°C, a 30-s annealing step at 58°C, and a 30-s extension at 72°C, finishing with a final extension step for 7 min at 72°C as previously described (9). The PCR products were visualized using UV light and documented with a digital imaging system (FluorChem; Alpha Innotech, Santa Clara, CA, USA) following electrophoresis on 1% (wt/vol) agarose gel (Phenix Research Products, Candler, NC, USA) containing 0.1 μl GelRed nucleic acid gel stain 10,000× (Biotium, Hayward, CA, USA) per ml of gel. The 1-kb Plus molecular weight ladder (Invitrogen, Grand Island, NY, USA) was used for the comparison of product sizes (Fig. 1).
FIG 1.
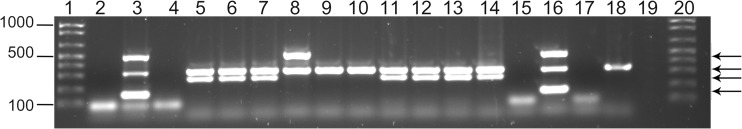
Multiplex PCR for aminoglycoside-modifying enzyme genes and mecA. The molecular size marker used in lanes 1 and 20 was the 1-kb Plus DNA ladder (Invitrogen, Grand Island, NY). The template DNAs used in the multiplex PCR were as follows: lanes 2 and 15, S. aureus ATCC 29213; lanes 3 and 16, S. aureus ATCC 43300; lanes 4, 17, and 19, no template DNA as a negative control; lanes 5 to 14 and 18, clinical S. pseudintermedius isolates (lane 5, isolate 13-089; lane 6, isolate 24-089, lane 7, isolate 29-086, lane 8, isolate 30-027, lane 9, isolate 30-076, lane 10, isolate 30-077, lane 11, isolate 31-094, lane 12, isolate 32-006, lane 13, isolate 32-010, lane 14, isolate 35-079; lane 18, isolate 30-077). Black arrows from top to bottom correspond to the PCR products, aac(6′)/aph(2″) (predicted 491 bp), mecA (predicted 314 bp), aph(3′)-IIIa (predicted 242 bp), and ant(4′)-Ia (predicted 135 bp), respectively.
Statistical analysis.
Data were summarized using cross-tabulations and analyzed using chi-square or Fisher's exact tests and logistic regression based on the binary outcome of amikacin resistance and binary outcomes of susceptibility for each of the other antimicrobials. Results of logistic regression were summarized as odds ratios (ORs) and 95% confidence intervals (CIs) for the ORs, estimated using maximum likelihood methods. Analysis was performed at the level of the isolate, ignoring the fact that some isolates originated from the same dog. Significance was set at a value of P <0.05, and all analyses were performed using S-PLUS statistical software (version 8.2; Tibco, Inc., Seattle, WA).
RESULTS
A total of 422 isolates were collected from 413 dogs. Of these 422 isolates, 369 (87%) were susceptible to amikacin and 53 (13%) were resistant to amikacin based on the CLSI interpretive criteria for amikacin (12). Of the 422 isolates, 338 (80%) were methicillin-susceptible S. pseudintermedius (MSSP) and 84 (20%) were MRSP. All of the MRSP isolates were positive for mecA by PCR. There were 316 isolates (75%) that were susceptible to both methicillin and to amikacin, 53 (13%) that were resistant to methicillin but susceptible to amikacin, 22 (5%) that were susceptible to methicillin but resistant to amikacin, and 31 (7%) that were methicillin and amikacin resistant. The odds of an isolate being methicillin resistant were significantly greater for isolates that were amikacin resistant rather than amikacin susceptible (Table 2). Resistance to each of the other antimicrobials tested was significantly associated with resistance to amikacin (Table 2). Because amikacin resistance was significantly associated with methicillin resistance, it was unclear whether resistance to the other antimicrobial drugs was associated with amikacin resistance alone or due to the association of amikacin resistance with methicillin resistance. Of the 422 S. pseudintermedius isolates, there were 106 isolates that were resistant to methicillin only (MR) (n = 53), resistant to amikacin only (AR) (n = 22), or resistant to both amikacin and methicillin (ARMR) (n = 31). Among these 106 resistant S. pseudintermedius isolates, those that were resistant to both amikacin and methicillin were significantly more likely to be resistant to chloramphenicol, clindamycin, enrofloxacin, erythromycin, gentamicin, marbofloxacin, and the trimethoprim-sulfonamide combination than the isolates that were either methicillin resistant or amikacin resistant alone (Table 3). Of resistance in the antimicrobials examined, only that to doxycycline and rifampin was not more likely among S. pseudintermedius isolates resistant to both amikacin and methicillin relative to isolates that were only resistant to 1 drug (Table 3). Isolates with an MIC of ≥8 μg/ml were considered resistant to doxycycline.
TABLE 2.
Association of resistance to amikacin with resistance to other antimicrobials for isolates of Staphylococcus pseudintermedius (n = 422), expressed as odds ratios derived by logistic regression analysis
| Antimicrobial agenta | No. (%) of isolates that were: |
Odds ratio (95% CI) | P | |
|---|---|---|---|---|
| Amikacin susceptible | Amikacin resistant | |||
| Oxacillin | ||||
| S | 316 (93) | 22 (7) | 1 (NAb) | |
| R | 53 (63) | 31 (37) | 8.4 (4.5–15.6) | <0.0001 |
| Chloramphenicol | ||||
| S | 325 (92) | 30 (8) | 1 (NA) | |
| R | 44 (66) | 23 (34) | 5.7 (3.0–10.6) | <0.0001 |
| Clindamycin | ||||
| S | 298 (96) | 11 (4) | 1 (NA) | |
| R | 71 (63) | 42 (37) | 16.0 (7.6–34.0) | <0.0001 |
| Doxycycline | ||||
| S | 251 (95) | 13 (5) | 1 (NA) | |
| R | 117 (75) | 39 (25) | 6.4 (3.3–12.5) | <0.0001 |
| Enrofloxacin | ||||
| S | 328 (95) | 18 (5) | 1 (NA) | |
| R | 41 (54) | 35 (46) | 15.6 (8.1–29.9) | <0.0001 |
| Erythromycin | ||||
| S | 299 (96) | 11 (4) | 1 (NA) | |
| R | 68 (62) | 42 (38) | 16.8 (8.2–34.3) | <0.0001 |
| Gentamicin | ||||
| S | 348 (100) | 0 (0) | 1 (NA) | |
| R | 21 (28) | 53 (72) | Inestimablec | <0.0001c |
| Marbofloxacin | ||||
| S | 331 (92) | 27 (8) | 1 (NA) | |
| R | 37 (59) | 26 (41) | 8.6 (4.6–16.3) | <0.0001 |
| Rifampin | ||||
| S | 359 (88) | 48 (12) | 1 (NA) | |
| R | 8 (62) | 5 (38) | 4.7 (1.5–14.9) | 0.0093 |
| Trimethoprim-sulfonamide | ||||
| S | 309 (96) | 14 (4) | 1 (NA) | |
| R | 58 (60) | 39 (40) | 14.8 (7.6–28.9) | <0.0001 |
S, susceptible; R, resistant.
NA, not applicable (reference category).
Inestimable because of complete separation (i.e., no isolate resistant to amikacin was susceptible to gentamicin); the P value was derived from the chi-square test with continuity correction.
TABLE 3.
Association of resistance to amikacin and/or methicillin relative to resistance to other antimicrobials in Staphylococcus pseudintermedius isolatesa
| Antimicrobial agentb | No. (%) of isolates that werec: |
Odds ratio (95% CI) | P | |
|---|---|---|---|---|
| AR and MR | AR or MR | |||
| Chloramphenicol | ||||
| S | 14 (22) | 50 (78) | 1 (NAd) | |
| R | 17 (40) | 25 (60) | 4.9 (1.2–4.9) | 0.0444 |
| Clindamycin | ||||
| S | 1 (5) | 21 (95) | 1 (NA) | |
| R | 30 (36) | 54 (64) | 11.6 (1.5–85.7) | 0.0177 |
| Doxycycline | ||||
| S | 5 (22) | 18 (78) | 1 (NA) | |
| R | 25 (31) | 56 (69) | 1.6 (0.5–4.8) | 0.3986 |
| Enrofloxacin | ||||
| S | 2 (5) | 35 (95) | 1 (NA) | |
| R | 29 (42) | 40 (58) | 12.7 (2.8–56.9) | 0.0012 |
| Erythromycin | ||||
| S | 1 (4) | 22 (96) | 1 (NA) | |
| R | 30 (37) | 51 (63) | 12.9 (1.8–95.0) | 0.0135 |
| Gentamicin | ||||
| S | 0 (0) | 38 (100) | 1 (NA) | |
| R | 31 (46) | 37 (54) | Inestimablee | <0.0001e |
| Marbofloxacin | ||||
| S | 8 (17) | 40 (83) | 1 (NA) | |
| R | 23 (40) | 34 (60) | 3.4 (1.3–8.5) | 0.0112 |
| Rifampin | ||||
| S | 1 (29) | 65 (71) | 1 (NA) | |
| R | 4 (31) | 5 (69) | 1.1 (0.3–3.8) | 0.9163 |
| Trimethoprim/sulfonamide | ||||
| S | 0 (0) | 26 (100) | 1 (NA) | |
| R | 31 (40) | 47 (60) | Inestimablee | 0.0003e |
n = 106.
S, susceptible; R, resistant.
AR and MR, isolates resistant to amikacin and oxacillin; AR or MR, isolates resistant to amikacin or oxacillin.
NA, not applicable (reference category).
Inestimable because of complete separation (no observations in one category); the P value was derived from the chi-square test with continuity correction.
There were 32 isolates from 32 unique dogs available for identification of amikacin resistance genes. A retrospective analysis of patient records associated with the isolates was performed. Among these 32 dogs, the majority were treated on an outpatient basis (59%, 19/32), while the remainder (41%, 13/32) were hospitalized. Of the 13 isolates from hospitalized dogs, 4 were MSSP. Of the 9 MRSP isolates, 4 were susceptible to marbofloxacin, clindamycin, or chloramphenicol, while 5 were resistant to all drugs tested except rifampin. The median time of culture upon entry into the hospital was 0 days (standard deviation [SD], 2.2 days; range, 0 to 12 days) with day 0 defined as entry into the hospital for either the patient appointment or clinical emergency. Most samples (69%, 22/32) were collected on day 0. Cultures were primarily taken from sources of skin disease (72%, 23/32). The majority of these samples were collected from dogs with pyoderma skin lesions (47%, 15/32) followed by skin wounds (12%, 4/32). The next most common samples collected were from urine and orthopedic implants, both seen in 3/32 cultures (9%).
Of the 32 dogs, 81% (26/32) had a history of prior antimicrobial administration, of which 53% (17/26) had received antimicrobials within 6 weeks. Of the 26 dogs with a history of antimicrobial administration, 54% (14/26) had received monotherapy, while 46% (12/26) had received multiple antimicrobials before their samples was cultured. Only 19% (6/32) had no history of prior antimicrobial use. There was no significant difference (P = 1.0000; Fisher's exact test) in whether dogs with amikacin-resistant isolates had a history of prior antimicrobial administration or whether their isolates were MSSP (82%, 9/11) or MRSP (81%, 17/21). The distribution of the three categories of prior antimicrobials (none, monotherapy, and multidrug) did not differ significantly (P = 0.7926; Fisher's exact test) between amikacin-resistant isolates that were MSSP and MRSP. Similarly, the proportion of dogs that received multiple drugs did not differ significantly (P = 0.6530) between isolates that were MSSP (27%, 3/11) and those that were MRSP (43%, 9/21). However, using logistic regression analysis, the odds of an MRSP isolate coming from a dog with a history of antimicrobials within the preceding 6 weeks of culture was significantly (P = 0.0498) greater than that for MSSP isolates (OR, 5.3; 95% CI, 1.1 to 26.6).
Among the amikacin-resistant isolates, 66% (21/32) were concurrently methicillin resistant, while 34% (11/32) were methicillin susceptible. Of the 32 amikacin-resistant isolates tested, the gene aac(6′)/aph(2″) was present in 12% (4/32) of isolates, the gene aph(3′)-IIIa was present in 75% (24/32) of isolates, and the gene ant(4′)-Ia was present in 3% (1/32) of isolates. There were four amikacin-resistant isolates in which none of these three genes was detected. Some isolates carried more than one gene. Representative PCR results are shown in Fig. 1. There was no association between methicillin resistance and carriage of a specific amikacin resistance gene. The aph(3′)-IIIa gene tended to be more prevalent among MRSP isolates (86%, 18/21) than among MSSP isolates (55%, 6/11); however, this difference was not significant (P = 0.0877; Fisher's exact test). The proportion of MRSP isolates that carried the aac(6′)/aph(2″) gene (5%, 1/21) was less than that of MSSP isolates (27%, 3/11); however, this difference was not significant (P = 0.1055; Fisher's exact test). The 1 isolate that carried the ant(4′)-Ia gene was MRSP.
DISCUSSION
Staphylococcus pseudintermedius is the most common bacterial pathogen associated with canine pyoderma and postoperative wound infections (1, 2). Treatment of these infections typically involves β-lactam antibiotics such as amoxicillin and cephalexin. The spread of methicillin-resistant S. pseudintermedius (MRSP) across Europe led to the widespread use of alternative antibiotics such as chloramphenicol (2, 5). This ultimately led to resistance to chloramphenicol and to virtually all classes of antibiotics approved for use in dogs (2, 5). Methicillin resistance has begun to emerge in the United States and over the past 5 years, resistance to chloramphenicol has become common (Table 1). This has led to reliance on other antimicrobial drugs such as amikacin to treat life-threatening MRSP infections. During the past 2 years, aminoglycoside-resistant MRSP infections have been identified among patients in our small animal hospital (Table 1). Aminoglycosides like amikacin are not routinely used to treat staphylococcal infections due to the potential nephrotoxic effects of these drugs and the inconvenient route of administration (3, 17). The increased prevalence of methicillin-resistant and multidrug-resistant S. pseudintermedius strains has left clinicians with few choices for antimicrobial therapy, sometimes making aminoglycosides the last available choice.
Aminoglycosides are bactericidal agents that bind irreversibly to the 30S ribosomal subunit of susceptible bacteria, thereby inhibiting protein synthesis (6). Drug inactivation by AMEs is the main mechanism of aminoglycoside resistance (7, 8, 10). In a study of S. aureus, the aac(6′)/aph(2″) gene was found in 66% of resistant S. aureus isolates followed by ant(4′)-Ia and aph(3′)-IIIa genes with frequencies of 24% and 8%, respectively (7). Similar results have previously been found (9). In contrast, we found that in S. pseudintermedius, the most common amikacin resistance gene was aph(3′)-IIIa, which was present in 75% (24/32) of the amikacin-resistant isolates of S. pseudintermedius, followed by aac(6′)/aph(2″) and ant(4′)-Ia genes at 12% (4/32) and 3% (1/32), respectively. The gene aph(3′)-IIIa has been demonstrated in the chromosomal DNA of S. pseudintermedius strains as well as on transposons carried on plasmids (18–20). It is unclear whether aph(3′)-IIIa is carried on the chromosome or on a plasmid and whether it is part of a transposable element in the strains in our study. Understanding which resistance genes are present and how they are transmitted has important clinical ramifications for infected dogs. Under antimicrobial selective pressure, antibiotic resistance genes can be transferred from one strain or species of Staphylococcus to another by plasmid conjugation, phage-mediated transduction, or transposon movement, which might result in widespread antibiotic resistance among staphylococci (20).
Our study is limited by its retrospective nature. Patient histories were obtained solely through the available medical records. Some cases may have had previous cultures or antimicrobial therapy that was not noted in the records. Due to the use of an antimicrobial susceptibility testing system that did not measure amikacin MICs, amikacin-resistant isolates may have been missed prior to 2010. Additionally, the exclusion of 21 amikacin-resistant isolates from genetic testing because they were not stored at the time of isolation may have affected the prevalence of certain amikacin resistance genes. Despite these limitations, we documented a 15% rise in aminoglycoside resistance at the VMTH over the past 2 years (Table 1) and determined that aph(3′)-IIIa was the most common gene in isolates from patients presented to our hospital. Another limitation of our study is that without genetic fingerprinting, spa typing, or another method to compare the genetic relatedness of the isolates, we cannot rule out the possibility that some of the isolates represent a nosocomial clone, particularly for the 5 MRSP isolates from inpatients that were resistant to all other drugs. Finally, this study found that doxycycline resistance was not more likely in isolates that were both amikacin resistant and methicillin resistant than in isolates that were either amikacin or methicillin resistant. In 2013, a strong case was made for the adoption of canine breakpoints for doxycycline (susceptible, ≤0.125 μg/ml; intermediate, 0.25 μg/ml; resistant, >0.5 μg/ml) for S. pseudintermedius isolates instead of using the human breakpoints (susceptible, ≤4 μg/ml; intermediate, 8 μg/ml; resistant, >16 μg/ml). The majority of isolates from this study (250/422) had an MIC of ≤2 μg/ml. It is possible that if we had used the more conservative breakpoints, we would have found that doxycycline resistance was more likely in isolates that were both methicillin and amikacin resistant. Unfortunately, we were unable to test this as the lowest concentration of doxycycline available for the commercial broth microdilution system used by our laboratory is 2 μg/ml, well above the proposed breakpoint for canine staphylococcal isolates and not all of the isolates tested by the laboratory were available for retesting.
This study also showed a significant association between amikacin resistance and methicillin resistance (Table 2). Similarly, a study in S. aureus and coagulase-negative staphylococci identified the presence of at least one AME gene associated with mecA in 72% of the methicillin-resistant staphylococci (7). One theory, aside from gene transfer from another source, as to why these genes seem to be commonly present together is that mecA and the AME genes may be located adjacent to each other on the bacterial chromosome (21, 22). Among the isolates in this study, resistance to other drugs was more likely to be found in isolates that were both amikacin and methicillin resistant (Table 3). This may reflect development of amikacin resistance as a result of treatment of multidrug-resistant MRSP with amikacin but does not explain the finding of amikacin-resistant isolates that were susceptible to all other drugs. Within the 422 S. pseudintermedius isolates, there were 22 isolates that were amikacin resistant but methicillin susceptible. In these isolates, it is possible, although unproven, that amikacin resistance may be plasmid mediated. If this is true, the association between amikacin and methicillin resistance may change over time, and future studies may find that amikacin resistance is not linked with methicillin resistance.
Aminoglycosides remain important antimicrobial drugs for the treatment of life-threatening infections in veterinary medicine even though resistance among species of staphylococci continues to be demonstrated worldwide (7, 23). Since AMEs can be carried on plasmids or on transposable elements, it may be important to monitor aminoglycoside resistance in staphylococci over time as transmission of resistance between bacterial strains or species (for example, from S. pseudintermedius to S. aureus) may represent a new nosocomial and zoonotic threat, particularly in the face of increased multidrug-resistant staphylococci (8, 20, 24).
ACKNOWLEDGMENTS
The members of the Texas A&M Veterinary Medical Teaching Hospital clinical microbiology laboratory performed the initial isolation and antimicrobial susceptibility testing on all isolates as part of the routine analysis of clinical samples submitted to the laboratory.
R. M. Gold was supported through a Texas A&M University College of Veterinary Medicine and Biomedical Sciences merit fellowship, and the experimental work was supported through a College of Veterinary Medicine and Biomedical Sciences postdoctoral research grant to R. M. Gold and Department of Veterinary Pathobiology support to S. D. Lawhon.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1.Morris DO, Boston RC, O'Shea K, Rankin SC. 2010. The prevalence of carriage of meticillin-resistant staphylococci by veterinary dermatology practice staff and their respective pets. Vet. Dermatol. 21:400–407. 10.1111/j.1365-3164.2010.00866.x [DOI] [PubMed] [Google Scholar]
- 2.Moodley A, Stegger M, Ben Zakour NL, Fitzgerald JR, Guardabassi L. 2009. Tandem repeat sequence analysis of staphylococcal protein A (spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet. Microbiol. 135:320–326. 10.1016/j.vetmic.2008.09.070 [DOI] [PubMed] [Google Scholar]
- 3.Papich MG. 2012. Selection of antibiotics for meticillin-resistant Staphylococcus pseudintermedius: time to revisit some old drugs? Vet. Dermatol. 23:352–360, e64. 10.1111/j.1365-3164.2011.01030.x [DOI] [PubMed] [Google Scholar]
- 4.Turutoglu H, Hasoksuz M, Ozturk D, Yildirim M, Sagnak S. 2009. Methicillin and aminoglycoside resistance in Staphylococcus aureus isolates from bovine mastitis and sequence analysis of their mecA genes. Vet. Res. Commun. 33:945–956. 10.1007/s11259-009-9313-5 [DOI] [PubMed] [Google Scholar]
- 5.Nienhoff U, Kadlec K, Chaberny IF, Verspohl J, Gerlach GF, Kreienbrock L, Schwarz S, Simon D, Nolte I. 2011. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet. Microbiol. 150:191–197. 10.1016/j.vetmic.2010.12.018 [DOI] [PubMed] [Google Scholar]
- 6.Davies J, Anderson P, Davis BD. 1965. Inhibition of protein synthesis by spectinomycin. Science 149:1096–1098. 10.1126/science.149.3688.1096 [DOI] [PubMed] [Google Scholar]
- 7.Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. 2006. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol. Res. 161:49–54. 10.1016/j.micres.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 8.Brown NM, Reeves DS. 1992. Mechanisms and epidemiology of aminoglycoside resistance. J. Med. Microbiol. 36:11–14 [Google Scholar]
- 9.Choi SM, Kim SH, Kim HJ, Lee DG, Choi JH, Yoo JH, Kang JH, Shin WS, Kang MW. 2003. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J. Korean Med. Sci. 18:631–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan W, Hu Q, Cheng H, Shang W, Liu N, Hua Z, Zhu J, Hu Z, Yuan J, Zhang X, Li S, Chen Z, Hu X, Fu J, Rao X. 2013. Cell wall thickening is associated with adaptive resistance to amikacin in methicillin-resistant Staphylococcus aureus clinical isolates. J. Antimicrob. Chemother. 68:1089–1096. 10.1093/jac/dks522 [DOI] [PubMed] [Google Scholar]
- 11.Casagrande Proietti P, Bietta A, Coletti M, Marenzoni ML, Scorza AV, Passamonti F. 2012. Insertion sequence IS256 in canine pyoderma isolates of Staphylococcus pseudintermedius associated with antibiotic resistance. Vet. Microbiol. 157:376–382. 10.1016/j.vetmic.2011.12.028 [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; second informational supplement. CLSI document VET01-S2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13.Bemis DA, Jones RD, Frank LA, Kania SA. 2009. Evaluation of susceptibility test breakpoints used to predict mecA-mediated resistance in Staphylococcus pseudintermedius isolated from dogs. J. Vet. Diagn. Invest. 21:53–58. 10.1177/104063870902100108 [DOI] [PubMed] [Google Scholar]
- 14.Schissler JR, Hillier A, Daniels JB, Cole LK, Gebreyes WA. 2009. Evaluation of Clinical and Laboratory Standards Institute interpretive criteria for methicillin-resistant Staphylococcus pseudintermedius isolated from dogs. J. Vet. Diagn. Invest. 21:684–688. 10.1177/104063870902100514 [DOI] [PubMed] [Google Scholar]
- 15.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala JL. 1995. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J. Clin. Microbiol. 33:2864–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, Qian Q. 2001. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J. Clin. Microbiol. 39:3946–3951. 10.1128/JCM.39.11.3946-3951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank LA, Loeffler A. 2012. Meticillin-resistant Staphylococcus pseudintermedius: clinical challenge and treatment options. Vet. Dermatol. 23:283−291, e256. 10.1111/j.1365-3164.2012.01047.x [DOI] [PubMed] [Google Scholar]
- 18.Boerlin P, Burnens AP, Frey J, Kuhnert P, Nicolet J. 2001. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 79:155–169. 10.1016/S0378-1135(00)00347-3 [DOI] [PubMed] [Google Scholar]
- 19.Schwarz S, Feßler A, Hauschild T, Kehrenberg C, Kadlec K. 2011. Plasmid-mediated resistance to protein biosynthesis inhibitors in staphylococci. Ann. N. Y. Acad. Sci. 1241:82–103. 10.1111/j.1749-6632.2011.06275.x [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resistance Updates 6:41–52. 10.1016/S1368-7646(03)00003-7 [DOI] [PubMed] [Google Scholar]
- 21.Dubin DT, Matthews PR, Chikramane SG, Stewart PR. 1991. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 35:1661–1665. 10.1128/AAC.35.8.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubukata K, Nonoguchi R, Matsuhashi M, Song MD, Konno M. 1989. Restriction maps of the regions coding for methicillin and tobramycin resistances on chromosomal DNA in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 33:1624–1626. 10.1128/AAC.33.9.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maple PA, Hamilton-Miller JM, Brumfitt W. 1989. World-wide antibiotic resistance in methicillin-resistant Staphylococcus aureus. Lancet i:537–540 [DOI] [PubMed] [Google Scholar]
- 24.Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115–3121. 10.1128/JCM.39.9.3115-3121.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]


