Abstract
The commutability of international reference standards is critical for ensuring quantitative agreement across different viral load assays. Here, we demonstrate the commutability of the Epstein-Barr virus (EBV) WHO international standard for the BamHI-W and artus EBV assays.
TEXT
Viral load testing for Epstein-Barr virus (EBV) is important for monitoring patients at risk for posttransplant lymphoproliferative disorder and provides prognostic information for nasopharyngeal carcinoma patients after therapy (1–3). Despite the widespread use and clinical utility of EBV DNA testing, a lack of quantitative agreement has been demonstrated when common specimens are tested using different methods (4–7). Multivariate analysis indicates that the material used for calibration accounts for a significant proportion of the variation in EBV quantitation (7).
The 1st WHO international standard for EBV was introduced to address variation attributed to assay calibration (8). While the availability of an international EBV standard provides an important first step toward the harmonization of quantitative EBV assays, the commutability of the reference material must also be taken into consideration. Commutability refers to the ability of a reference material to have interassay properties comparable to the properties demonstrated by authentic clinical samples (9). Critically, the use of reference materials that lack commutability may reduce quantitative agreement (10–12). We therefore evaluated the commutability of the EBV WHO standard across two common real-time PCR assays, the laboratory-developed BamHI and the commercial artus EBV QIAsymphony Rotor-Gene Q (QS-RGQ) assays.
The BamHI assay was performed as previously described (13) with the following modifications, (i) the probe was used at a final concentration of 100 nM and contained a black hole quencher, (ii) the FastStart TaqMan Probe Master (Roche Applied Science, Indianapolis, IN) was used in a 25-μl reaction mixture, (iii) and cycling was performed under the following conditions: an initial hold at 95°C for 10 min, then 45 cycles at 95°C for 15 s, and 56°C for 30 s. Calibration was performed using DNA extracted from the diploid Namalwa cell line that contains two integrated EBV genomes per cell using the conversion factor 6.6 pg of DNA/diploid cell, as previously described (13). The artus EBV QS-RGQ assay (Qiagen, Germantown, MD) was performed according to the manufacturer's recommendations except the reaction mixtures were scaled to 25 μl. Calibration was performed by using DNA standards provided by the manufacturer. The BamHI and artus protocols were performed on the RGQ real-time PCR instrument, and for all experiments, DNA was isolated from 1.0 ml of plasma collected in K2 EDTA tubes (BD Diagnostics, Franklin Lakes, NJ) using the virus/bacteria midi kit on the QIAsymphony SP (Qiagen, Germantown, MD). The purified DNA was eluted into a final volume of 90 μl, and each PCR utilized 10 μl. An internal control was added to each primary sample prior to extraction, and amplification was performed with specific primers and hydrolysis probes contained in the artus master mix to ensure adequate extraction efficiency and the absence of inhibitors. Statistical analyses were performed using Prism v.6.0 (GraphPad, La Jolla, CA), XLSTAT (Addinsoft USA, New York, NY), and Excel (Microsoft, Redmond, WA).
The 1st WHO international standard for EBV was obtained from the National Institute for Biological Standards and Control (Hertfordshire, United Kingdom) and was diluted to 5.0, 4.7, 4.0, 3.7, and 3.0 log10 international units (IU)/ml in EBV-negative EDTA plasma (SeraCare, Milford, MA). Six replicates at each concentration were tested using the two assays on 4 separate days (24 total replicates per assay). Within-run, between-run, and total imprecision was calculated at each concentration level (Table 1). The difference in variance at each level was evaluated using the right-tailed F test, which revealed that the total imprecision of the BamHI assay was greater than the total imprecision of the artus assay (P<0.001, levels 2 to 5), except at the lowest concentration, where there was no statistical difference (P = 0.13). In addition, the means of the observed log10 copies/ml concentrations were plotted against the nominal log10 IU/ml values, and ordinary least-squares regression was performed (Fig. 1). This analysis revealed the linear regression equations for BamHI (Y = 0.9699 X + 0.7070; R2 = 0.95) and artus (Y = 0.9538 X + 0.8630; R2 = 0.99) and the 95% confidence intervals for the slopes (BamHI, 0.9295 to 1.010; artus, 0.9386 to 0.9689) and the intercepts (BamHI, 0.5396 to 0.8744; artus, 0.8004 to 0.9256). These data show that there is a similar relationship between BamHI copies and IUs and artus copies and IUs.
TABLE 1.
Precision of the BamHI and artus assays using the 1st WHO international standard for EBV
| Assay | Level | Average observed EBV DNA titer (log10 copies/ml) | Within-run SD | Between-run SD | Total SD |
|---|---|---|---|---|---|
| BamHI | 1 | 3.633 | 0.088 | 0.077 | 0.117 |
| 2 | 4.275 | 0.252 | 0.121 | 0.279 | |
| 3 | 4.567 | 0.082 | 0.089 | 0.121 | |
| 4 | 5.316 | 0.097 | 0.000 | 0.097 | |
| 5 | 5.532 | 0.130 | 0.000 | 0.130 | |
| artus | 1 | 3.735 | 0.071 | 0.057 | 0.091 |
| 2 | 4.367 | 0.052 | 0.033 | 0.062 | |
| 3 | 4.689 | 0.035 | 0.016 | 0.039 | |
| 4 | 5.349 | 0.045 | 0.000 | 0.045 | |
| 5 | 5.632 | 0.033 | 0.044 | 0.055 |
FIG 1.
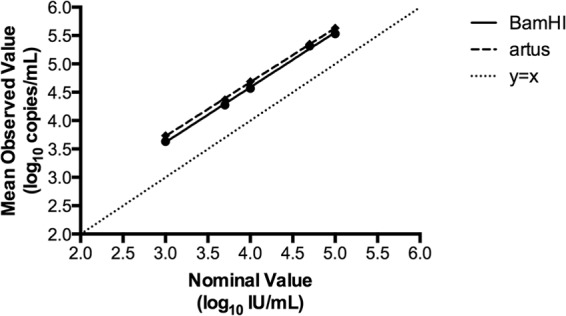
Quantitation of the EBV WHO international standard using the BamHI and artus assays. The mean of the observed log10 copies/ml for each dilution of the EBV WHO international standard was plotted against the nominal log10 IU/ml values. Linear regression lines are shown for the BamHI (solid) and artus (dashed) assays. The line of identity (y = x) is represented as a dotted line.
To investigate the quantitative agreement between the BamHI and artus assays in clinical samples, 40 EBV DNA-positive plasma specimens were tested by each of the two methods. These leftover specimens were deidentified and would otherwise have been discarded. The Stanford University Institutional Review Board waived the review. The log10 concentrations measured by the BamHI and artus assays were plotted against one another, and Passing-Bablok regression was performed (Fig. 2A), which resulted in a regression line of Y = 1.177 X − 0.851. Passing-Bablok regression was used in this case because it required no assumptions regarding the distribution of samples and measurement errors. The 95% confidence intervals of the slope (1.018 to 1.302) and intercept (−1.312 to −0.248) did not include 1 or 0, respectively, indicating that the BamHI assay showed slight positive proportional bias and negative systematic bias. Next, the differences in log10 concentrations were plotted against the average values to generate a Bland-Altman plot (Fig. 2B). This analysis revealed a bias of −0.063 log10 copies/ml (BamHI − artus), although the mean of the differences between the paired data was not statistically significant (P = 0.30, paired t test, two sided). Altogether, these results indicate that using the respective laboratory calibrators, the BamHI assay will give, on average, slightly lower values than will the artus assay. Nevertheless, this bias is not clinically significant, and overall, these assays display good quantitative agreement.
FIG 2.
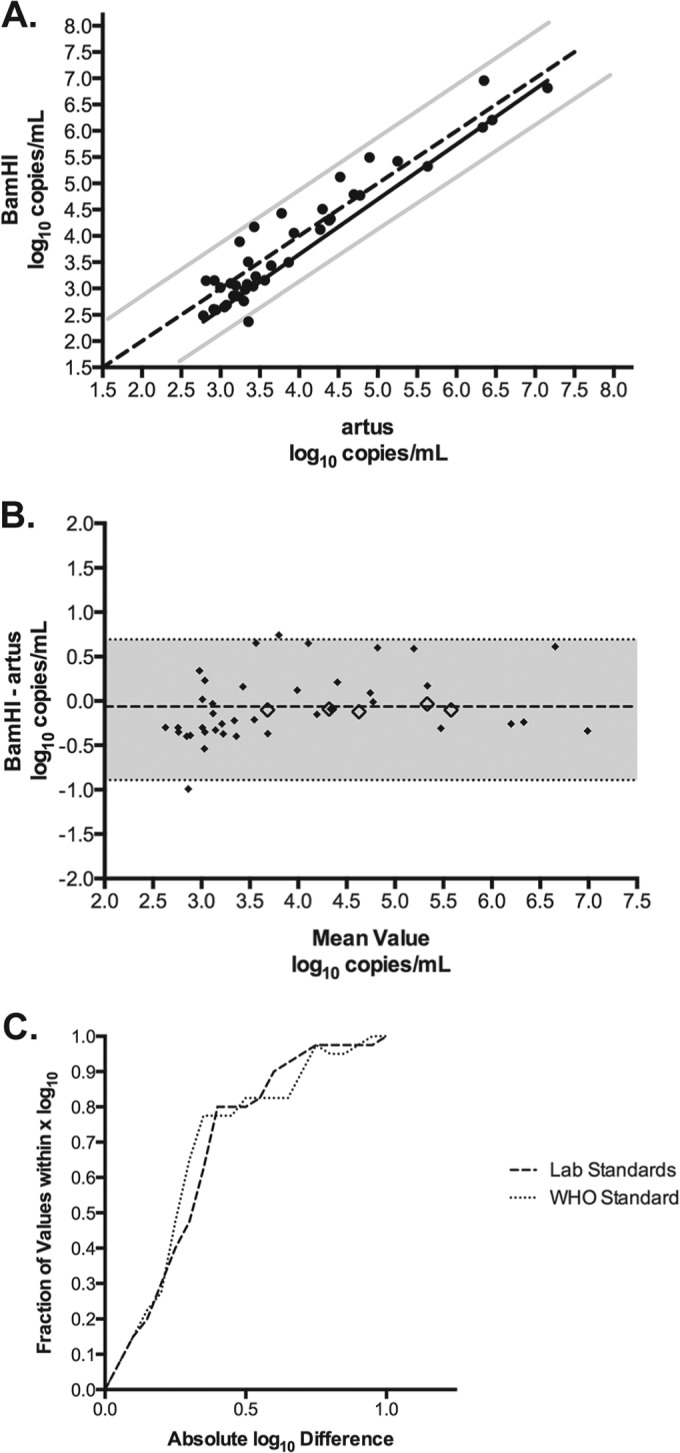
Quantitative comparison of clinical specimens tested with the BamHI and artus assays. (A) Passing-Bablok regression analysis of 40 clinical specimens tested by the BamHI and artus assays. The regression line (black solid line) and the 95% confidence intervals (gray solid lines) are displayed. The line of identity (y = x) is represented as a dotted line. (B) Bland-Altman plot comparing the BamHI assay with the artus method (solid diamonds). The bias (dashed line) was −0.063 log10 copies/ml; the shaded area indicates the 95% limits of agreement. The differences in the log10 copies/ml concentrations of the mean WHO standard values at each concentration level plotted against the average of the two values are shown as open diamonds. (C) The absolute difference in log10 values between assays was calculated using the laboratory standards and the WHO standard, and the percentages of specimens that fell within various ranges were plotted.
To determine the commutability of the WHO standard, the difference in the observed log10 copies/ml concentrations of the mean WHO standard values at each concentration level were plotted against the average of the two values (Fig. 2B, open diamonds). These values fall within the 95% confidence interval of the clinical specimens and have a mean difference of −0.09 log10 copies/ml. This indicates that the WHO standard behaved similarly to clinical specimens in these assays and is therefore commutable.
To evaluate the affect that recalibration of the BamHI and artus assays to the WHO standard had on quantitative agreement, the observed copies/ml were converted to IU/ml according to the linear regression equations described in Fig. 1. Next, the absolute difference in log10 values between assays was calculated before and after recalibration. Finally, the percentages of specimens that fell within various ranges were plotted (e.g., 15% of the samples were within 0.1 log10 and 100% of the samples were within 1.0 log10), as previously described (Fig. 2C) (12). This analysis revealed that the quantitative agreement between assays was unchanged when calibrated to the WHO standard.
The collaborative study to evaluate the 1st WHO international standard for EBV determined potency and demonstrated the suitability of candidate preparation for use as the international standard, although commutability was not specifically addressed (8). Here, we show that the EBV WHO standard is commutable across the BamHI and artus EBV assays. Because the BamHI assay targets the variably repeated BamHI-W sequence (14), it has been suggested that this assay is not suitable for EBV viral load measurement (8, 15). However, compared to the artus EBV assay, which targets a single-copy sequence in the Epstein-Barr nuclear antigen 1 (EBNA-1) gene, the BamHI assay demonstrated good quantitative agreement. This finding is consistent with previous reports (8, 16) and is independent of harmonization to the WHO standard.
Future studies will be required to determine the commutability of the WHO standard and secondary EBV standards across the large number of quantitative EBV assays currently in use in clinical laboratories worldwide.
ACKNOWLEDGMENTS
We thank the staff of the Stanford Clinical Virology Laboratory for their diligent work and dedication to patient care.
We declare no conflicts of interest in the publication of this research.
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1.Chan AT, Lo YM, Zee B, Chan LY, Ma BB, Leung SF, Mo F, Lai M, Ho S, Huang DP, Johnson PJ. 2002. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J. Natl. Cancer Inst. 94:1614–1619. 10.1093/jnci/94.21.1614 [DOI] [PubMed] [Google Scholar]
- 2.Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, Jiang RS. 2004. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N. Engl. J. Med. 350:2461–2470. 10.1056/NEJMoa032260 [DOI] [PubMed] [Google Scholar]
- 3.Gulley ML, Tang W. 2010. Using Epstein-Barr viral load assays to diagnose, monitor, and prevent posttransplant lymphoproliferative disorder. Clin. Microbiol. Rev. 23:350–366. 10.1128/CMR.00006-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH., Jr 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J. Clin. Microbiol. 46:157–163. 10.1128/JCM.01252-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preiksaitis JK, Pang XL, Fox JD, Fenton JM, Caliendo AM, Miller GG. 2009. Interlaboratory comparison of Epstein-Barr virus viral load assays. Am. J. Transplant. 9:269–279. 10.1111/j.1600-6143.2008.02514.x [DOI] [PubMed] [Google Scholar]
- 6.Abbate I, Zanchetta M, Gatti M, Gabrielli L, Zanussi S, Milia MG, Lazzarotto T, Tedeschi R, Ghisetti V, Clementi M, De Rossi A, Baldanti F, Capobianchi MR. 2011. Multicenter comparative study of Epstein-Barr virus DNA quantification for virological monitoring in transplanted patients. J. Clin. Virol. 50:224–229. 10.1016/j.jcv.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 7.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J. Clin. Microbiol. 50:337–345. 10.1128/JCM.01287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fryer JF, Heath AB, Wilkinson DE, Minor PD. 2011. Collaborative study to evaluate the proposed 1st WHO international standard for Epstein-Barr virus (EBV) for nucleic acid amplification technology (NAT)-based assays World Health Organization Expert Committee on Biological Standardization. WHO/BS/11.2172. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/handle/10665/70781 [Google Scholar]
- 9.Vesper HW, Miller WG, Myers GL. 2007. Reference materials and commutability. Clin. Biochem. Rev. 28:139–147 [PMC free article] [PubMed] [Google Scholar]
- 10.Eckfeldt JH, Copeland KR. 1993. Accuracy verification and identification of matrix effects. The College of American Pathologists' Protocol. Arch. Pathol. Lab Med. 117:381–386 [PubMed] [Google Scholar]
- 11.Cattozzo G, Franzini C, Melzi d'Eril GM. 2001. Commutability of calibration and control materials for serum lipase. Clin. Chem. 47:2108–2113 [PubMed] [Google Scholar]
- 12.Hayden RT, Shahbazian MD, Valsamakis A, Boonyaratanakornkit J, Cook L, Pang XL, Preiksaitis JK, Schonbrunner ER, Caliendo AM. 2013. Multicenter evaluation of a commercial cytomegalovirus quantitative standard: effects of commutability on interlaboratory concordance. J. Clin. Microbiol. 51:3811–3817. 10.1128/JCM.02036-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo YM, Chan LY, Lo KW, Leung SF, Zhang J, Chan AT, Lee JC, Hjelm NM, Johnson PJ, Huang DP. 1999. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 59:1188–1191 [PubMed] [Google Scholar]
- 14.Allan GJ, Rowe DT. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489–498. 10.1016/0042-6822(89)90561-8 [DOI] [PubMed] [Google Scholar]
- 15.Stevens SJ, Pronk I, Middeldorp JM. 2001. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J. Clin. Microbiol. 39:1211–1216. 10.1128/JCM.39.4.1211-1216.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le QT, Jones CD, Yau TK, Shirazi HA, Wong PH, Thomas EN, Patterson BK, Lee AW, Zehnder JL. 2005. A comparison study of different PCR assays in measuring circulating plasma Epstein-Barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin. Cancer Res. 11:5700–5707. 10.1158/1078-0432.CCR-05-0648 [DOI] [PubMed] [Google Scholar]


