Abstract
Aspergillus section Fumigati contains 12 clinically relevant species. Among these Aspergillus species, A. fumigatus is the most frequent agent of invasive aspergillosis, followed by A. lentulus and A. viridinutans. Genealogical concordance and mating experiments were performed to examine the relationship between phylogenetic distance and mating success in these three heterothallic species. Analyses of 19 isolates from section Fumigati revealed the presence of three previously unrecognized species within the broadly circumscribed species A. viridinutans. A single mating type was found in the new species Aspergillus pseudofelis and Aspergillus pseudoviridinutans, but in Aspergillus parafelis, both mating types were present. Reciprocal interspecific pairings of all species in the study showed that the only successful crosses occurred with the MAT1-2 isolates of both A. parafelis and A. pseudofelis. The MAT1-2 isolate of A. parafelis was fertile when paired with the MAT1-1 isolates of A. fumigatus, A. viridinutans, A. felis, A. pseudoviridinutans, and A. wyomingensis but was not fertile with the MAT1-1 isolate of A. lentulus. The MAT1-2 isolates of A. pseudofelis were fertile when paired with the MAT1-1 isolate of A. felis but not with any of the other species. The general infertility in the interspecies crossings suggests that genetically unrelated species are also biologically incompatible, with the MAT1-2 isolates of A. parafelis and A. pseudofelis being the exception. Our findings underscore the importance of genealogical concordance analysis for species circumscription, as well as for accurate species identification, since misidentification of morphologically similar pathogens with differences in innate drug resistance may be of grave consequences for disease management.
INTRODUCTION
Invasive aspergillosis (IA) is one of the major mold infections, and it results in high fatality rates among severely immunocompromised patients, especially those with profound neutropenia (1). Aspergillus fumigatus, the major etiologic agent of IA, can be readily distinguished from other, less common agents, such as Aspergillus flavus, Aspergillus terreus, Aspergillus niger, and Aspergillus nidulans, by the characteristic morphology of its conidial structures (2). Aspergillus fumigatus has been reported to be a variable species (3, 4). Variations are commonly observed in colony growth, the robustness of the production of conidia, conidial surface markings, the presence or absence of septation in phialides, and maximum growth temperatures (4–6). Many strains showing such phenotypical variations were considered atypical A. fumigatus because they showed conidiogenous structure morphology indistinguishable from that typical of A. fumigatus (4). Using multilocus sequence data, several genetically diverse clades that qualify as phylogenetic species (4, 7, 8) were found among the atypical A. fumigatus strains, and they have been described as separate species. Examples of such species are Aspergillus lentulus and Aspergillus felis.
Recently, heterothallic sexual cycles were discovered in A. lentulus (9) and A. felis (10). Although the sexual state of Aspergillus viridinutans has yet to be found, the presence of the MAT1-1 (11) mating type homolog in the type strain NRRL 4365 suggests heterothallic sexuality. In an outbreeding system, it is possible to compare the phylogenetic distance with biological relatedness through interspecific crosses. Such comparisons between biological species recognition (BSR) and phylogenetic species recognition (PSR) have been done with Neurospora species (12, 13). Mating between numerous Neurospora strains revealed several reproductively isolated biological species. Using genealogical concordance of four independent nuclear loci to identify phylogenetic species in these strains, congruence was observed between the results of the two species identification systems, BSR and PSR. Increased genetic distance between parents was associated with decreased sexual reproduction. These results suggest that genealogical concordance PSR can be reliably used to recognize species in fungi that are not candidates for BSR (12).
The need for correct identification of the etiologic agents of IA that resemble A. fumigatus, such as A. lentulus, A. viridinutans, and A. felis, is underscored by the fact that these species tend to be refractory to antifungal treatment and produce more chronic infection than A. fumigatus (10, 14, 15). Since they are all heterothallic species, we attempted to determine whether the phylogenetic and reproductive methods concurred in species identification and whether reproductive success between different species reflected their genetic distance. While comparing the phylogenetic relationship between strains previously identified as A. lentulus, A. viridinutans, and A. felis and a reference strain of A. fumigatus (16), it was observed that the species designated A. viridinutans contained several cryptic species. These species are described here, and the MIC values for common antifungals and virulence testing in model systems for the new species are reported.
MATERIALS AND METHODS
Isolates.
The isolates used in this study are listed in Table 1. The type strains A. lentulus NRRL 35552 and A. viridinutans NRRL 4365 and NRRL 6106 were obtained from the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA. Aspergillus wyomingensis isolates CCF 4417 and CCF 4416 were kindly provided by V. Hubka of Charles University, Prague, Czech Republic. Isolates were cultured at 37°C on Aspergillus minimal medium agar (MMA) (17), malt extract agar (MEA), Czapek's solution agar (CZA), or oatmeal agar (OA) (18).
TABLE 1.
Aspergillus isolates, mating types, and originsa
| Species | Designation | ARS accession no. | Mating type | Source | Origin |
|---|---|---|---|---|---|
| A. viridinutans | NRRL 4365T | NRRL 4365T | MAT1-1 | Rabbit dung | Australia |
| A. felis | CBS 130245T | ND | MAT1-2 | Cat | Australia |
| A. felis | CBS 130246 | ND | MAT1-1 | Cat | Australia |
| A. fumigatus | AFB62 | ND | MAT1-1 | Human | USA |
| A. fumigatus | AFB623 | ND | MAT1-1 | Human | USA |
| A. fumigatus | AFIR928 | ND | MAT1-2 | Environmental | Ireland |
| A. wyomingensis | CCF 4417T | ND | MAT1-1b | Environmental | USA |
| A. wyomingensis | CCF 4416 | ND | MAT1-2b | Environmental | USA |
| A. lentulus | NRRL 35552T | NRRL 35552T | MAT1-2 | Human | USA |
| A. lentulus | CM-4428 | ND | MAT1-1 | Human, skin | Spain |
| A. lentulus | CM-4843 | ND | MAT1-1 | Human, sputum, | Spain |
| A. lentulus | CM-5027 | ND | MAT1-2 | Human, sputum | Spain |
| A. lentulus | CM-5563 | ND | MAT1-1 | Human, BAS | Spain |
| A. parafelis (former A. viridinutans/A. felis) | CM-3147T | NRRL 62900 | MAT1-2 | Human, OPE | Spain |
| A. parafelis (former A. viridinutans/A. felis) | CM-5623 | NRRL 62901 | MAT1-1 | Human, lungs | Portugal |
| A. pseudofelis (former A. viridinutans) | CM-6087T | NRRL 62903 | MAT1-2 | Human, sputum | Spain |
| A. pseudofelis (former A. viridinutans/A. felis) | CM-4518 | NRRL 62902 | MAT1-2 | Human, nail | Spain |
| A. pseudoviridinutans (former A. viridinutans) | NIHAV1T | NRRL 62904 | MAT1-1 | Human, lymph node | USA |
| A. pseudoviridinutans (former A. viridinutans) | NRRL 6106 | NRRL 6106 | MAT1-1 | Unknown | Unknown |
Superscript T, type strain; ARS, Agricultural Research Service Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA; ND, not deposited in ARS; BAS, bronchoalveolar secretion; OPE, oropharyngeal exudate.
Mating type reported in Nováková et al. (11).
Phylogenetic analysis.
DNA isolation, sequencing, and phylogenetic analysis were carried out as described elsewhere (19). Briefly, sequences from each locus were aligned using Clustalw, the best fit model was determined using ModelTest, phylogeny was calculated using likelihood, and clade credibility was determined from 500 bootstrap samples, all with routines contained in Mega5.2. Routines were also run in Mega to determine whether these loci were suitable for determining evolutionary distance as a function of the molecular clock (20). The partition homogeneity test in PAUP* was used to determine the suitability of combining data (21). Highly credible clades were compared between five loci, encoding beta-tubulin (BT2), calmodulin (CF), minichromosome maintenance factor (Mcm7), the second-largest subunit of RNA polymerase II (RPB2), and a pre-rRNA processing protein (Tsr1). Majority rule was used to determine the species according to genealogical concordance analysis (12, 22). Loci were amplified using primersMcm7-709for and Mcm7-1348rev for Mcm7, fRPB2-5F and fRPB2-7cR for RPB2, Tsr1-1453for and Tsr1-2308rev for Tsr1, BT2a and BT2b for BT2, and cf-1L and cf-4 for CF, as described elsewhere (23–26).
Mating type.
Mating type was determined by sequencing amplicons generated with primers AVN-9F (CTAGGCCACTTGATATCGAATG) and AVN-12R (TTGCTCAAGTAACAACAACAGAAG). The template was genomic DNA isolated as described elsewhere (19).
Growth.
To determine growth, strains were inoculated onto either MEA or CZA plates and incubated at 37, 42, and 50°C for 2 to 7 days.
Drug resistance.
Antifungal MICs were determined according to the Clinical and Laboratory Standards Institute reference method for broth dilution (M38-A2) (27). The drugs tested were voriconazole (Pfizer), itraconazole (Sigma, St. Louis, MO), and amphotericin B (Sigma, St. Louis, MO).
Virulence.
Galleria mellonella larvae in the final larval stage (Vanderhorst Wholesale, Inc., St. Marys, OH) were inoculated on the same day of their arrival. Larvae (15 per strain) were inoculated with 5 μl of conidial suspension (2 × 107 conidia/ml) as described elsewhere (28, 29) and incubated at 37°C. Mortality was scored based on color change (from pale beige to dark brown or black) and unresponsiveness to tactile stimulus. Two strains of mice were also used for virulence studies, BALB/c mice (Division of Cancer Treatment, National Cancer Institute, NIH, USA) immunosuppressed with hydrocortisone acetate (Sigma, St. Louis, MO) and gp91phox-deficient mice (B6.129S6-Cybbtm1Din/J) (The Jackson Laboratory, USA). Hydrocortisone acetate treatment was carried out as described elsewhere (18). BALB/c and gp91phox-deficient mice were inoculated with 30 μl of conidial suspension containing 3 × 107 and 3 × 105 conidia/ml, respectively. All mice were inoculated via pharyngeal aspiration (30). Groups of 6 and 10 mice per strain were used for gp91phox-deficient and BALB/c mice, respectively. Survival was monitored for 15 days, and survival curves were compared using the log-rank test. Studies were performed under a protocol approved by the Institutional Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases at the U.S. National Institutes of Health (NIH).
Mating.
Strain crosses were carried out as described elsewhere (18). Briefly, strains of the opposite mating type were inoculated pairwise onto OA plates, which were then sealed with Parafilm and incubated at 30°C. The mating plates were monitored weekly for the formation of cleistothecia and viable ascospores. Ascospore viability assays were carried out as described previously (18). Briefly, ascospores were released from cleistothecia and suspended in water at concentrations adjusted to 1 × 103 ascospores/ml by hemocytometer counts. Aliquots of 100 μl were incubated at 70°C for 30 min to eliminate contaminating conidia and then inoculated onto MEA plates. The plates were incubated at 37°C for 48 h. The number of growing colonies was scored as the number of viable ascospores. Single colonies were then transferred to MMA slants, incubated at 37°C for 7 days, and stored at 26°C for further analysis. To confirm that the heat treatment at 70°C eliminated all contaminating conidia from the parental strains, the following control was prepared: conidia from the parental strains were harvested from the mating plate, suspended in water, heat treated as described above, plated on MEA, and incubated at 37°C for 2 to 3 days. No growth was observed in any of the controls, indicating efficient killing of the parental conidia.
Hygromycin resistance.
Conidia were inoculated onto MEA with or without hygromycin (Sigma, St. Louis, MO) (200 μg/ml) and incubated at 37°C for 6 days. Colonies growing on MEA supplemented with hygromycin were scored as resistant to the drug.
SEM.
Conidia and cleistothecia were suspended in 6% aqueous glutaraldehyde solution (Electron Microscopy Sciences, Hatfield, PA) before processing for scanning electron microscopy (SEM). Samples were fixed for 2 h and then allowed to settle on silicon chips for 1 h. Subsequent postfixation with 1% OsO4 was performed with microwave irradiation (Pelco 3451 microwave processor) in cycles of 2 min on, 2 min off, and 2 min on at 250 W under a vacuum of 15 in. of Hg (Ted Pella, Redding, CA). Specimens were dehydrated in a graded ethanol series for 1 min under vacuum. Samples were then critical point dried (CPD) in a Bal-Tec CPD 030 drier (Balzer, Bal-Tec AG, Balzers, Liechtenstein). Cells were then coated with 75 Å of iridium in an ion beam sputter (IBS) system (South Bay Technology, Inc., San Clemente, CA) and imaged on a Hitachi SU-8000 SEM (Hitachi, Pleasanton, CA).
Accession numbers.
The sequences of the genes encoding the mating-type proteins of the MAT locus are deposited in GenBank under accession numbers KJ858505 (A. parafelis CM-3147), KJ858506 (A. parafelis CM-5623), KJ858507 (A. pseudofelis CM-6087), KJ858508 (A. pseudofelis CM-4518), KJ858509 (A. pseudoviridinutans NIHAV1), KJ858510 (A. pseudoviridinutans NRRL 6106), and KJ858511 (A. viridinutans NRRL4365). Sequences of the protein coding loci used in the phylogenetic analysis are deposited in GenBank under the following accession numbers: EF661252, EF669805, EF669808, EF669825, KJ572796, and KJ914684 to KJ914697 (BT2); EF661266, EF669874, EF669877, EF669895, KJ572797, and KJ914698 to KJ914711 (CF); KJ572799 and KJ914712 to KJ914728 (Mcm7); EF661238, EF669734, EF669738, EF669756, KJ572800, and KJ914729 to KJ914742 (RPB2); and KJ572801 and KJ914743 to KJ914760 (Tsr1). The scientific names and descriptions of the novel species were deposited in MycoBank under accession numbers MB 808634, MB 808636, and MB 808637.
RESULTS
Phylogenetic analysis.
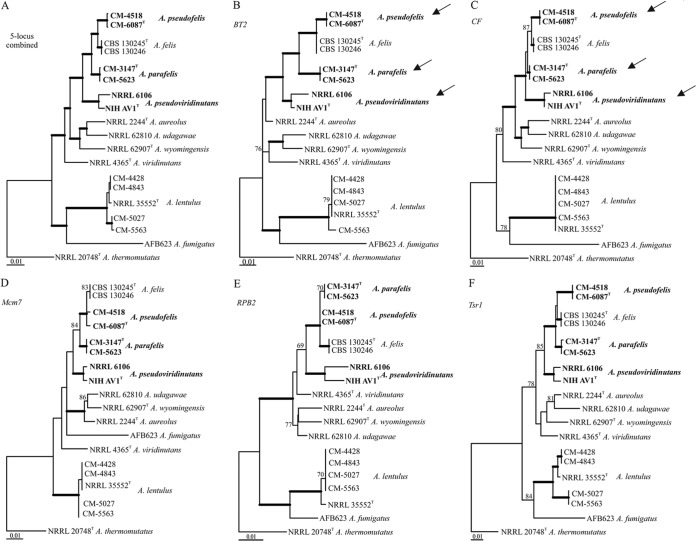
A total of 19 strains were employed for mating, construction of phylogenetic trees, and phenotypic analysis (Table 1). Of those, nine clinical strains (one isolated in the United States, one in Portugal, and seven in Spain) were originally identified as A. lentulus or A. viridinutans (15, 31–33; also M. Cuenca-Estrella and E. Mellado, unpublished data). Since three of these strains were recently reclassified as A. felis (10), a phylogenetic reexamination was carried out to confirm species identity. Strains CM-4428, CM-4843, CM-5027, CM-5563, CM-3147, CM-5623, CM-4518, CM-6087, and NIHAV1 were chosen for the phylogenetic analysis. The trees were constructed on the basis of genealogical concordance analysis using the sequences of BT2, CF, RPB2, Mcm7, and Tsr1. Genealogical concordance analysis based on gene sequences from all five loci showed that CM-4428, CM-4843, CM-5027, and CM-5563 form a monophyletic cluster with the A. lentulus type strain, confirming their original identification (Fig. 1). The other strains (formerly identified as A. viridinutans or A. felis) formed three clusters that separated from the type strains of A. viridinutans and A. felis based on the sequences of BT2 and CF (Fig. 1B and C, arrows). The partition homogeneity test showed that the loci could be combined, and the tree based on the combined sequences of all five loci strongly supported the presence of three groups distinct from A. viridinutans or A. felis, i.e., strains CM-3147 and CM-5623 in Aspergillus parafelis, strains CM-4518 and CM-6087 in Aspergillus pseudofelis, and strains NIHAV1 and NRRL 6106 in Aspergillus pseudoviridinutans (Fig. 1A), which could be identified as novel phylogenetic species.
FIG 1.
Maximum-likelihood trees based on DNA sequences from the five loci combined and each locus separately. (A) Combined data from five loci, BT2, CF, Mcm7, RPB2, and Tsr1; (B) beta-tubulin gene (BT2); (C) calmodulin gene (CF); (D) minichromosome maintenance factor 7 gene (Mcm7); (E) gene encoding the second-largest subunit of RNA polymerase II (RPB2); (F) preribosomal protein gene (Tsr1). Branches with thick lines have 90 to 100% bootstrap support. Bootstrap support of 69 to 89% is indicated by numerals. The validity of the new species is based on the congruence of a majority of statistically supported nodes in the single-locus trees (12, 13).
Species descriptions.
Aspergillus parafelis (Sugui, S. W. Peterson, et Kwon-Chung) sp. nov., Aspergillus pseudofelis (Sugui, S. W. Peterson, et Kwon-Chung) sp. nov., and Aspergillus pseudoviridinutans (Sugui, S. W. Peterson, et Kwon-Chung) sp. nov. were submitted to MycoBank under accession numbers MB 808634, MB 808636, and MB 808637, respectively.
A. parafelis. (i) Holotype.
The holotype of A. parafelis is NRRL 62900 (= CM-3147), a culture permanently preserved in a metabolically inactive state as a freeze-dried preparation. The culture was isolated in 2004 from a human oropharyngeal exudate specimen at the National Center for Microbiology, Spain.
(ii) Etymology.
The name is taken from the Greek root “para,” or “beside,” to indicate that the species is beside A. felis phylogenetically.
(iii) Description.
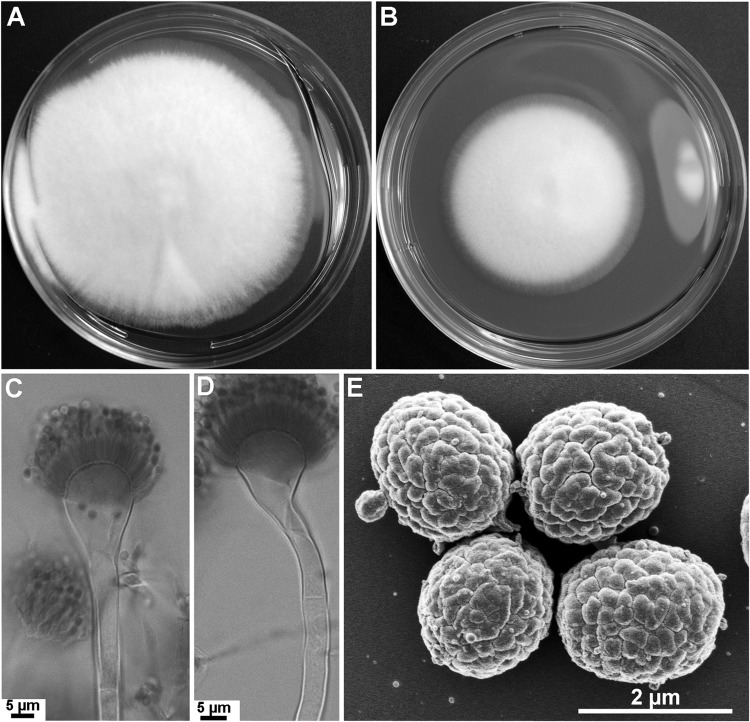
Colonies attained a diameter of 5.5 to 6.5 cm on MEA, as well as on CZA, in 5 days at 37°C (Fig. 2A and B). On MEA, colonies are white with dense aerial hyphae and scanty conidiation. The colony reverse is yellow. Conidiophores are hyaline, bearing conidial heads consisting of flask-shaped vesicles (10 to 20 μm in diameter) with uniseriate phialides producing long chains of conidia (Fig. 2C). Some conidial heads are slightly nodding, while the majority of conidial heads are straight (Fig. 2C and D). Conidia are subglobose (1.5 to 1.7 by 1.8 to 2.2 μm) and green in color, with delicate surface roughness created by shallow grooves (Fig. 2E). Cleistothecia are globose, white, 400 to 800 μm in diameter, and contain numerous 8-spored asci. Ascospores are lenticular, 3.5 to 4.5 by 4 to 4.75 μm, and have two equatorial crests and echinulate convex surfaces (Fig. 2F and G).
FIG 2.
Aspergillus parafelis CM-3147 T (T, type strain). (A, B) Colonies grown on malt extract agar (MEA) (A) and Czapek's solution agar (CZA) (B) for 5 days at 37°C; (C, D) variation in conidiophores: straight (C) and slightly nodding (D); (E) SEM of conidia; (F, G) ascospores viewed by light microscopy (F) and SEM (G). (F) White arrow points to conidia; black arrows point to ascospores.
A. pseudofelis. (i) Holotype.
The holotype of A. pseudofelis is NRRL 62903 (= CM-6087), a culture permanently preserved in a metabolically inactive state as a freeze-dried preparation. The culture was isolated in 2010 from a human sputum sample at the National Center for Microbiology, Spain.
(ii) Etymology.
The name is taken from the Greek root “pseudo,” or “false,” indicating that the species is not A. felis.
(iii) Description.
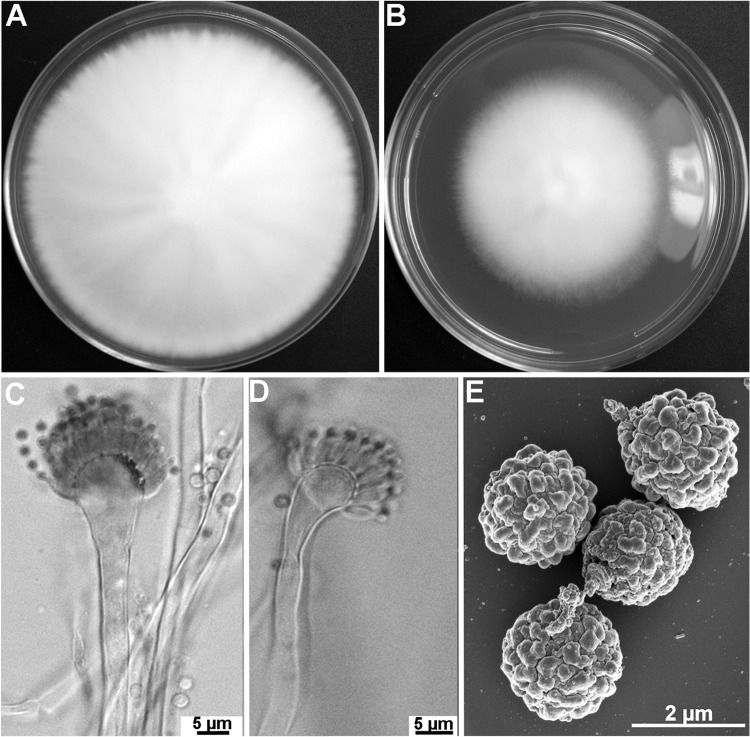
Colonies attained a diameter of 7.5 to 8 cm on MEA or 5 to 5.5 cm on CZA in 5 days at 37°C (Fig. 3A and B). On MEA, colonies are whitish, with dense aerial hyphae and poor sporulation. The colony reverse is yellow. Conidiophores are hyaline, straight or slightly sinuous, and bear columnar conidial heads composed of flask-shaped vesicles (12 to 16 μm diameter) with uniseriate phialides producing long conidial chains (Fig. 3C and D). Conidia are green and subglobose (1.7 to 1.9 by 1.9 by 2.2 μm), with delicate surface roughness and shallow grooves (Fig. 3E). All the strains included in this study were of one mating type, MAT1-2, and thus, cleistothecium formation was not observed.
FIG 3.
Aspergillus pseudofelis CM-6087 T. (A, B) Colonies grown on malt extract agar (MEA) (A) and Czapek's solution agar (CZA) (B) for 5 days at 37°C; (C, D) variation in conidiophores: straight (C) and slightly nodding (D); (E) SEM of conidia.
A. pseudoviridinutans. (i) Holotype. The holotype of A. pseudoviridinutans is NRRL 62904 (= NIHAV1), a culture permanently preserved in a metabolically inactive state as a freeze-dried preparation. The culture was isolated in 2004 from a human mediastinal lymph node specimen at the U.S. National Institutes of Health.
(ii) Etymology.
The name is taken from the Greek root “pseudo,” or “false,” indicating that the species is not A. viridinutans.
(iii) Description.
Colonies attained a diameter of 9 to 9.5 cm on MEA or 6 to 6.5 cm on CZA in 5 days at 37°C (Fig. 4A and B). On MEA, colonies are white with dense aerial hyphae and scant conidiation. The colony reverse becomes yellow with age. Conidiophores are hyaline and straight or slightly wavy. The conidial heads are columnar and consist of round, flask-shaped vesicles (7.5 to 10 μm diameter) with uniseriate phialides that produce conidial chains (Fig. 4C and D). Conidia are green and globose (1.75 to 2 μm), with a rough surface texture and uneven waviness (Fig. 4E). The two strains belonging to this species are of the MAT1-1 mating type, and thus, sexual reproduction could not be evaluated.
FIG 4.
Aspergillus pseudoviridinutans NIHAV1T. (A, B) Colonies grown on malt extract agar (MEA) (A) and Czapek's solution agar (CZA) (B) for 5 days at 37°C; (C, D) variation in conidiophores: straight (C) and slightly nodding (D); (E) SEM of conidia.
Phenotypical analysis.
Phenotypical analyses were carried out to evaluate the biotypes of the novel strains. Aspergillus fumigatus strain B-5233 and the type strains of A. lentulus, A. felis, and A. viridinutans were included in the assays for purposes of comparison.
Conidial morphology.
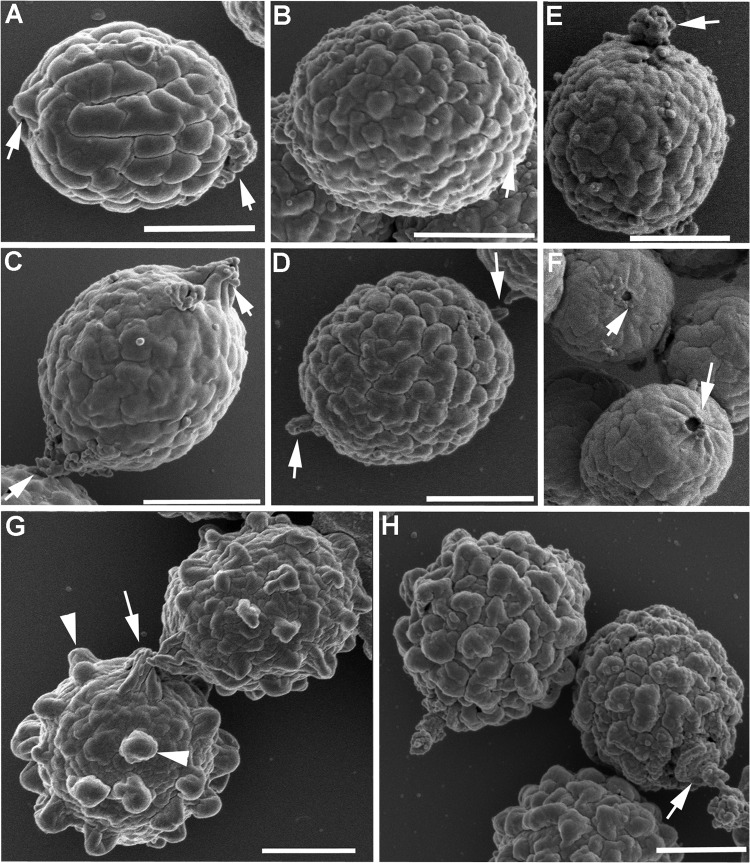
SEM revealed that the conidial topographies of A. parafelis and A. pseudofelis were distinct from those of A. felis and A. viridinutans. The surface texture of A. felis conidia resembled a walnut shell, with smooth waviness and deep grooves that were widely spaced (Fig. 5A), whereas the conidial surface in A. viridinutans was less wavy and had shallower grooves (Fig. 5C). The surface textures of A. parafelis and A. pseudofelis conidia were similar to each other, with jagged waviness and narrowly spaced grooves arranged in irregular reticulation (Fig. 5B and D). The surface roughness observed in A. pseudoviridinutans conidia resembled that of A. fumigatus conidia except that in A. fumigatus conidia, the texture was due to prominent spikes (Fig. 5G, arrowheads), whereas in A. pseudoviridinutans conidia, the surface roughness resulted from an accentuated topographic unevenness (Fig. 5H). The conidia of A. parafelis appeared to be linked to each other by a snap-and-lock type of mechanism, with a connecting pin and a docking orifice located in opposite poles of each conidium (Fig. 5E and F, arrows). The conidia of A. felis, A. pseudofelis, A. pseudoviridinutans, A. viridinutans, and A. fumigatus show appendages, often seen bipolarly, which appear to function as bridges connecting the conidia (Fig. 5A, C, D, G, and H, arrows). Whether the connection between conidia is species specific is unclear. Further examination is required to analyze the connectivity in detail.
FIG 5.
SEM of conidia. (A) Aspergillus felis; (B, E, F) A. parafelis; (C) A. viridinutans; (D) A. pseudofelis; (G) A. fumigatus; (H) A. pseudoviridinutans. Arrowheads point to spikes (G), and arrows point to appendagelike structures (A, C, D, E, G, H) or round orifices (F) located on the conidial surface. Bars = 1 μm.
Growth.
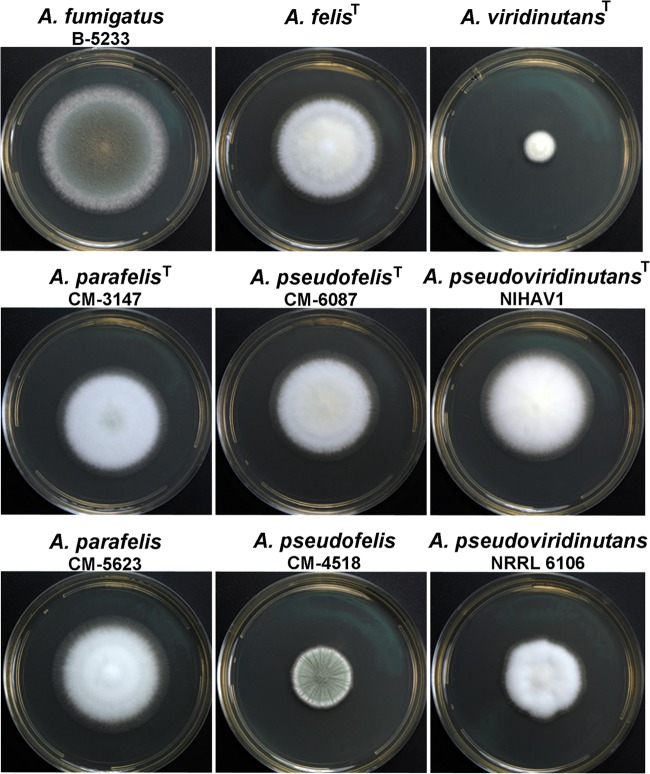
At 37°C, A. parafelis, A. pseudofelis, and A. pseudoviridinutans showed an intermediate growth rate between those of A. fumigatus (fast) and A. viridinutans (slow) (Fig. 6). Of note, the growth of the two A. pseudofelis strains, CM-6087 and CM-4518, was distinct. The colonies of CM-6087 attained a larger diameter than those of CM-4518 but sporulated poorly, whereas the colonies of CM-4518 produced abundant conidia. However, the conidia produced by these strains exhibited identical surface topographies as observed by SEM (data not shown). At 42°C, all the strains showed reduced growth but the growth pattern was similar to that observed at 37°C. At 50°C, only A. fumigatus was able to grow.
FIG 6.
Colonies grown on malt extract agar (MEA). Plates were inoculated and then incubated at 37°C for 3 days.
Drug resistance.
Antifungal susceptibilities were assessed by determining the MIC values (27) for amphotericin B, itraconazole, and voriconazole (Table 2). Among the three drugs, susceptibility to amphotericin B varied the least, with the A. viridinutans type strain being the most susceptible species. With voriconazole, the highest MIC values were observed in A. felis, A. parafelis, A. pseudofelis, and A. pseudoviridinutans NIHAV1. Aspergillus lentulus showed an intermediate susceptibility, while A. fumigatus and A. viridinutans were the most susceptible to voriconazole. The greatest variation of MIC values was observed with itraconazole. Aspergillus fumigatus, A. lentulus, A. viridinutans, and A. pseudoviridinutans NRRL 6106 showed MICs of ≦2 μg/ml, while the other species showed MICs of ≧8 μg/ml. Of note, A. parafelis and A. pseudofelis strains were able to grow at 16 μg/ml, the highest concentration of the drug tested, suggesting significant resistance to itraconazole.
TABLE 2.
MICs of amphotericin B, itraconazole, and voriconazole
| Strain | MIC (μg/ml) of: |
||
|---|---|---|---|
| Amphotericin B | Itraconazole | Voriconazole | |
| A. fumigatus B-5233 | 1 | 2 | 2 |
| A. lentulusT | 1 | 1 | 4 |
| A. viridinutansT | 0.25 | 1 | 1 |
| A. felisT | 1 | 8 | 8 |
| A. parafelis CM-3147 | 2 | >16 | 8 |
| A. parafelis CM-4518 | 2 | >16 | 8 |
| A. pseudofelis CM-5623 | 2 | >16 | 8 |
| A. pseudofelis CM-6087 | 2 | >16 | 8 |
| A. pseudoviridinutans NIHAV1 | 2 | >16 | 8 |
| A. pseudoviridinutans NRRL 6106 | 2 | 2 | 2 |
Virulence.
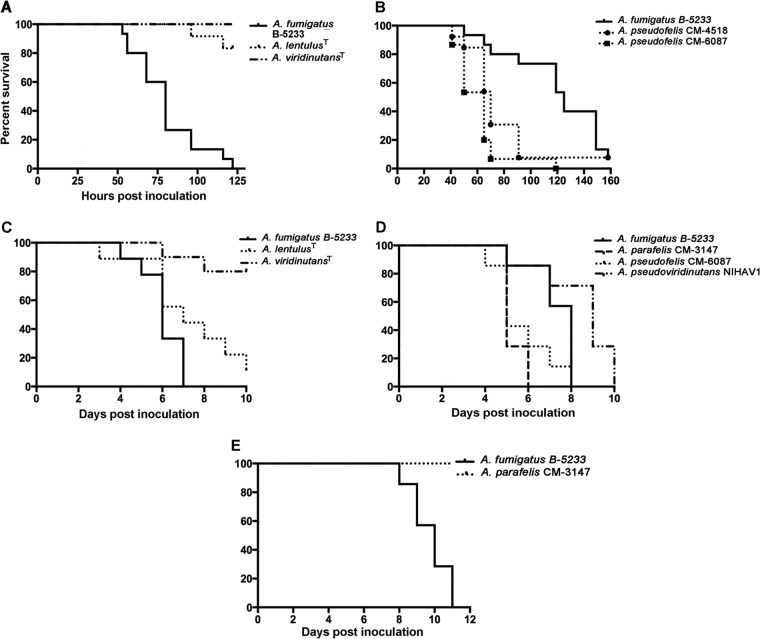
Two models were used for the virulence assays. In the first model, larvae of G. mellonella were used as the host. In this model, A. fumigatus was significantly more virulent than the A. lentulus and A. viridinutans type strains (Fig. 7A). Strains of Aspergillus pseudofelis and the A. felis type strain showed significantly higher virulence than A. fumigatus B-5233 (P ≤ 0.0001) (Fig. 7B and data not shown), while A. parafelis and A. pseudoviridinutans showed virulence similar to that of A. fumigatus B-5233 (data not shown). The survival curves also showed that the two isolates representing A. parafelis possessed similar virulence. The same concordance in virulence was found in the isolates of A. pseudofelis and A. pseudoviridinutans.
FIG 7.
Virulence of Aspergillus species in larvae and murine models. (A, B) Larvae of G. mellonella; (C, D) BALB/c mice immunosuppressed with hydrocortisone; (E) CGD mice. Hosts were inoculated with A. fumigatus (B-5233), A. parafelis, A. pseudofelis, A. pseudoviridinutans, and type strains of A. lentulus and A. viridinutans. G. mellonella larvae were inoculated with 2 × 107 conidia/ml and incubated at 37°C. Mice were inoculated via postpharyngeal aspiration with 3 × 107 (BALB/c) or 3 × 105 (CGD) conidia/ml.
Virulence comparisons were then performed in two murine hosts, BALB/c mice immunosuppressed with hydrocortisone and mice with chronic granulomatous disease (CGD). Strains CM-3147 (A. parafelis), CM-6087 (A. pseudofelis), and NIHAV1 (A. pseudoviridinutans), along with the type strains of A. viridinutans, A. lentulus, and A. felis, were chosen for the virulence assays. In immunosuppressed BALB/c mice, A. parafelis, A. pseudofelis, and A. felis showed slightly higher virulence than A. fumigatus (P ≤ 0.04) (Fig. 7D and data not shown), whereas A. pseudoviridinutans and A. lentulus displayed virulence similar or lower to that of A. fumigatus (Fig. 7C and D). The Aspergillus viridinutans type strain was the least virulent of all the strains and was the only strain that showed significantly reduced virulence compared to that of A. fumigatus (P = 0.0001) (Fig. 7C).
In the CGD model, while A. fumigatus caused 100% fatality in 11 days, none of the other species tested caused death in the same period of time. Figure 7E shows a representative survival curve of CGD mice inoculated with either A. fumigatus or A. parafelis CM-3147. The results of the virulence assays in hydrocortisone acetate-treated mice suggested that, similar to A. fumigatus, the novel species are capable of causing fatal infection in severely immunosuppressed hosts. However, dissimilar to A. fumigatus, the novel species are less virulent in the host that is deficient in the production of reactive oxygen species. Whether the novel species are isolated more often from patients with any particular type of immunodeficiency is unknown.
Mating.
Sexual crosses included A. fumigatus, A. lentulus, A. viridinutans, A. felis, A. wyomingensis, A. parafelis, A. pseudofelis, and A. pseudoviridinutans (Table 3). Intraspecific mating was observed between several A. lentulus strains of the opposite mating type, confirming the functional heterothallism of this species (9). Analysis of 8-week-old crosses between CM-5027 and CM-5563, the most fertile A. lentulus isolates, revealed abundant cleistothecia in the junction zones of the two opposite mating type strains. The ascospore viability was 8% on average, and equal ratios of MAT1 to MAT2 mating types were observed among the progeny. A. parafelis isolates CM-3147 (MAT1-2) and CM-5623 (MAT1-1) also completed their sexual cycle in 8 weeks. Isolates of the other species used in this study (A. viridinutans, A. pseudofelis, and A. pseudoviridinutans) were either all MAT1-1 or all MAT1-2 mating type, and thus, their intraspecific fertility could not be assessed.
TABLE 3.
Intra- and interspecific mating among sister species of section Fumigati
| MAT1-2 strain | Fertility of MAT1-1 strain |
|||||||
|---|---|---|---|---|---|---|---|---|
| A. fumigatus AFB623 | A. lentulus CM-5563 | A. felis CBS130246 | A. parafelis CM-5623 | A. viridinutans NRRL 35552T |
A. pseudoviridinutans |
A. wyomingensis CCF 4417 | ||
| NIHAV1 | NRRL6106 | |||||||
| A.fumigatus AFIR928 | + | − | − | − | − | − | − | − |
| A. lentulus CM-5027 | − | + | − | − | − | − | − | − |
| A. felis CBS 130245T | − | − | + | − | − | − | − | − |
| A. parafelis CM-3147 | + | − | + | + | + | + | + | + |
| A. pseudofelis CM-6087 | − | − | + | − | − | − | − | − |
| A. pseudofelis CM-4518 | − | − | + | − | − | − | − | − |
| A. wyomingensis CCF 4416 | − | − | − | − | − | − | − | + |
Two cases of unanticipated fertility were observed among the interspecific crosses (Table 3). Aspergillus parafelis isolate CM-3147 (MAT1-2) mated successfully with MAT1-1 isolates of A. fumigatus, A. pseudoviridinutans, A. viridinutans, A. felis, and A. wyomingensis. The MAT1-1 isolate of A. parafelis CM-5623, however, did not mate successfully in any interspecific crosses. In contrast to A. parafelis CM-3147, the A. pseudofelis MAT1-2 isolates (CM-4518 and CM-6087) were only fertile when crossed with A. felis CBS130246 (MAT1-1). In general, interspecific crosses produced cleistothecia with viable ascospores in 4 to 8 weeks. The average viability of the hybrid ascospores harvested from 8-week-old crosses was less than 1%, significantly lower than that from conspecific pairings of A. parafelis (4%), A. lentulus (8%), and A. fumigatus (40%) (18).
Interspecies genetic recombination.
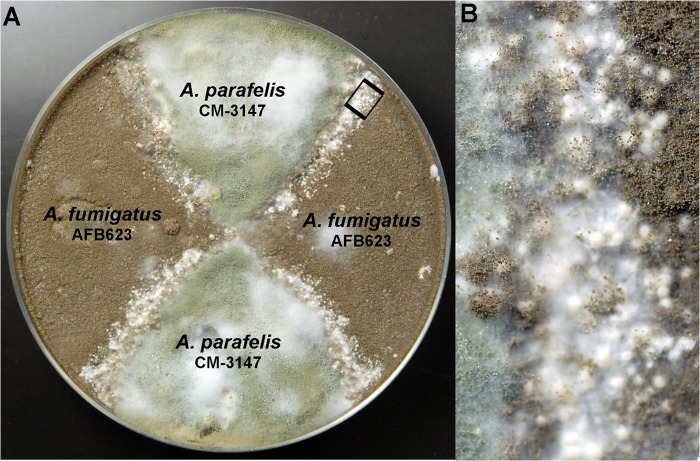
Progeny from the cross between A. parafelis and A. fumigatus were chosen for recombinational analysis of the genetic markers. The mutant AFB623 (18), derived from A. fumigatus supermater AFB62 (18), was used as a mating partner to facilitate genetic recombination analysis (Fig. 8). This mutant, a MAT1-1 mating type strain, produces brown conidia due to deletion of the abr2 gene (AFUA_2g17530) (18) and harbors a hygromycin resistance cassette used for the deletion of the gene. The genetic markers analyzed for recombination included conidial color, hygromycin resistance, thermotolerance at 50°C, and mating type. For recombinational analysis, eight offspring from an 8-week-old mating plate were randomly chosen at an early growth stage, before conidiation. The selected progeny included parental ditypes and recombinants (Table 4). Progeny P2 was brown, hygromycin resistant, thermotolerant to 50°C, and of the MAT1-1 mating type, all from the AFB623 parent. Progeny P3 and P8 were green, susceptible to hygromycin, unable to grow at 50°C, and of the MAT1-2 mating type, all from the A. parafelis parent. Progeny P1 displayed a mixed phenotype with brown conidia, hygromycin resistance, MAT1-2 mating type, and inability to grow at 50°C. Progeny P4, P5, P6, and P7 also showed a mixed phenotype. They produced green conidia that were hygromycin resistant and thermotolerant. However, the conidia harbored MAT1-1and MAT1-2 mating types. Since their conidial sizes were slightly larger than that of either parent, these may be diploids or aneuploids, which could have resulted from chromosome missegregation during meiosis due to the genetic diversity between the parental strains. Among these progeny, only P5 and P7 were fertile; they produced a complete sexual cycle with parent A. fumigatus AFB623 but not with parent A. parafelis CM-3147. Interestingly, only P5 underwent self-mating with production of cleistothecia and viable ascospores. The fertility of P1, P2, P3, and P8 was also tested by backcrosses with parental strains. The MAT1-2 progeny, P1, P3, and P8, were crossed with A. fumigatus AFB623, whereas the MAT1-1 progeny, P2, was crossed with A. parafelis CM-3147. At 6 weeks, all four crosses showed cleistothecia with viable ascospores at the junction zone.
FIG 8.
Mating between A. parafelis CM-3147 and A. fumigatus AFB623. (A) An 8-week-old cross with an abundant production of cleistothecia; (B) magnification of inset in panel A showing cleistothecia produced in the junction zone of A. parafelis (green conidia) and A. fumigatus AFB623 (brown conidia).
TABLE 4.
Interspecies genetic recombination
| Parental strain or progeny | Mating type(s) | Conidial color | Growth at 50°C | Hygromycin resistance |
|---|---|---|---|---|
| Parents | ||||
| A. fumigatus AFB623 | MAT1-1 | Brown | Yes | Yes |
| A. parafelis CM-3147 | MAT1-2 | Green | No | No |
| Progeny | ||||
| P1 | MAT1-2 | Brown | No | Yes |
| P2 | MAT1-1 | Brown | Yes | Yes |
| P3 | MAT1-2 | Green | No | No |
| P4 | MAT1-1 and MAT1-2 | Green | Yes | Yes |
| P5 | MAT1-1 and MAT1-2 | Green | Yes | Yes |
| P6 | MAT1-1 and MAT1-2 | Green | Yes | Yes |
| P7 | MAT1-1 and MAT1-2 | Green | Yes | Yes |
| P8 | MAT1-2 | Green | No | No |
Ultrastructure comparisons.
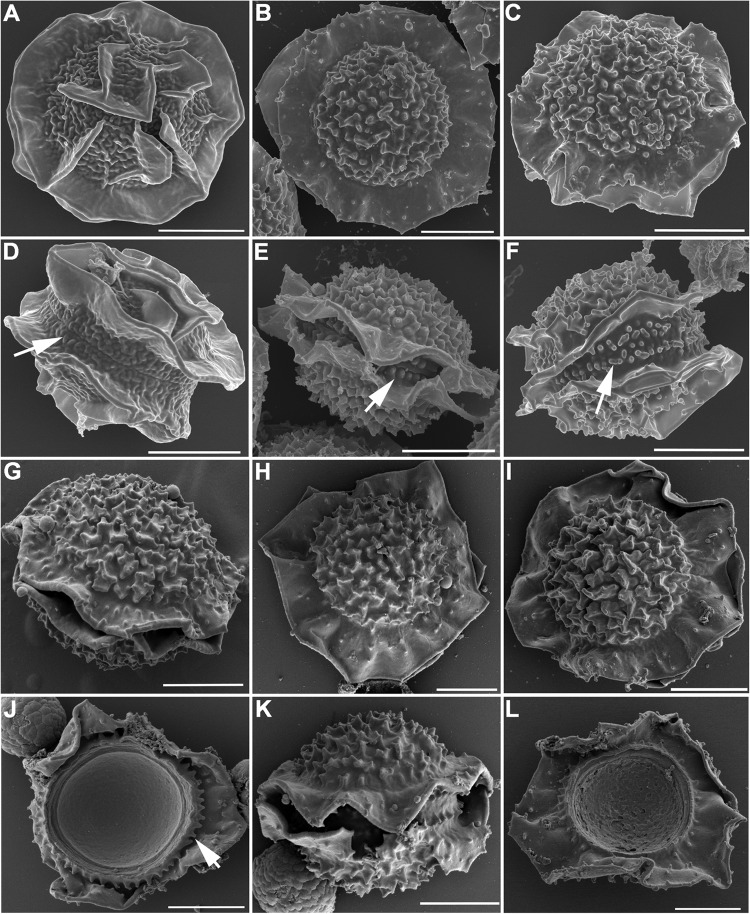
Scanning electron microscopy revealed that the ascospores produced by A. parafelis are lenticular, with two prominent equatorial crests and echinulate convex surfaces (Fig. 9B and E). The groove between the crests was ornamented with blunt toothlike projections arranged in double rows (Fig. 9E, arrow). Ascospores from A. fumigatus are also lenticular with two equatorial crests, but their convex surface is ornamented with unorganized tissue flaps and the groove between the crests is smoother and without the toothlike projections (Fig. 9A and D, arrow). The surface markings on the hybrid ascospores produced by A. parafelis CM-3147 and A. fumigatus AFB623 resembled those of A. parafelis more than those of A. fumigatus, except that the toothlike projections between the equatorial crests were not as homogeneous in size and shape as those of A. parafelis (Fig. 9C and F, arrow). The hybrid ascospores produced by mating of A. parafelis CM-3147 with A. viridinutans, A. felis, or A. pseudoviridinutans (Fig. 9G to L) or by mating of A. pseudofelis with A. felis showed the same general morphology: a lenticular spore body with two equatorial crests, toothlike projections in the groove between the crests, and echinulate convex surfaces. Figure 9J shows the toothlike structures aligned on the outer edge of one of the ascospore hemispheres (arrow). A similar view is shown in Fig. 9L.
FIG 9.
SEM of ascospores. (A, D) A. fumigatus AFIR928 × AFB623; (B, E) A. parafelis CM-3147 × CM-5623; (C, F) A. parafelis CM-3147 × A. fumigatus AFB623; (G, J) A. parafelis CM-3147 × A. viridinutans type strain; (H, K) A. parafelis CM-3147 × A. felis CBS 130246; (I, L) A. parafelis CM-3147 × A. pseudoviridinutans NIHAV1. Bars = 2 μm.
DISCUSSION
The identification of Aspergillus species is a multifaceted process that involves morphological, physiological, and phylogenetic parameters. Until DNA sequencing became readily available, several clinical strains were misidentified as A. fumigatus based solely on the morphological resemblance of their conidiogenous structures to those of A. fumigatus. The use of unlinked sequences from polymorphic loci was instrumental in distinguishing the isolates that resembled A. fumigatus from the true A. fumigatus (6). Since then, there has been a steady increase in the number of species in section Fumigati, which includes A. fumigatus (Table 5). The concordance of multiple-gene genealogies has been used previously to examine the phylogenetic relationship in other species. For instance, phylogeny based on three unlinked loci confirmed the prediction that the Neurospora discreta complex included several separate species, which correlated with the morphological variability and reproductive behavior in that complex (13).
TABLE 5.
List of Aspergillus species included in section Fumigati
| Speciesa | Reference strain |
|---|---|
| A. brevipes (37) | CBS 118.53T |
| A. caatingaensis (44) | IFM 61335T |
| A. duricaulis (37) | CBS 481.65T |
| A. felis* (10) | CBS 130245T |
| A. fumigatiaffinis* (37) | IBT12703T |
| A. fumigatus* (37) | CBS 133.61T |
| A. fumisynnematus* (37) | IFM 42277T |
| A. huiyaniae (45) | IFM57847T |
| A. lentulus* (37) | NRRL 35552T |
| A. marvanovae (46) | CCM8003T |
| A. novofumigatus* (37) | IBT 16806T |
| A. parafelis* (this study) | CM-3147T |
| A. pernambucoensis (44) | IFM 61342T |
| A. pseudofelis* (this study) | CM-6087T |
| A. pseudoviridinutans* (this study) | NIHAV1T |
| A. siamensis (47) | KUFC 6349 |
| A. turcosus (37) | KACC 4209T |
| A. unilateralis (37) | CBS 126.56T |
| A. viridinutans* (37) | NRRL 4365T |
| A. waksmanii (46) | NRRL 179T |
| A. wyomingensis (11) | CCF 4417T |
| N. assulata (37) | KACC 4169T |
| N. aurata (37) | CBS 466.65T |
| N. aureola (37) | CBS 105.55T |
| N. australensis (37) | CBS 112.55T |
| N. coreana (37) | KACC 41659T |
| N. denticulata (37) | CBS 652.73T |
| N. fennelliae (37) | CBS 598.74T |
| N. ferenczii (37) | CBS 121594T |
| N. fischeri* (37) | CBS 544.65T |
| N. galapagensis (37) | CBS 117522T |
| N. glabra (37) | CBS 111.55T |
| N. hiratsukae* (37) | CBS 294.93T |
| N. indohii (48) | CBM-FA-934T |
| N. laciniosa* (37) | KACC 41657T |
| N. multiplicata (37) | CBS 646.95T |
| N. nishimurae (37) | CBS 116047 |
| N. papuensis (37) | CBS 841.96T |
| N. pseudofischeri* (37) | NRRL 20748T |
| N. quadricincta (37) | CBS 135.52T |
| N. shendaweii (49) | IFM 5761T |
| N. spathulata (37) | CBS 408.89T |
| N. spinosa (37) | CBS 483.65T |
| N. stramenia (37) | CBS 498.65T |
| N. sublevispora (50) | IFM 53598T |
| N. takakii (51) | CBM-FA-884T |
| N. tatenoi (37) | CBS 407.93T |
| N. tsunodae (49) | IFM57609T |
| N. tsurutae (48) | CBM-FA-93T |
| N. udagawae* (37) | CBS 114217T |
| N. warcupii (37) | NRRL 35723T |
The novel species described in this study are highlighted in boldface. Human pathogens are marked with an asterisk (52).
In this study, the genealogical analysis carried out with five loci provided evidence that the five strains previously identified as A. viridinutans contained novel phylogenetic species. Three of these strains (CM-3147, CM-5623, and CM-4518) (Table 1) were recently reclassified as A. felis (10) based on the DNA sequence of either BT2 or CF. Barrs et al. reported that among the 24 strains identified as A. felis, CM-3147, CM-5623, and CM-4518 belonged to a small group (five strains) isolated from humans (10). Further phylogenetic analysis performed in this study, using the sequences of RPB2, Mcm7, andTsr1 in addition to those of BT2 or CF, demonstrated that in at least three of the five loci, these three strains clustered in separate monophyletic clades from A. felis. Thus, although these strains are biologically very close to A. felis, they should be considered different species. The phylogenetic analysis based on all five loci established the identity of a third novel species, Aspergillus pseudoviridinutans, formed by strains NIHAV1 and NRRL 6106. The morphological, physiological, and genetic variability within the A. viridinutans complex has been an issue since the late 1990s (34, 35). Phylogenetic analyses based on the sequences of BT2, an alkaline protease gene (Alp), the hydrophobin gene, and CF showed multiple clades among strains identified as A. viridinutans (4, 36). Recently, a revision based on phylogenetic analysis using the BT2 or CF sequences, as well as the biotypical characteristics, recognized the existence of five species within this complex, Aspergillus aureolus, Aspergillus udagawae, A. felis, A. viridinutans, and A. wyomingensis (11).
Phenotypic and morphological analysis corroborated the identification of the novel species identified in this study. Growth, thermotolerance, and drug resistance were similar among the three novel species but dissimilar to those of A. viridinutans and A. fumigatus. Furthermore, virulence assays in G. mellonella larvae and mice showed that A. viridinutans was markedly less virulent than the three novel species. The Aspergillus viridinutans type strain was isolated from rabbit dung (37), whereas the strains used in this study were mostly isolated from human patients, which may explain the differences in virulence. SEM revealed that (i) the surface topographies of A. parafelis and A. pseudofelis conidia were similar to each other but different from that of A. felis and (ii) the conidial morphology of A. pseudoviridinutans was markedly different from those of A. viridinutans, A. parafelis, and A. pseudofelis; both findings support the phylogenetic analysis (Fig. 5).
No differences in the biotypes of A. parafelis and A. pseudofelis were found within the parameters tested. However, a comparison of the MAT1-2 locus of the two A. pseudofelis strains, CM-6087 and CM-4518, with that of three other aspergilli (A. parafelis CM-3147, A. felis CBS130245, and A. fumigatus AF293) showed 100% identity between the two strains of A. pseudofelis and 10 single-nucleotide polymorphisms (SNPs) unique to the A. pseudofelis isolates. One SNP was found in the open reading frame of one of the mating proteins of the MAT locus, and five SNPs were present in the other mating protein. While the sequence of the MAT locus is not traditionally used for species identification, the unique SNPs of A. pseudofelis reinforce the hypothesis that they are separate species.
The phylogenetic distinction between members of section Fumigati is well established. However, their biological relatedness remains unclear. Crossbreeding between species was used as a tool to study the correlation between phylogenetic distance and biological reproductive success (or isolation). Mating between A. fumigatus and the related species included in this study revealed that reproductive compatibility varied among phylogenetically close sibling species. A. fumigatus and A. lentulus, though phylogenetically closer to each other than to the other sibling species, were reproductively isolated, but A. pseudofelis and A. felis were both phylogenetically close and reproductively compatible. In addition, mating between the MAT1-2 isolate of A. parafelis and the MAT1-1 isolates of A. fumigatus, A. viridinutans, A. felis, A. pseudoviridinutans, and A. wyomingensis (Fig. 8) suggests that phylogenetic divergence might have preceded the reproductive isolation in some of the related species. Disagreement between mating success and genealogical concordance is not an unprecedented phenomenon; a lack of correlation was also observed in the ancestral nature of the mating type genes in the Fusarium graminearum complex (38, 39). The finding of promiscuous mating seen in some A. parafelis and A. pseudofelis isolates demonstrates a mechanism for interspecific (horizontal) gene flow that could result in the variable phylogenies found at the different loci used in this study and the rejection of a molecular clock in the sequences as a consequence. This is apparently a rare occurrence, as to the best of our knowledge, no other promiscuous maters were described in section Fumigati. However, there is no reason to assume that such interspecies mating does not occur from time to time. In fact, interspecies mating has been shown to occur in Aspergillus section Flavi (40). In mating between Aspergillus flavus and Aspergillus minisclerotigenes, which were designated as two distinct species according to beta-tubulin and calmodulin sequences (41), not only were they able to undergo complete sexual cycles but their progeny were fertile when backcrossed with the parental strain of the opposite mating type (40).
Phenotype analysis of hybrid ascospores produced by mating of A. fumigatus with A. parafelis suggested that genetic recombination similar to that found in intraspecific mating occurred in half of the progeny. The presence of both mating loci in the other half of the progeny suggested the occurrence of chromosomal missegregation resulting in diploids or aneuploids. Such events occur frequently in matings between genetically diverse strains, e.g., in mating between the two varieties (var. neoformans and var. grubii) of Cryptococcus neoformans or between C. neoformans and its sister species C. gattii (42). The rate of aneuploid and diploid clone production was higher among the progeny isolated from mating between C. neoformans and C. gattii. The genetic diversity between the two Cryptococcus species is considerably higher than that between the two varieties within C. neoformans (43).
The findings in this study indicate that mating between species may not be reliable to determine species boundaries among the members of section Fumigati, since A. parafelis was able to mate with several of its sister species. In cases such as section Fumigati, majority rule concordance analysis should be used to recognize species boundaries. This study also reinforces the need of multiple loci for a higher resolution of the phylogenetic species boundary and accurate species identification. In cases where analysis of multiple loci is not feasible, either due to time constraints or availability of resources, the sequences of the beta-tubulin (BT2) and calmodulin (CF) genes, the two most commonly found sequences in public databases, can be used for the initial identification of the Aspergillus species. It is essential, however, to confirm the identity of the species with a third locus. Erroneous identification of clinical isolates might result in grave consequences for patients. For instance, the three novel species described here showed considerably higher resistance to antifungal drugs than did the type strain of A. viridinutans; thus, disease caused by these species is likely to be refractory to routine antifungal therapies and might require more aggressive therapy than is commonly used for aspergillosis due to A. fumigatus.
ACKNOWLEDGMENTS
This work was supported by the Intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1.Lin SJ, Schranz J, Teutsch SM. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358–366. 10.1086/318483 [DOI] [PubMed] [Google Scholar]
- 2.Raper KB, Fennell DI. 1965. The genus Aspergillus. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 3.Samson RA, Frisvad JC. 1990. Chemotaxonomy and morphology of Aspergillus fumigatus and related taxa, p 201–208 In Samson RA, Pitt JI. (ed), Modern concepts in Penicillium and Aspergillus classification, 185Springer, New York, NY [Google Scholar]
- 4.Katz ME, Dougall AM, Weeks K, Cheetham BF. 2005. Multiple genetically distinct groups revealed among clinical isolates identified as atypical Aspergillus fumigatus. J. Clin. Microbiol. 43:551–555. 10.1128/JCM.43.2.551-555.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon-Chung KJ. 1975. A new pathogenic species of Aspergillus in the Aspergillus fumigatus series. Mycologia 67:770–779. 10.2307/3758338 [DOI] [PubMed] [Google Scholar]
- 6.Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625–632. 10.1128/EC.4.3.625-632.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- 8.Balajee SA, Nickle D, Varga J, Marr KA. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705–1712. 10.1128/EC.00162-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swilaiman SS, O'Gorman CM, Balajee SA, Dyer PS. 2013. Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryot. Cell 12:962–969. 10.1128/EC.00040-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrs VR, van Doorn TM, Houbraken J, Kidd SE, Martin P, Pinheiro MD, Richardson M, Varga J, Samson RA. 2013. Aspergillus felis sp. nov., an emerging agent of invasive aspergillosis in humans, cats, and dogs. PLoS One 8:e64871. 10.1371/journal.pone.0064871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nováková A, Hubka V, Dudová Z, Matsuzawa T, Kubátová A, Yaguchi T, Kolaøík M. 2014. New species in Aspergillus section Fumigati from reclamation sites in Wyoming (U.S.A.) and revision of A. viridinutans complex. Fungal Divers. 64:253–274. 10.1007/s13225-013-0262-5 [DOI] [Google Scholar]
- 12.Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57:2721–2741. 10.1554/03-074 [DOI] [PubMed] [Google Scholar]
- 13.Dettman JR, Jacobson DJ, Taylor JW. 2006. Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia 98:436–446. 10.3852/mycologia.98.3.436 [DOI] [PubMed] [Google Scholar]
- 14.Zbinden A, Imhof A, Wilhelm MJ, Ruschitzka F, Wild P, Bloemberg GV, Mueller NJ. 2012. Fatal outcome after heart transplantation caused by Aspergillus lentulus. Transpl. Infect. Dis. 14:E60–E63. 10.1111/j.1399-3062.2012.00779.x [DOI] [PubMed] [Google Scholar]
- 15.Vinh DC, Shea YR, Jones PA, Freeman AF, Zelazny A, Holland SM. 2009. Chronic invasive aspergillosis caused by Aspergillus viridinutans. Emerg. Infect. Dis. 15:1292–1294. 10.3201/eid1508.090251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, Galvez EM, Mullbacher A, Gallin JI, Simon MM, Kwon-Chung KJ. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562–1569. 10.1128/EC.00141-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugui JA, Losada L, Wang W, Varga J, Ngamskulrungroj P, Abu-Asab M, Chang YC, O'Gorman CM, Wickes BL, Nierman WC, Dyer PS, Kwon-Chung KJ. 2011. Identification and characterization of Aspergillus fumigatus “supermater” pair. mBio 2:e00234–11. 10.1128/mBio.00234-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugui JA, Peterson SW, Clark LP, Nardone G, Folio L, Riedlinger G, Zerbe CS, Shea Y, Henderson CM, Zelazny AM, Holland SM, Kwon-Chung KJ. 2012. Aspergillus tanneri sp. nov., a new pathogen that causes invasive disease refractory to antifungal therapy. J. Clin. Microbiol. 50:3309–3317. 10.1128/JCM.01509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromham L, Penny D. 2003. The modern molecular clock. Nat. Rev. Genet. 4:216–224. 10.1038/nrg1020 [DOI] [PubMed] [Google Scholar]
- 21.Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods), version 4, Sinauer Associates, Sunderland, MA, USA [Google Scholar]
- 22.Degnan JH, DeGiorgio M, Bryant D, Rosenberg NA. 2009. Properties of consensus methods for inferring species trees from gene trees. Syst. Biol. 58:35–54. 10.1093/sysbio/syp008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K, Nelsen MP, Nelson NA, Rivas-Plata E, Shimp AD, Widhelm T, Lumbsch HT. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23:35–40. 10.3767/003158509X470602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16:1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- 25.Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226. 10.3852/mycologia.100.2.205 [DOI] [PubMed] [Google Scholar]
- 27.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed. Approved standard M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28.Jackson JC, Higgins LA, Lin X. 2009. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PLoS One 4:e4224. 10.1371/journal.pone.0004224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves EP, Messina CG, Doyle S, Kavanagh K. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158:73–79. 10.1023/B:MYCO.0000038434.55764.16 [DOI] [PubMed] [Google Scholar]
- 30.Rao GV, Tinkle S, Weissman DN, Antonini JM, Kashon ML, Salmen R, Battelli LA, Willard PA, Hoover MD, Hubbs AF. 2003. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J. Toxicol. Environ. Health A 66:1441–1452. 10.1080/15287390306417 [DOI] [PubMed] [Google Scholar]
- 31.Mellado E, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2011. Role of Aspergillus lentulus 14-alpha sterol demethylase (Cyp51A) in azole drug susceptibility. Antimicrob. Agents Chemother. 55:5459–5468. 10.1128/AAC.05178-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2008. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob. Agents Chemother. 52:1244–1251. 10.1128/AAC.00942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alastruey-Izquierdo A, Mellado E, Pelaez T, Peman J, Zapico S, Alvarez M, Rodriguez-Tudela JL, Cuenca-Estrella M, FILPOP Study Group 2013. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob. Agents Chemother. 57:3380–3387. 10.1128/AAC.00383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varga J, Rinyu E, Kiss I, Botos B, Kozakiewicz Z. 1997. Carbon source utilization and isoenzyme analysis as taxonomic aids for toxigenic Neosartorya species and their relatives. Acta Microbiol. Immunol. Hung. 44:1–11 [PubMed] [Google Scholar]
- 35.Varga J, Toth B, Rigo K, Debets F, Kozakiewicz Z. 2000. Genetic variability within the Aspergillus viridinutans species. Folia Microbiol. (Praha) 45:423–428 [DOI] [PubMed] [Google Scholar]
- 36.Yaguchi T, Horie Y, Tanaka R, Matsuzawa T, Ito J, Nishimura K. 2007. Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Nippon Ishinkin Gakkai Zasshi 48:37–46. 10.3314/jjmm.48.37 [DOI] [PubMed] [Google Scholar]
- 37.Samson RA, Hong S, Peterson SW, Frisvad JC, Varga J. 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud. Mycol. 59:147–203. 10.3114/sim.2007.59.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donnell K, Ward TJ, Geiser DM, Corby Kistler H, Aoki T. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41:600–623. 10.1016/j.fgb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 39.Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, Geiser DM, Suga H, Toth B, Varga J, O'Donnell K. 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 44:1191–1204. 10.1016/j.fgb.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 40.Damann K, DeRobertis C. 2013. Mating of Aspergillus flavus × Aspergillus minisclerotigenes hybrids: are they functionally mules? Phytopathology 103:S2.32–S2.33 [Google Scholar]
- 41.Varga J, Frisvad JC, Samson RA. 2011. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 69:57–80. 10.3114/sim.2011.69.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon-Chung KJ, Varma A. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574–587. 10.1111/j.1567-1364.2006.00088.x [DOI] [PubMed] [Google Scholar]
- 43.Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4:e5862. 10.1371/journal.pone.0005862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzawa T, Takaki GMC, Yaguchi T, Okada K, Gonoi T, Horie Y. 2014. Two new species of Aspergillus section Fumigati isolated from caatinga soil in the State of Pernambuco, Brazil. Mycoscience 55:79–88. 10.1016/j.myc.2013.04.001 [DOI] [Google Scholar]
- 45.Matsuzawa T, Horie Y, Abliz P, Gonoi T, Yaguchi T. 2014. Aspergillus huiyaniae sp. nov., a teleomorphic species in section Fumigati isolated from desert soil in China. Mycoscience 55:213–220. 10.1016/j.myc.2013.08.007 [DOI] [Google Scholar]
- 46.Hubka V, Peterson SW, Frisvad JC, Yaguchi T, Kubatova A, Kolarik M. 2013. Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. Int. J. Syst. Evol. Microbiol. 63:783–789. 10.1099/ijs.0.047076-0 [DOI] [PubMed] [Google Scholar]
- 47.Eamvijarn A, Manoch L, Chamswarng C, Piasai O, Visarathanonth N, Luangsa-ard, Kijjoa JJA. 2013. Aspergillus siamensis sp. nov. from soil in Thailand. Mycoscience 54:401–405. 10.1016/j.myc.2013.01.005 [DOI] [Google Scholar]
- 48.Horie Y, Abliz P, Fukushima K, Okada K, Takaki GMC. 2003. Two new species of Neosartorya from Amazonian soil, Brazil. Mycoscience 44:397–402. 10.1007/S10267-003-0132-1 [DOI] [Google Scholar]
- 49.Yaguchi T, Matsuzawa T, Tanaka R, Abliz P, Hui Y, Horie Y. 2010. Two new species of Neosartorya isolated from soil in Xinjiang, China. Mycoscience 51:253–262. 10.1007/S10267-010-0037-8 [DOI] [Google Scholar]
- 50.Someya A, Yaguchi T, Udagawa S. 1999. Neosartorya sublevispora, a new species of soil-borne Eurotiales. Mycoscience 40:405–409. 10.1007/BF02464395 [DOI] [Google Scholar]
- 51.Horie Y, Abliz P, Fukushima K, Okada K, Gusmão NB. 2001. Neosartorya takakii, a new species from soil in Brazil. Mycoscience 42:91–95. 10.1007/BF02463980 [DOI] [Google Scholar]
- 52.Sugui JA, Kwon-Chung KJ, Juvvadi P, Latgé JP, Steinbach WJ. Aspergillus fumigatus and related species. Cold Spring Harbor Perspect. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]