Abstract
From June to September 2012, 500 urine samples were recovered from patients with urinary tract infections (UTI) due to Gram-negative bacilli (≥104 leukocytes/ml and ≥105 Gram-negative isolates/ml) who visited the University hospital Bicêtre (France). They were challenged with extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae (ESBL-E) using the rapid diagnostic ESBL NDP test. Results of the ESBL NDP test were compared to the results of the double-disc susceptibility test (DDST) performed on solid-agar plates and molecular identification of the β-lactamase genes. Among the 450 nonduplicate urine samples, 11.3% were positive for ESBL-E by using the DDST, the ESBL determinants being mostly of the CTX-M type (CTX-M-15) according to molecular testing. Results of the ESBL NDP test were obtained within 15 min. The sensitivity and specificity of the ESBL NDP test were 98% and 99.8%, respectively, whereas the positive and negative predictive values of this test were 98% and 99.8%, respectively. A perfect correlation between cefotaxime resistance and positivity of the ESBL NDP test was observed. Therefore, the ESBL NDP test offers a powerful tool for a rapid identification of ESBL-E and associated resistance to expanded-spectrum cephalosporins. It may be useful in particular for guiding first-line antibiotic therapy.
INTRODUCTION
Urinary tract infections (UTIs) are the most prevalent infectious diseases, with an estimated overall incidence of 18/1,000 persons per year in the United States (1). According to the Centers for Disease Control and Prevention, UTIs that are mostly due to Escherichia coli account for more than 8.6 million visits to health care professionals each year in the United States (1, 2). In addition, multidrug resistance is now emerging worldwide among Gram-negative organisms, which are mostly responsible for UTIs (2).
One of the most important emerging resistance traits corresponds to resistance to broad-spectrum β-lactams in Enterobacteriaceae, which is mainly associated with acquired ESBLs of the CTX-M type (ESBL-E) (3, 4). Hence, ESBL-E are usually resistant to most β-lactams except cefoxitin and carbapenems. Therefore, efficient treatment of those infections is becoming challenging due a concomitant and rapid increase of the prevalence rate of ESBL-E worldwide and the perspective of a paucity of novel anti-Gram-negative molecules (3, 4).
The ESBL NDP test has been developed recently for rapid identification of ESBL-E (5). This test, based on the detection of hydrolysis of the β-lactam ring of cefotaxime, an extended-spectrum cephalosporin, is rapid, sensitive, and specific (5). Results are obtained within less than 1 h, long before any antibiotic testing results are obtained (ca. 6 to 24 h). The ESBL NDP test has previously been validated using cultured bacteria (5). Here, the test has been evaluated prospectively for its ability to detect ESBL-E responsible for UTIs directly from urine samples. Results showed that this test used directly with infected urine samples might be a useful guide for choosing a first-line antibiotic therapy.
(This work was presented to the European Congress of Clinical Microbiology and Infectious Diseases [ECCMID] in 2013 in Berlin, Germany.)
MATERIALS AND METHODS
Patient samples.
Over a 4-month period (June to November 2012), all urine samples sent to the Microbiology laboratory of the hospital Bicêtre, a 950-bed university hospital in a suburb of Paris, France, were tested. Urine samples were analyzed for the presence of leukocytes and bacteria. First, leukocytes were manually counted using Kova slides (Hycor Biomedical, Penicuit, United Kingdom) and interpreted as recommended (UTI = more than 104 leukocytes/ml [6]). Gram staining and microscope reading were done on all urine samples with more than 104 leukocytes/ml by examining 50 fields. The presence of 1 or more microorganisms per field after observation of at least 20 fields was considered a positive result corresponding to more than 105 Gram-negative bacteria/ml. The precise number of bacteria present in the urine sample was subsequently obtained by serial dilutions and plating on UriSelect 4 medium (Bio-Rad, Marnes-la-Coquette, France). CFU were counted after 24 h of incubation at 37°C. Only urine samples recovered from UTI due to Gram-negative bacilli (≥104 leukocytes/ml and positive Gram-negative staining) were included in the study.
ESBL NDP test using urine samples.
The ESBL NDP test was performed with 500 urine samples during the study period. The protocol of the ESBL NDP test for detection of the ESBL-E in urine samples was adapted from the original protocol published previously (5). First, 4.5 ml of urine specimen was transferred into three 1.5-ml Eppendorf tubes (tubes A, B, and C) (each tube containing 1.5 ml of urine). After a centrifugation at 13,000 × g during 2 min, the supernatant was discarded and the bacterial pellet was resuspended in 500 μl of distilled water. After a second centrifugation at 13,000 × g for 2 min, the supernatant was discarded and the bacterial pellet was resuspended in 100 μl of Tris-HCl lysis buffer (B-PER II bacterial protein extraction reagent; Pierce/Thermo Scientific, Villebon-sur-Yvette, France) (20 mM). Then, 10 μl of a concentrated tazobactam solution (40 mg/ml) was added to tube C. In tube A (internal control), 100 μl of the revealing solution containing a pH indicator (phenol red) was added. In tubes B and C (test tubes), 100 μl of an extemporaneously prepared revealing solution supplemented with cefotaxime at 6 mg/ml was added. Tubes A, B, and C were incubated at 37°C for a maximum of 15 min. Optical reading of the color change of each tube was used. The ESBL activity was detected through the transformation of cefotaxime into a carboxylic form, leading to a pH decrease revealed by a color change (red to yellow/orange) in tube B, and inhibition of this reaction by tazobactam leading to no color change in tube C. The results of the ESBL NDP test were interpreted as follows: (i) when tubes A, B, and C were red, a non-ESBL-producing isolate was present; (ii) when tube A was red, tube B was yellow/orange, and tube C was red, an ESBL-producing isolate was present; (iii) when tube A was red, tube B was yellow/orange, and tube C was yellow/orange, a cephalosporinase-producing strain or a cephalosporinase-plus-ESBL-producing strain was present; and (iv) when tubes A, B, and C were yellow/orange, the result was not interpretable. The phenol red revealing solution was prepared as previously described (5). A series of 10 negative and 10 positive isolates (CTX-M-producing strains) were included in the study.
ESBL NDP test on colonies.
After 24 h of incubation, the ESBL NDP test was performed on bacterial cultures recovered on UriSelect 4 nonselective chromogenic medium (Bio-Rad). Briefly, 100 μl of Tris-HCl lysis buffer (B-PERII) (20 mM) was added in three 1.5-ml Eppendorf tubes (A, B, and C). Then, one-fourth to one-third of a single calibrated inoculation loop (10 μl) of bacterial colonies was resuspended in each of those 100 μl of Tris-HCl lysis buffer (20 mM). The final step of the protocol was identical to that described above.
Antibiotic susceptibility testing.
Antibiotic susceptibility testing of bacterial colonies grown on UriSelect 4 (Bio-Rad) was performed by the disc diffusion method following the EUCAST recommendations. Results of the antibiogram (obtained at day 2) were interpreted according to the EUCAST breakpoints (www.eucast.org/clinical_breakpoints/), as updated in 2014. Since no EUCAST breakpoints have been determined for temocillin, those of the British Society for Antimicrobial Chemotherapy (susceptible, ≤8 mg/liter in systemic infections and ≤32 mg/liter in UTIs) were used. MICs of cefotaxime, ceftazidime, and cefepime were determined on Muller-Hinton (MH) agar and MH agar supplemented with 4 μg/ml of tazobactam, respectively.
ESBL phenotypic detection.
The double-disk synergy test (DDST) was performed for the phenotypic detection of ESBL producers according to the EUCAST recommendations, using cefotaxime, ceftazidime, cefixime, and cefepime disks on the one hand and a disk containing ticarcillin plus clavulanate on the other hand. For species naturally producing inducible AmpC (Enterobacter spp., Serratia spp., Morganella spp., Providentia spp., and Citrobacter freundii), the DDST was also performed on cloxacillin (150 μg/ml)-containing MH agar plates (bioMérieux). The production of ESBLs was inferred by a synergy image as previously described (7, 8). A DDST was performed in parallel to the antibiogram.
Molecular characterization of ESBLs.
ESBL producers that have been also detected with the DDST were submitted to molecular characterization of the ESBL-encoding genes. Whole-cell DNAs were extracted using a QIAmp minikit (Qiagen, Les Ulis, France). PCR amplifications followed by sequencing of the entire β-lactamase genes were performed for blaTEM and blaSHV and for blaCTX-M and blaOXA using previously described primers (9).
RESULTS
Epidemiology of UTIs caused by ESBL-producing Enterobacteriaceae.
Among the urine samples that were sent to the laboratory, 500 were positive for ≥104 leukocytes/ml and positive for Gram-negative isolates after Gram staining (meaning ≥105 Gram-negative isolates/ml after culture). Among those 500 urine samples corresponding to Gram-negative bacilli, 450 were considered nonduplicate specimens. Among those 450 samples, 51 (11.3%) were positive for ESBL-E using the DDST (confirmed by PCR amplification and sequencing) as the reference technique. The percentage of UTI caused by ESBL-E was significantly higher for males (16.9%) than for females (8.9%) (P of <0.02) (Table 1). However, no significant difference could be observed between the three classes of age (0 to 15 years, 16 to 65 years, and >65 years) (Table 1). In addition, the percentages of ESBL-E isolated from urine recovered from micturition (9.2%) and from urinary catheters (15.2%) did not differ significantly (Table 1). The rates of UTIs caused by ESBL-E were similar (average = 11.3%) regardless of the hospital setting in which the patients were hospitalized, except for the nephrology unit, where a significantly higher proportion was observed (35.7%, P of <0.0001), and the gynecology/obstetric unit, where a significantly lower rate was observed (0%, P of <0.05) (Table 1).
TABLE 1.
Characteristics of patient origins and ESBL producersa
| Category and characteristic | n (all) | Non-ESBL producer |
ESBL producer |
P | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Patients | ||||||
| Total | 450 | 399 | 88.7 | 51 | 11.3 | |
| Sex | ||||||
| Male | 136 | 113 | 83.1 | 23 | 16.9 | <0.02 |
| Female | 314 | 286 | 91.1 | 28 | 8.9 | |
| Age (yr) | ||||||
| Total | ||||||
| 0–15 | 32 | 31 | 96.9 | 1 | 3.1 | ns |
| 15–65 | 218 | 196 | 89.9 | 22 | 10.1 | |
| >65 | 200 | 172 | 86.0 | 28 | 14.0 | |
| Male | ||||||
| 0–15 | 11 | 11 | 100 | ns | ||
| 15–65 | 66 | 52 | 78.8 | 14 | 21.2 | |
| >65 | 59 | 50 | 84.7 | 9 | 15.3 | |
| Female | ||||||
| 0–15 | 21 | 20 | 95.2 | 1 | 4.8 | ns |
| 15–65 | 152 | 144 | 94.7 | 8 | 5.3 | |
| >65 | 141 | 122 | 86.5 | 19 | 13.5 | |
| Department of hospitalization/consultation | ||||||
| Adult emergency | 167 | 156 | 93.4 | 11 | 6.6 | ns |
| Children emergency | 28 | 27 | 96.4 | 1 | 3.6 | ns |
| Intensive care | 30 | 25 | 83.3 | 5 | 16.7 | ns |
| Nephrology | 42 | 27 | 64.3 | 15 | 35.7 | <0.0001 |
| Hepatology | 4 | 3 | 75 | 1 | 25 | ns |
| Urology | 22 | 17 | 77.3 | 5 | 22.7 | ns |
| Geriatrics | 6 | 4 | 66.7 | 2 | 33.3 | ns |
| Orthopedic surgery | 9 | 8 | 88.9 | 1 | 11.1 | ns |
| Rheumatology | 7 | 7 | 100 | ns | ||
| Infectious diseases | 40 | 35 | 87.5 | 5 | 12.5 | ns |
| Gynecology/obstetrics | 33 | 33 | 100 | <0.05 | ||
| Endocrinology | 8 | 8 | 100 | ns | ||
| Cardiology | 9 | 8 | 88.9 | 1 | 11.1 | ns |
| Pneumatology | 5 | 4 | 80 | 1 | 20 | ns |
| Neurology | 21 | 21 | 100 | ns | ||
| Other | 19 | 16 | 84.2 | 3 | 15.8 | ns |
| Urine specimen | ||||||
| Urine sample source | ||||||
| Micturition | 325 | 293 | 90.2 | 32 | 9.2 | ns |
| Urinary catheter | 125 | 106 | 84.8 | 19 | 15.2 | |
n, number of urine samples; ns, no significant difference.
Among the 51 ESBL-E-related UTIs, 33 E. coli, 10 Klebsiella pneumoniae, 5 Enterobacter cloacae, and 2 Citrobacter freundii isolates and 1 Citrobacter koseri isolate were identified (Table 2). The majority of the ESBLs identified were of the CTX-M type (49/51, 96.1%), with CTX-M-15 being predominant (35/51, 68.6%). A single (E. coli) isolate produced two ESBLs (SHV-12 and CTX-M-3). All ESBL-E were resistant to cefotaxime (the substrate used in the ESBL NDP test) except a single E. coli isolate producing TEM-24 (Table 2). These isolates were also resistant to other non-β-lactam antibiotics used for treating UTIs such as gentamicin (64.7%), cotrimoxazole (83.3%), fosfomycin (27.5%), ciprofloxacin (83.3%), amoxicillin plus clavulanate (29.4%), piperacillin plus tazobactam (13.7%), cefoxitin (15.7%), and nitrofurantoin (9.8%) and remained susceptible to all carbapenems (imipenem, ertapenem, and meropenem) and to temocillin.
TABLE 2.
Results of the ESBL NDP test performed with urine samples and from isolated colonies of EBSL producersa
| Species | CFU/ml | β-Lactamase content | ESBL NDP test result for urine samples |
ESBL NDP test result for bacterial colonies |
MIC (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CTX + TZB | CTX | CTX + TZB | CTX | CTX + TZB | CAZ | CAZ + TZB | FEP | FEP + TZB | |||
| E. coli | 1.2 · 106 | CTX-M-1 | + | − | + | − | 8 | 0.125 | 2 | 0.016 | 3 | 0.023 |
| E. coli | 2 · 106 | CTX-M-1 | + | − | + | − | 24 | 0.032 | 1.5 | 0.016 | 3 | 0.016 |
| E. coli | 3.8 · 107 | CTX-M-1 | + | − | + | − | 32 | 0.25 | 2 | 0.25 | 12 | 0.023 |
| E. coli | 2.8 · 107 | CTX-M-1 + TEM-1 | + | − | + | − | 4 | 0.25 | 48 | 0.094 | 16 | 0.094 |
| E. coli | 1.1 · 107 | CTX-M-1 + TEM-1 | + | − | + | − | 48 | 0.19 | 2 | 0.125 | 6 | 0.032 |
| E. coli | 1.2 · 106 | CTX-M-1 + TEM-1 | + | − | + | − | 24 | 0.064 | 2 | 0.064 | 6 | 0.032 |
| E. coli | 1.1 · 107 | CTX-M-3 + SHV-12 | + | − | + | − | 48 | 0.125 | 24 | 0.19 | 12 | 0.047 |
| E. coli | 9.4 · 107 | CTX-M-14 | + | − | + | − | 24 | 0.047 | 1 | 0.032 | 4 | 0.023 |
| E. coli | 2.8 · 108 | CTX-M-14 | + | − | + | − | 32 | 0.047 | 4 | 0.094 | 3 | 0.032 |
| E. coli | 6 · 107 | CTX-M-14 + TEM-1 | + | − | + | − | 8 | 0.25 | 0.75 | 0.047 | 2 | 0.032 |
| E. coli | 1.3 · 106 | CTX-M-15 | + | − | + | − | >256 | 0.19 | 8 | 0.125 | 12 | 0.032 |
| E. coli | 3.3 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.19 | 8 | 0.064 | 8 | 0.032 |
| E. coli | 9.8 · 107 | CTX-M-15 | + | − | + | − | 192 | 0.25 | 32 | 0.125 | 12 | 0.19 |
| E. coli | 5.2 · 108 | CTX-M-15 | + | − | + | − | >256 | 0.125 | 32 | 0.016 | 24 | 0.064 |
| E. coli | 4.1 · 107 | CTX-M-15 | + | − | + | − | 192 | 0.094 | 16 | 0.064 | 12 | 0.047 |
| E. coli | 2.3 · 108 | CTX-M-15 | + | − | + | − | 96 | 0.094 | 16 | 0.023 | 12 | 0.047 |
| E. coli | 2.8 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.032 | 6 | 0.016 | 6 | 0.023 |
| E. coli | 6.5 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.047 | 32 | 0.19 | 16 | 0.032 |
| E. coli | 5.3 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.047 | 12 | 0.047 | 12 | 0.032 |
| E. coli | 9.4 · 106 | CTX-M-15 | + | − | + | − | >256 | 0.064 | 16 | 0.047 | 12 | 0.047 |
| E. coli | 1.6 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.047 | 12 | 0.094 | 12 | 0.023 |
| E. coli | 1.2 · 108 | CTX-M-15 | + | − | + | − | 64 | 0.064 | 12 | 0.047 | 6 | 0.047 |
| E. coli | 1.8 · 107 | CTX-M-15 | + | − | + | − | >256 | 0.032 | 128 | 0.064 | 128 | 0.047 |
| E. coli | 9.5 · 106 | CTX-M-15 + TEM-1 | + | − | + | − | 16 | 0.19 | 3 | 0.19 | 1.5 | 0.023 |
| E. coli | 2.1 · 108 | CTX-M-15 + TEM-1 | + | − | + | − | 64 | 0.19 | 24 | 0.125 | 12 | 0.032 |
| E. coli | 6.3 · 107 | CTX-M-15 + TEM-1 | + | − | + | − | 96 | 0.047 | 8 | 0.016 | 8 | 0.032 |
| E. coli | 6 · 108 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 0.047 | 32 | 0.094 | 16 | 0.032 |
| E. coli | 4 · 107 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 0.047 | 24 | 0.125 | 8 | 0.016 |
| E. coli | 6.2 · 108 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 0.38 | 64 | 0.25 | 32 | 0.094 |
| E. coli | 4 · 107 | CTX-M-15 + TEM-1 | + | − | + | − | 192 | 0.064 | 24 | 0.25 | 16 | 0.047 |
| E. coli | 3.4 · 107 | CTX-M-15 + TEM-1 | NI | NI | + | − | 24 | 0.032 | 1 | 0.032 | 2 | 0.016 |
| E. coli | 9 · 108 | CTX-M-27 + TEM-1 | + | − | + | − | 2 | 0.064 | >256 | 0.125 | 6 | 0.047 |
| E. colib | 8.9 · 106 | TEM-24 | − | − | − | − | 1 | 0.032 | >256 | 0.047 | 0.38 | 0.016 |
| K. pneumoniae | 3.1 · 106 | CTX-M-1 | + | − | + | − | 16 | 0.064 | 2 | 0.064 | 3 | 0.016 |
| K. pneumoniae | 1.1 · 108 | CTX-M-15 + SHV-1 | + | − | + | − | 32 | 0.064 | 4 | 0.125 | 3 | 0.023 |
| K. pneumoniae | 2.3 · 108 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | >256 | 0.064 | 64 | 0.125 | 24 | 0.047 |
| K. pneumoniae | 4.3 · 108 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | >256 | 1.5 | 48 | 0.5 | 48 | 0.25 |
| K. pneumoniae | 3.7 · 107 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | 48 | 0.19 | 8 | 0.047 | 3 | 0.032 |
| K. pneumoniae | 2.7 · 108 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | 192 | 0.19 | 24 | 0.19 | 6 | 0.023 |
| K. pneumoniae | 1.3 · 108 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | 48 | 0.032 | 24 | 0.032 | 6 | 0.047 |
| K. pneumoniae | 4.3 · 107 | CTX-M-15 + SHV-1 + TEM-1 | NI | NI | + | − | >256 | 0.047 | 8 | 0.064 | 6 | 0.023 |
| K. pneumoniae | 3.9 · 108 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | 48 | 0.032 | 12 | 0.125 | 3 | 0.023 |
| K. pneumoniae | 3.1 · 107 | CTX-M-15 + SHV-1 + TEM-1 | + | − | + | − | 192 | 0.125 | 96 | 0.75 | 12 | 0.094 |
| C. freundii | 2.3 · 108 | CTX-M-15 | + | − | + | − | >256 | 0.032 | 24 | 0.023 | 8 | 0.023 |
| C. freundii | 1.8 · 107 | CTX-M-3 | + | − | + | − | >256 | 0.032 | 16 | 0.023 | 128 | 0.016 |
| C. koseri | 2.1 · 107 | CTX-M-1 | + | − | + | − | 16 | 0.023 | 1.5 | 0.016 | 32 | 0.016 |
| E. cloacae | 5.1 · 107 | CTX-M-15 | + | − | + | − | >256 | 16 | >256 | 8 | 48 | 1 |
| E. cloacae | 5 · 107 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 192 | >256 | 64 | 64 | 1.5 |
| E. cloacae | 1.5 · 108 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 1.5 | 48 | 0.75 | 48 | 0.19 |
| E. cloacae | 1.1 · 107 | CTX-M-15 + TEM-1 | + | − | + | − | >256 | 0.38 | >256 | 0.064 | 96 | 0.19 |
| E. cloacae | 1.4 · 108 | SHV-12 + TEM-1 | + | ± | + | ± | 6 | 0.19 | >256 | 0.119 | 2 | 0.047 |
CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; TZB, tazobactam. ESBLs are indicated in bold. +, yellow/orange; −, red; NI, not interpretable. EUCAST breakpoints were ≤1 and >2 μg/ml for cefotaxime, ≤1 and >4 μg/ml for ceftazidime, and ≤1 and >4 μg/ml for cefepime.
This E. coli isolate produced TEM-24, an ESBL that does not hydrolyze cefotaxime.
Results of the ESBL NDP test performed directly on urine samples.
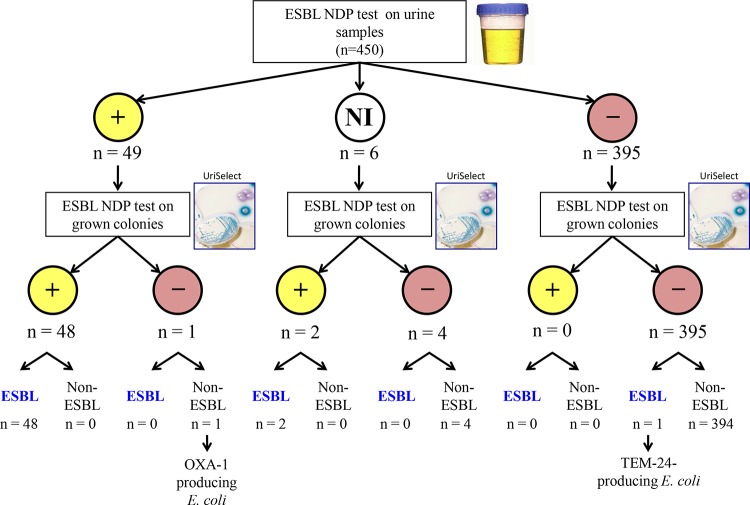
Among the 450 nonduplicate urine specimens tested, the ESBL NDP test gave interpretable results with 444 samples (98.7%). A positive result was obtained for 49 samples, and 48 were confirmed as representing ESBL-E (Fig. 1 and Table 2). A single urine sample containing an E. coli isolate producing narrow-spectrum-clavulanic-acid-resistant OXA-1 gave a false-positive result. The ESBL NDP test gave negative results for 395 urine specimens that were all negative for ESBL-E (Fig. 1). Among the six noninterpretable results, two results corresponded to urine samples containing ESBL-E whereas four corresponded to urine samples without ESBL-E. When performed directly on clinical urine samples with ≥104 leukocytes/ml and positive for Gram-negative isolates after Gram staining (and including only interpretable results), the ESBL NDP test had a sensitivity of 98%, a specificity of 99.8%, a positive predictive value (PPV) of 98%, and a negative predictive value (NPV) of 99.8%. Considering ESBL-E that were not detected because of the classification of noninterpretable results as false negative, the ESBL NDP test had a sensitivity of 94.1%, a specificity of 99.8%, a PPV of 98%, and a NPV of 99.3%.
FIG 1.
Strategy of ESBL-E detection using the ESBL NDP test directly on urine samples and on cultured bacteria grown on selective culture medium. NI, noninterpretable.
Performance of the ESBL NDP test on cultured bacteria.
When performed on grown colonies recovered from urine samples at day 1, interpretable results were obtained for all specimens with the ESBL NDP test. A total of 50 of the 51 ESBL-E specimens were correctly detected using the ESBL NDP test. A TEM-24 producer in E. coli isolates which did not confer resistance to cefotaxime was not detected (Table 2). No false-positive result was obtained. When performed directly on cultured bacteria on UriSelect medium, the ESBL NDP test showed a sensitivity of 98%, a specificity of 100%, a PPV of 100%, and a NPV of 99.8%.
DISCUSSION
During this prospective study, 11.3% of those urine samples were positive for ESBL-E. This rate is similar to the rate obtained in a French survey of multidrug-resistant bacteria showing a rising prevalence rate of ESBL-E (10). This rate correlates with the rate of fecal carriage of ESBL-producing E. coli among healthy subjects reported recently from the Paris area (11). The positivity rate for ESBL-E that was observed for males was higher than that observed for females, likely resulting from the nature of the UTI. In fact, UTIs in males are usually associated with underlying diseases and multiple episodes of treatment or/and hospitalization, which may in turn result in higher multidrug resistance of the UTI isolates.
Although a rapid and global spread of CTX-M-type ESBL-E has been observed since the 2000s, most of the ESBL-E recovered in this study accordingly produced a CTX-M-type β-lactamase, in particular, CTX-M-15.
Here, we showed the technical feasibility of the use of the ESBL-NDP test for determinations directly from urine samples. The protocol of the ESBL NDP test first devised with cultured bacteria has been modified here and led to a shorter period of detection, which was reduced from 60 min (5) to 15 min. Another chromogenic test called the Cica-β-test or βLacta test (Bio-Rad) has been recently developed for the rapid (15-min) detection of Enterobacteriaceae resistant to expanded-spectrum cephalosporins from cultures grown on agar media (12–14). A prospective evaluation performed with the βLacta test on enterobacterial isolates found sensitivity, specificity, PPV, and NPV values of 87.7%, 99.6%, 97.1%, and 98.2%, respectively (13). However, the βLacta test cannot differentiate between resistance to expanded-spectrum-generation cephalosporins due to ESBL production and resistance due to overexpressed (chromosomally encoded, plasmid-acquired) AmpC. In addition, the βLacta test may also detect several types of carbapenemases, raising the possibility of potential confusion between detection of carbapenemases and detection of ESBLs. The most recent study published using the βLacta test reported on false-negative results obtained with AmpC overproducers, producers of ESBL at low levels, and producers of VIM-type carbapenemase (14). In addition, the βLacta test has not been evaluated for detection of ESBL-E directly from clinical samples.
The rate of noninterpretable results with the ESBL NDP test is very low (1.3%), making this test adequate for routine use. A single negative result was obtained with a TEM-24 ESBL producer (Table 2). Interestingly, this TEM-24 ESBL producer was susceptible to cefotaxime. Therefore, all cefotaxime-resistant isolates which were ESBL producers were perfectly detected by using the ESBL NDP test. The unique false-positive result was obtained with an OXA-1-producing E. coli isolate; however, this enzyme is not an ESBL, sensu stricto, since its activity is not inhibited by clavulanic acid whereas it hydrolyzes cefotaxime and cefepime when overproduced (15). Of note, in our study, Gram staining and microscope reading were done systematically on all urine samples with more than 104 leukocytes/ml, leading to inclusion of only those urine samples recovered from true UTI due to Gram-negative bacilli. Accordingly, since several laboratories do not do Gram staining on urine samples, the results of the ESBL NDP test performed directly on all urine samples might differ slightly.
Detection of those ESBL-E that are resistant to cefotaxime by using the ESBL NDP test is perfect. Therefore, by using the ESBL NDP test, we may not only detect ESBL producers for epidemiological purposes as recommended by the EUCAST and CLSI but also guide the first choice of antibiotherapy. For instance, temocillin might be an alternative to carbapenems for treatment as well as β-lactam/β-lactam inhibitors such as the promising combination of ceftazidime plus avibactam. Its use may also reduce the median length of hospitalization due to the delay in introducing the appropriate therapy for treating infections due to ESBL-E (16–19).
In areas with a high prevalence of carbapenemase producers, the rapid Carba NP test could also be performed, for instance, for detection of KPC producers that may give positive results for both the ESBL NDP test and the Carba NP test; since those special carbapenemases hydrolyze both carbapenems and cefotaxime, with this hydrolytic activity being inhibited by clavulanic acid (20). The ESBL NDP test will give negative results in the presence of a carbapenemase with metallo-β-lactamase (hydrolysis of cefotaxime but no inhibition by clavulanic acid) as well as with a carbapenemase of the OXA-48 type (no hydrolysis of cefotaxime).
Fortunately, a low prevalence of carbapenemase producers as a source of UTIs is observed in the United States and many European countries. The latest study performed in France reported a 0.01% incidence of carbapenemase-producing E. coli infections (21).
The use of the ESBL NDP test directly on clinical samples provides useful guidance for implementation of hygiene control measures at the early stage of the hospitalization (at day 0 and not 48 h later) in cases in which it shows positivity in particular for ESBL producers with respect to K. pneumoniae, which is the main species responsible for ESBL-E outbreaks (22).
As a conclusion, we may be at the edge of a change of paradigm by detecting biochemical activity (ESBL or carbapenemase) directly from clinical samples and then waiting for confirmatory results from susceptibility testing.
ACKNOWLEDGMENTS
This work was funded by grants from the INSERM (UMR914), France, and the University of Fribourg, Switzerland.
An international patent form for the ESBL NDP test has been filed on behalf of INSERM Transfer (Paris, France).
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1.Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital. Health Stat. 13 2011:1–38 [PubMed] [Google Scholar]
- 2.Hooton TM. 2012. Clinical practice. Uncomplicated urinary tract infection. N. Engl. J. Med. 366:1028–1037. 10.1056/NEJMcp1104429 [DOI] [PubMed] [Google Scholar]
- 3.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. 2001. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 32:1162–1171. 10.1086/319757 [DOI] [PubMed] [Google Scholar]
- 4.Ramphal R, Ambrose PG. 2006. Extended-spectrum β-lactamases and clinical outcomes: current data. Clin. Infect. Dis. 42(Suppl 4):S164–S172. 10.1086/500663 [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Dortet L, Poirel L. 2012. Rapid detection of extended-spectrum-β-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 50:3016–3022. 10.1128/JCM.00859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pezzlo M, York MK. 2004. Urine cultures, p 1–31 In Isenberg HD. (ed), Clinical microbiology procedure handbook. American Society of Microbiology, Washington, DC [Google Scholar]
- 7.Drieux L, Brossier F, Sougakoff W, Jarlier V. 2008. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14(Suppl 1):90–103 [DOI] [PubMed] [Google Scholar]
- 8.Leclercq R, Cantón R, Brown DFJ, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy C-J, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 19:141–160. 10.1111/j.1469-0691.2011.03703.x [DOI] [PubMed] [Google Scholar]
- 9.Bonnin RA, Nordmann P, Poirel L. 2013. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: a state of the art. Expert Rev. Anti Infect. Ther. 11:571–583. 10.1586/eri.13.38 [DOI] [PubMed] [Google Scholar]
- 10.Carbonne A, Arnaud I, Maugat S, Marty N, Dumartin C, Bertrand X, Bajolet O, Savey A, Fosse T, Eveillard M, Sénéchal H, Coignard B, Astagneau P, Jarlier V, MDRB Surveillance National Steering Group (BMR-Raisin) 2013. National multidrug-resistant bacteria (MDRB) surveillance in France through the RAISIN network: a 9 year experience. J. Antimicrob. Chemother. 68:954–959. 10.1093/jac/dks464 [DOI] [PubMed] [Google Scholar]
- 11.Nicolas-Chanoine MH, Gruson C, Bialek-Davenet S, Bertrand X, Thomas-Jean F, Bert F, Moyat M, Meiller E, Marcon E, Danchin N, Noussair L, Moreau R, Leflon-Guibout V. 2013. Ten-fold increase (2006–11) in the rate of healthy subjects with extended-spectrum β-lactamase-producing Escherichia coli faecal carriage in a Parisian check-up centre. J. Antimicrob. Chemother. 68:562–568. 10.1093/jac/dks429 [DOI] [PubMed] [Google Scholar]
- 12.Livermore DM, Warner M, Mushtaq S. 2007. Evaluation of the chromogenic Cica-β-Test for detecting extended-spectrum, AmpC and metallo-β-lactamases. J. Antimicrob. Chemother. 60:1375–1379. 10.1093/jac/dkm374 [DOI] [PubMed] [Google Scholar]
- 13.Renvoisé A, Decré D, Amarsy-Guerle R, Huang TD, Jost C, Podglajen I, Raskine L, Genel N, Bogaerts P, Jarlier V, Arlet G. 2013. Evaluation of the βLacta test, a rapid test detecting resistance to third-generation cephalosporins in clinical strains of Enterobacteriaceae. J. Clin. Microbiol. 51:4012–4017. 10.1128/JCM.01936-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morosini IA, García-Castillo M, Tato M, Gijón D, Valverde A, Ruiz-Garbajosa P, Cantón R. 26 February 2014. Rapid detection of β-lactamase-hydrolyzing extended-spectrum cephalosporins in Enterobacteriaceae by use of the new chromogenic β-Lacta test. J. Clin. Microbiol. 10.1128/JCM.03614-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beceiro A, Maharjan S, Gaulton T, Doumith M, Soares NC, Dhanji H, Warner M, Doyle M, Hickey M, Downie G, Bou G, Livermore DM, Woodford N. 2011. False extended-spectrum β-lactamase phenotype in clinical isolates of Escherichia coli associated with increased expression of OXA-1 or TEM-1 penicillinases and loss of porins. J. Antimicrob. Chemother. 66:2006–2010. 10.1093/jac/dkr265 [DOI] [PubMed] [Google Scholar]
- 16.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect. Dis. 10:43–50. 10.1016/S1473-3099(09)70325-1 [DOI] [PubMed] [Google Scholar]
- 17.Bazaz R, Chapman AL, Winstanley TG. 2010. Ertapenem administered as outpatient parenteral antibiotic therapy for urinary tract infections caused by extended-spectrum-β-lactamase-producing Gram-negative organisms. J. Antimicrob. Chemother. 65:1510–1513. 10.1093/jac/dkq152 [DOI] [PubMed] [Google Scholar]
- 18.Hawkey PM, Livermore DM. 2012. Carbapenem antibiotics for serious infections. BMJ 344:e3236. 10.1136/bmj.e3236 [DOI] [PubMed] [Google Scholar]
- 19.Schwaber MJ, Carmeli Y. 2007. Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. J. Antimicrob. Chemother. 60:913–920. 10.1093/jac/dkm318 [DOI] [PubMed] [Google Scholar]
- 20.Nordmann P, Dortet L, Poirel L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert J, Pantel A, Mérens A, Lavigne JP, Nicolas-Chanoine MH, on behalf of ONERBA's Carbapenem Resistance Study Group 16 June 2014. Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011–12. J. Antimicrob. Chemother. 10.1093/jac/dku208 [DOI] [PubMed] [Google Scholar]
- 22.Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, Frank U, Kahlmeter G, Pan A, Petrosillo N, Rodríguez-Baño J, Singh N, Venditti M, Yokoe DS, Cookson B, European Society of Clinical Microbiology 12 December 2013. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 10.1111/1469-0691.12427 [DOI] [PubMed] [Google Scholar]