Abstract
Streptococcus pneumoniae (pneumococcus) produces hydrogen peroxide as a by-product of metabolism and provides a competitive advantage against cocolonizing bacteria. As pneumococci do not produce catalase or an inducible regulator of hydrogen peroxide, the mechanism of resistance to hydrogen peroxide is unclear. A gene responsible for resistance to hydrogen peroxide and iron in other streptococci is that encoding nonheme iron-containing ferritin, dpr, but previous attempts to study this gene in pneumococcus by generating a dpr mutant were unsuccessful. In the current study, we found that dpr is in an operon with the downstream genes dhfr and clpX. We generated a dpr deletion mutant which displayed normal early-log-phase and mid-log-phase growth in bacteriologic medium but survived less well at stationary phase; the addition of catalase partially rescued the growth defect. We showed that the dpr mutant is significantly more sensitive to pH, heat, iron concentration, and oxidative stress due to hydrogen peroxide. Using a mouse model of colonization, we also showed that the dpr mutant displays a reduced ability to colonize and is more rapidly cleared from the nasopharynx. Our results thus suggest that Dpr is important for pneumococcal resistance to stress and for nasopharyngeal colonization.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) resides primarily in the human nasopharynx, which is exposed to an oxygen-rich environment on the airway surface. During in vitro growth, pneumococci can generate hydrogen peroxide (H2O2) in the millimolar range. Despite this, S. pneumoniae lacks (H2O2-destroying) catalase, NADH peroxidase, and an H2O2-responsive regulator, such as PerR, all mechanisms that are used by other Gram-positive organisms for protection against H2O2 (1, 2). The mechanisms by which S. pneumoniae is able to survive and grow under oxygenated conditions without many of the defenses commonly found in aerobic and aerotolerant organisms are still unknown.
Hydrogen peroxide can interact with ferrous ions (Fe2+) and form a highly reactive hydroxyl radical (·OH) through the Fenton reaction, causing DNA damage and increased toxicity to the cells (3). At the same time, iron is an essential nutrient for S. pneumoniae. Pneumococci can utilize ferric and ferrous ions, as well as hemoglobin and hemin, as iron sources, as demonstrated in studies with iron-deficient media (4–6). Two iron transporter operons, piuBCDA and piaABCD, appear to be important for acquisition of iron from the environment by pneumococci. Mutation of both operons led to severe attenuation in virulence (5, 7, 8). Therefore, intracellular free iron concentration in pneumococcus must be balanced to both support growth and minimize the damage caused by the Fenton reaction.
In Escherichia coli, a DNA-binding protein from starved cells (Dps) can capture free iron and protect DNA from oxidative damage (9). Dps is a ferritin-like protein and considered functionally similar to classical ferritins. Dps forms a spherical hollow assembly of 12 monomeric proteins and can hold up to 500 molecules of free iron per complex (10). A homolog of Dps, Dpr (Dps-like peroxide resistance), is conserved in Gram-positive bacteria, such as Streptococcus mutans, Streptococcus pyogenes, and Streptococcus suis (11–13). The structure of Dpr from S. suis is similar to that of Dps, and it can bind ferrous ions as well as other divalent cations, such as Cu2+, Mn2+, and Zn2+ (14, 15). Dpr is essential for aerobic survival of S. mutans and also prevents iron-dependent hydroxyl radical formation in vitro (16). Further studies showed that protection against H2O2 damage in S. suis by Dpr is dependent on its iron-binding capacity (17).
A dpr homolog is present in the pneumococcal genome. dpr expression is controlled by an orphan two-component regulator, RitR, which also controls several genes associated with oxidative stress (18). We hypothesized that Dpr is an important factor for resistance of oxidative stress caused by H2O2 in pneumococcus, by chelating free iron. To investigate the role of this gene in resistance to stress, a dpr mutant was generated by homologous recombination, and the phenotype of this mutant in response to stress and in animal models was assessed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae was grown on tryptic soy agar (TSA) with 5% sheep blood (BAP) or in Todd-Hewitt medium plus 0.5% yeast extract (THY) at 37°C in a 5% CO2 incubator. Bovine liver catalase was purchased from Worthington Biochemical, Freehold, NJ. The hydrogen peroxide assay kit was purchased from Thermo Fisher Scientific (Rockland, IL). Other chemicals and reagents were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO).
Iron binding assay.
His-tagged Dpr was purified as described previously (19). Iron staining was performed as described previously (20, 21). Briefly, 1 mg/ml of Dpr and bovine serum albumin (BSA) were incubated in phosphate-buffered saline (PBS) with or without 1 mM Fe(NH4)2(SO4)2 · 6H2O on ice for 30 min and then run on a 3 to 8% Tris-acetate gel under nondenaturing conditions. Iron-bound Dpr was stained with 1 mM 3-(2-pyridyl)-5,6-bis(2-[5-furyl sulfonic acid])-1,2,4-triazine (Ferene S) and 15 mM thioglycolic acid in 2% (vol/vol) acetic acid. Proteins were visualized with Coomassie brilliant blue staining.
Construction of the dpr mutants.
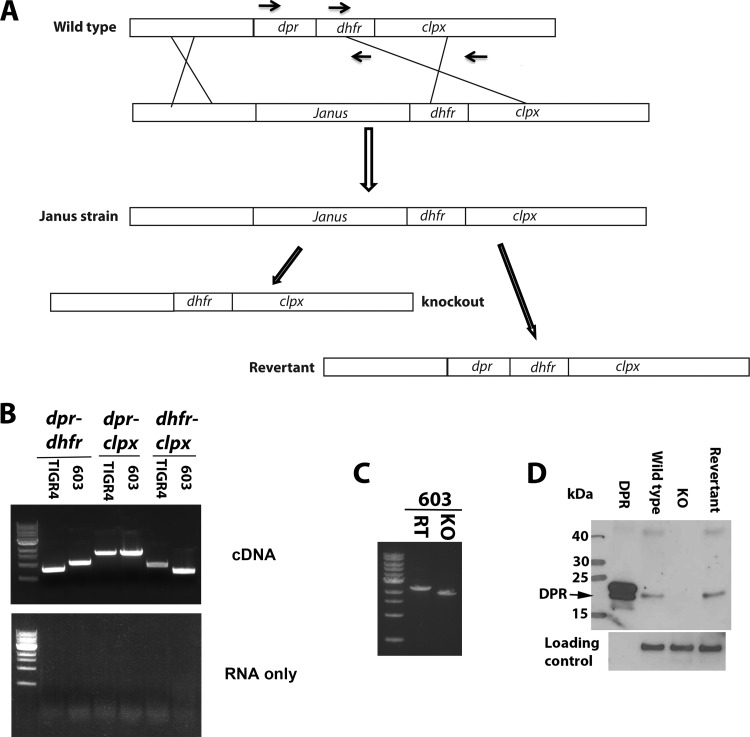
The dpr mutants were constructed as illustrated in Fig. 1A. Primers are listed in Table 1. Genomic DNA was obtained using an extraction kit (Qiagen), following the manufacturer's protocol. Fragments of 800 bp upstream (using primers P1 and P2) and 800 bp downstream (using P5 and P6) of dpr and the Janus cassette (P3 and P4) were amplified from genomic DNA by PCR. An overlapping PCR was performed using these three PCR products as the template and primer sets P1 and P6. Bacteria were grown to an optical density at 600 nm (OD600) of 0.05, and transformation was done as described previously (22). Transformants were identified on BAP supplemented with 600 μg kanamycin/ml and then sequenced to confirm that the Janus cassette was appropriately inserted. The Janus cassette was then replaced by recombination with either knockout (KO) or revertant constructs as illustrated in Fig. 1A. DNA for the KO strain was generated by overlapping PCR using the primer pairs P1 and P6 and products of P1 to P8 and P6 to P9 as a template. The DNA for the revertant strain was generated with primers P1 and P6 with wild-type genomic DNA as the template. Bacteria were selected on BAP with 1,000 μg streptomycin/ml and then sequenced using primers P0 and P7, both outside the cloning region. To generate a mutant in the TIGR4 background in which a chloramphenicol cassette was introduced to replace dpr, three DNA fragments were generated with P1 plus CM-P2, CM-P3 plus CM-P4, and CM-P5 plus P6 and then connected using overlapping PCR. Transformation was performed as described above. The full genome sequences of 603 and TIGR4 revertant (RT) and KO strains were obtained by Illumina HiSeq platform using a protocol described previously (23).
FIG 1.
Generation of dpr mutants. (A) Schematic diagram of the generation of the dpr mutants and revertants using a bicistronic (Janus) cassette. Black arrows indicate the position of primers for reverse transcription-PCR in Fig. 1B. (B) dpr, dhfr, and clpX are in a single transcript. RNA was extracted from pneumococcal TIGR4 and 603 strains, and cDNA was generated by reverse transcription. PCR products using cDNA (top) or RNA (bottom) were amplified using primers listed in Table 1. (C) PCR amplification of genomic DNA from revertant and KO strains using two outside primers. (D) Western blotting was performed to confirm the phenotypes of the dpr KO and revertant strains in the 603 strain background. Pneumolysin was used as a loading control.
TABLE 1.
Primers used in the current study

Numbers in parentheses are the distance (bp) from the first base to the start codon of dpr.
Operon detection.
RNA was extracted from mid-log-phase bacteria using the RNeasy kit (Qiagen, Valencia, CA). cDNA was generated from RNA using reverse transcriptase and random primers. cDNA was then used as templates to amplify possible operons, and RNA was used as a control. Primers used for amplification are listed in Table 1.
Western blotting.
Dpr antiserum was obtained by immunizing New Zealand White rabbits with purified Dpr protein (19). Mid-log-phase bacteria were pelleted and resuspended in SDS loading buffer. After being heated to 100°C for 10 min, samples were loaded onto 4 to 12% bis-Tris SDS gel and separated under denaturing conditions. Western blotting was performed as described before (24). Briefly, proteins were transferred to nitrocellulose membranes and blocked with 5% skim milk in PBS containing 0.05% Tween 20 (PBST) at room temperature for 1 h. The membrane was incubated with anti-Dpr (1:5,000 dilution) or anti-pneumolysin antisera (1:10,000) in 1% skim milk in PBST for 1 h at room temperature. The membrane was then washed three times and further incubated with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG for 1 h at room temperature, and then it was developed using a chemiluminescent substrate. Purified Dpr was used as a positive control; pneumolysin was used as a loading control and probed with an antibody directed against pneumolysin.
Bacterial survival assays.
Mid-log-phase bacteria (OD600 = 0.5) were used for bacterial survival assays. Bacteria were washed with PBS, subjected to different stress conditions as described below, and then plated on BAP: bacteria were incubated in THY with different concentrations of hydrogen peroxide (1 mM, 5 mM, or 10 mM for 30 min at room temperature) or at different pHs (pH 4 for 2 h, pH 7 for 2 h or pH 11 for 75 min) or subjected to elevated temperature (42°C for 2 h). An incubation time for each treatment was chosen such that about 99% of the KO bacteria would be killed. For iron sensitivity assays, mid-log-phase bacteria were diluted into ferrous chloride-containing THY medium and further incubated at 37°C.
Iron concentration measurements.
RT and dpr mutant strains were grown in 1 liter of THY medium. Cells were collected when the OD600 reached 0.5 and then washed twice with ice-cold PBS. The cell pellets were then resuspended in 5 ml of PBS. Bacteria were lysed using a bead beater, following which the supernatant fraction was passed through a 30-kDa Amicon tube (Millipore). Retained fractions, the flowthrough, and whole bacteria were analyzed for iron content by inductively coupled plasma-mass spectrometry in the laboratory of J. Shine at the Harvard School of Public Health. The total iron concentration was normalized to total protein content of the bacterial lysates, which was measured by a micro-bicinchoninic acid (BCA) protein assay (Pierce).
Macrophage and neutrophil killing assays.
Macrophage killing assays were performed as described previously with minor modifications (25, 26). Briefly, 5 × 105 immortalized wild-type murine macrophages (a gift from Douglas Golenbock, University of Massachusetts) per well were plated in 24-well cell culture plates for 16 h, and 2 × 106 CFU of either strain was added into each well. Cells were incubated at 37°C for 30 min and then washed with PBS three times. To quantify intracellular bacteria, cells were incubated in medium with 100 μg gentamicin/ml for 30 min at 37°C to kill all extracellular bacteria. Fresh medium was added to the culture, and the concentration of intracellular surviving bacteria at each time point was determined by plating cell lysates. Total CFU without addition of gentamicin at different time points were determined by plating dilutions on BAP after lysing the cells. Neutrophil surface killing assays were performed as described previously using HL-60 cell-derived neutrophils (27).
Nasal colonization model.
To determine the ability of the dpr mutant to establish nasopharyngeal (NP) colonization in mice, mice were challenged with live encapsulated pneumococci as described previously (28) with 107 CFU of either RT or KO strains in 10 μl of PBS. Mice were euthanized, and nasal washes were collected at 6 h and 1, 2, 4, and 7 days after inoculation and plated on BAP supplemented with gentamicin for assessment of density of bacterial colonization. The competition between RT and KO strain in vivo was performed as described previously (29). Mice were challenged with 107 CFU of a 1:1 mixture of RT and KO strains in 10 μl of PBS at day 0, and samples from live animals were collected by applying 20 μl of ice-cold PBS to either nostril of a mouse and collecting droplets discharged by the animal at day 1, 3, and 7. Samples were cultured on blood agar plates (supplemented with 2 μg/ml of gentamicin) overnight, and the resulting colonies were harvested for genomic DNA extraction using a DNeasy blood and tissue kit (Qiagen, Valencia, CA). The absolute quantity of strain-specific genomic DNA in a sample was determined using strain-specific primer sets and then used to calculate the ratio of the two strains in the NP. RT and KO strain primers were designed around the dpr region. The upper and lower detection limits were set as total CFU and 1/total CFU, respectively.
Statistical analysis.
PRISM (version 5.0, GraphPad Software, Inc.) was used for statistical calculations. Differences in cell killing assay were analyzed by two-way analysis of variance (ANOVA). NP colonization densities were analyzed by Mann-Whitney U tests. For all comparisons, a P value of <0.05 was considered significant.
RESULTS
Pneumococcal Dpr binds iron.
The pneumococcal Dpr belongs to the ferritin-like protein family. A homolog of Dpr in S. suis binds iron and shares 62% identity and 80% similarity with the pneumococcal Dpr. To evaluate whether pneumococcal Dpr similarly binds iron, we purified Dpr from E. coli. As shown in Fig. 2 (left), Dpr can be stained by Ferene S (a colorimetric reagent for iron) in the presence of 1 mM Fe2+, whereas no such band was seen when BSA was used as a control. The majority of Dpr that bound to iron remained in the same position as the non-iron-binding Dpr; interestingly, a small portion of Dpr shifted to a lower position, as revealed by Coomassie brilliant blue staining (Fig. 2, right), which suggests the possibility that Dpr may have two conformations when incubated with iron.
FIG 2.
Binding of iron to Dpr. Purified Dpr and control BSA proteins were added to PBS with or without 1 mM Fe2+ and incubated on ice for 30 min. Proteins were separated with nondenaturing gel. Iron-bound proteins were visualized with Ferene S (left), and protein bands were visualized with Coomassie brilliant blue (right).
dpr is in an operon with dhfr and clpX.
As shown in Fig. 1A, two genes, dhfr (encoding dihydrofolate reductase) and clpX, are located downstream of dpr in the TIGR4 pneumococcal genome. These two genes have been previously shown to be essential in pneumococcus (30–32). In two published studies, attempts to generate a dpr mutant failed (1, 18). The details of previous attempts were not fully described, but we hypothesized that the inability to generate a dpr mutant may have been because of potential polar effects on the expression levels of downstream essential genes. To test the hypothesis that dpr is part of an operon, RNA was purified from pneumococcal strains TIGR4 and 603 (a serotype 6B clinical isolate) (28, 33). cDNA was generated using reverse transcriptase and used as the template to amplify PCR products covering the dpr-dhfr, dhfr-clpX, and dpr-clpX regions. All three amplifications resulted in readily detectable bands of PCR products for both strains (Fig. 1B, top), which were not seen in the absence of reverse transcriptase (Fig. 1B, bottom). These results clearly suggest that the genes dpr, dhfr, and clpX form part of an operon.
Generating dpr mutants.
Having confirmed that these three genes are in an operon, we attempted to generate dpr mutants using a Janus bicistronic cassette (whose insertion in the genome confers kanamycin resistance and streptomycin susceptibility [34]) or with the simple insertion of a chloramphenicol (CM) resistance marker with terminator after the CM resistance cassette. Both approaches are based on homologous recombination, and the PCR products used for transformation share the same upstream and downstream sequences. We favored using the bicistronic Janus cassette for the following reasons: (i) in the absence of a terminator, the downstream genes are expressed under the promoter in the Janus cassette, and (ii) a clean deletion strain or a revertant with wild-type DNA can be generated, to control for any potential effect of transformation on bacterial growth and virulence, which allows a comparison of the phenotypes of deletion strains and a strain that has undergone similar transformation processes but still contains the gene of interest. TIGR4 and 603 strains were transformed using the same concentration of PCR products derived from the two strategies. Kanamycin-resistant colonies were obtained using the bicistronic Janus cassette strategy, but no CM-resistant colony was obtained when the CM cassette strategy was used. These results suggest that the inability to generate dpr mutant was likely due to a polar effect of the construct upon the downstream genes.
Subsequently, clean knockout (KO) and revertant (RT) strains were generated using homologous recombination, as illustrated in Fig. 1A. RT strains were used as comparators for the KO strains in all subsequent experiments. Candidate mutant strains were confirmed by using the primer pair P0 and P7 (which do not span the cloning region for DNA amplification) and sequencing the PCR product (Fig. 1C). Western blotting was performed by using cultures of 603 wild-type, KO, and revertant strains and probing with anti-Dpr rabbit antibody (Fig. 1D).
Growth phenotype of dpr mutant.
The dpr mutant in the pneumococcal strain 603 background had no apparent growth defect on either blood agar plates or in THY medium, as shown in Fig. 3A. Both the OD600 and the number of CFU of the KO strain are similar to those of the RT strain. The growth curves of the dpr mutants in TIGR4 background, however, showed important growth defects in THY compared to TIGR4 RT or WT strains. Comparison of whole-genome sequences of the KO and the parent strain to identify potential additional mutations did not reveal any apparent explanation for this phenotype (data not shown). Due to the growth inhibition of the KO in the TIGR4 background, which could confound our examination of the role of dpr, we continued our studies using only the mutants generated in the 603 background.
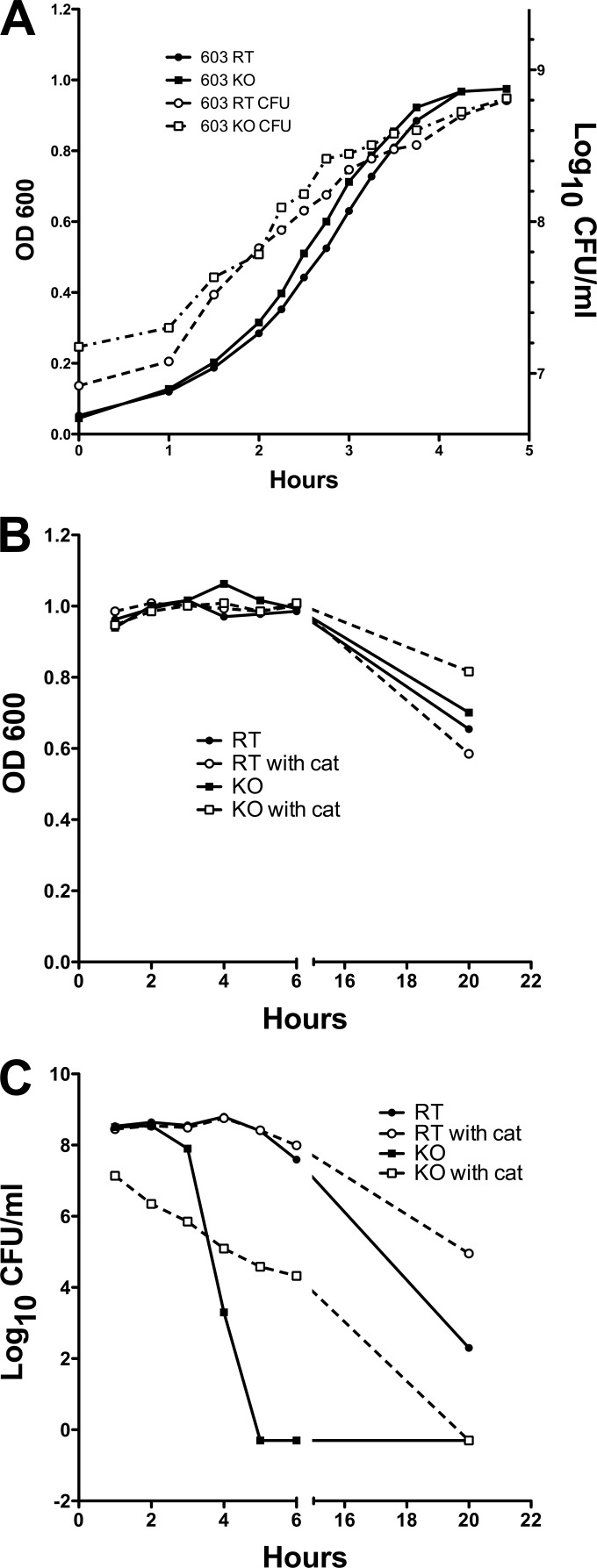
FIG 3.
Growth curve of dpr mutant and revertant strains. Bacteria were grown to an OD600 of 0.5 and diluted in THY medium to start the culture. (A) OD600 was monitored until growth of bacteria reached stationary phase, and the number of CFU at each time point was determined by serial dilution. (B) Optical density of the RT and KO strains in the presence (solid lines) or absence (dashed lines) of catalase (cat) after cells reached stationary phase. (C) Cell viability of RT and KO strains in the presence (solid lines) or absence (dashed lines) of catalase after cells reached stationary phase.
Decreased survival of the dpr mutant in stationary phase.
The production of H2O2 by S. pneumoniae has been shown to mediate pneumococcal cell death (35). We measured H2O2 production by RT and KO strains and found they were similar at every stage of growth (both strains produced over 100 μM at stationary phase). Based on the function of Dpr homologs in other bacteria, we hypothesized that the KO strain would have significantly decreased survival compared to the RT stain at stationary phase. As shown in Fig. 3A, while there were no differences in viability of the RT and KO strains at mid-log phase, important differences were noted at stationary phase: in contrast to the RT strain, the CFU counts in the KO strain decreased rapidly 3 h into stationary phase, while their OD600s were not different from each other (Fig. 3B and C). No viable KO bacteria could be detected after 5 h (Fig. 3C). The addition of catalase increased survival of the KO strain (Fig. 3C) but did not fully reverse the phenotype, suggesting that while H2O2 plays an important role, it may not be the only reason for the increased susceptibility of the KO strain at stationary phase.
Role of Dpr in resistance to stress.
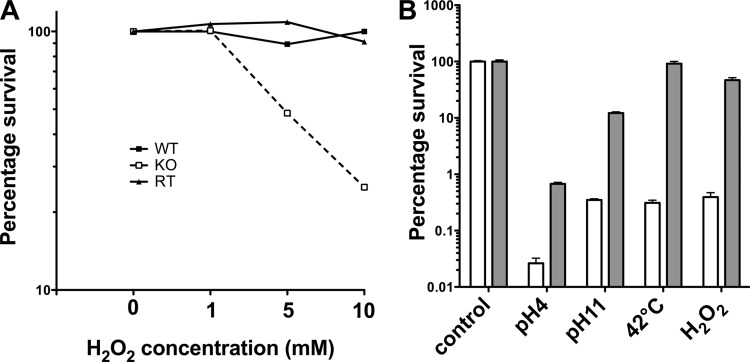
Dpr is thought to protect S. pyogenes under acid and alkaline stress conditions (13). We tested whether deletion of dpr would similarly affect the stress responses to pH changes in S. pneumoniae. Mid-log-phase bacteria were washed and resuspended in THY medium at pH 4, 7, or 11 at room temperature. No cell death was observed for either KO or RT at pH 7. As shown in Fig. 4, the survival rate of the KO strain at pH 4 or pH 11 for 120 and 75 min, respectively, was 3% that of the RT strain. This result thus confirmed the importance of Dpr in conferring resistance to extreme pH conditions. Our data also show that Dpr mediates resistance to lethal heat stress, as the KO strain incubated at 42°C for 2 h had a >2-log reduction in numbers of CFU compared to the RT strain.
FIG 4.
Sensitivity of the dpr deletion mutant to hydrogen peroxide, pH, and temperature stress conditions. (A) Bacteria were treated with different concentrations of H2O2 for 30 min. (B) Bacteria were treated at different pHs (pH 4 for 2 h or pH 11 for 75 min), subjected to elevated temperature (42°C for 2 h), or treated with 10 mM H2O2 for 2 h. Numbers of CFU were determined and normalized to the starting number of CFU. Open bar, KO strain; gray bar, RT strain. Results are from one experiment that is representative of at least three independent experiments.
To evaluate whether Dpr also plays a role in resistance to oxidative stress, RT and KO strains were tested for survival at a range of concentrations of H2O2. Whereas no killing was observed for the RT strain, viability of the KO strain was reduced in the presence of 5 mM or 10 mM H2O2 for 30 min, respectively (Fig. 4A; results are representative of at least three independent experiments). When incubated with 10 mM H2O2 for 2 h, viability was again reduced for the KO strain compared to the RT strain (Fig. 4B; results are representative of at least three independent experiments). These results strongly suggest that Dpr plays an important role in resistance to oxidative stress in pneumococcus, and the protection is likely due to its iron binding ability, as demonstrated in S. suis (17).
Sensitivity of dpr mutant to iron.
Since Dpr mediates storage of iron inside cells in other bacteria, we tested the iron concentration in dpr mutant and RT strains. As shown in Table 2, the total iron content in the RT strain was 1,197.6 ng/mg protein, which is similar to what has been reported previously (1). In contrast, the total iron content in the KO strain was only 304.51 ng/mg protein, significantly lower than that of the RT strain (P = 0.0079 by the Mann-Whitney U test). We also examined whether the dpr mutation specifically lowered the iron content in the cytoplasmic fraction. Cell lysates were separated into fraction 1 (components larger than 30 kDa) and fraction 2 (components <30 kDa) using Amicon tubes (Millipore, MA). The iron content in fraction 1 from the RT strain was also much higher than that of the KO strain (P = 0.0079 by the Mann-Whitney U test), whereas the iron content of fraction 2 from both RT and KO strains was the same.
TABLE 2.
Iron content in RT and dpr mutant strains
| Fraction | Iron content (ng/mg of protein) ina: |
|
|---|---|---|
| RT strain | KO strain | |
| Total bacteria | 1,197.60 ± 38.17 | 304.51 ± 223.05 |
| Lysate fraction 1b | 666.11 ± 114.00 | 109.79 ± 25.82 |
| Lysate fraction 2b | 681.16 ± 360.24 | 649.81 ± 283.67 |
Total iron content was determined in washed cells by inductively coupled plasma mass spectrometry. Values are the means ± standard deviations from five determinations. Results are representative of three independent experiments.
Washed cells were lysed with a bead beater, and the soluble fraction was separated using a 30-kDa-cutoff concentrator. Fractions larger than 30 kDa are designated fraction 1, and fractions smaller than 30 kDa are designated fraction 2.
Dpr forms a dodecameric complex (15), resulting in a molecular mass of 240 kDa, and thus will not flow through a 30-kDa membrane. Therefore, the differences in iron content between the RT and the KO strains in fraction 1 (>30 kDa) can most likely be ascribed to the lack of Dpr in the KO strain.
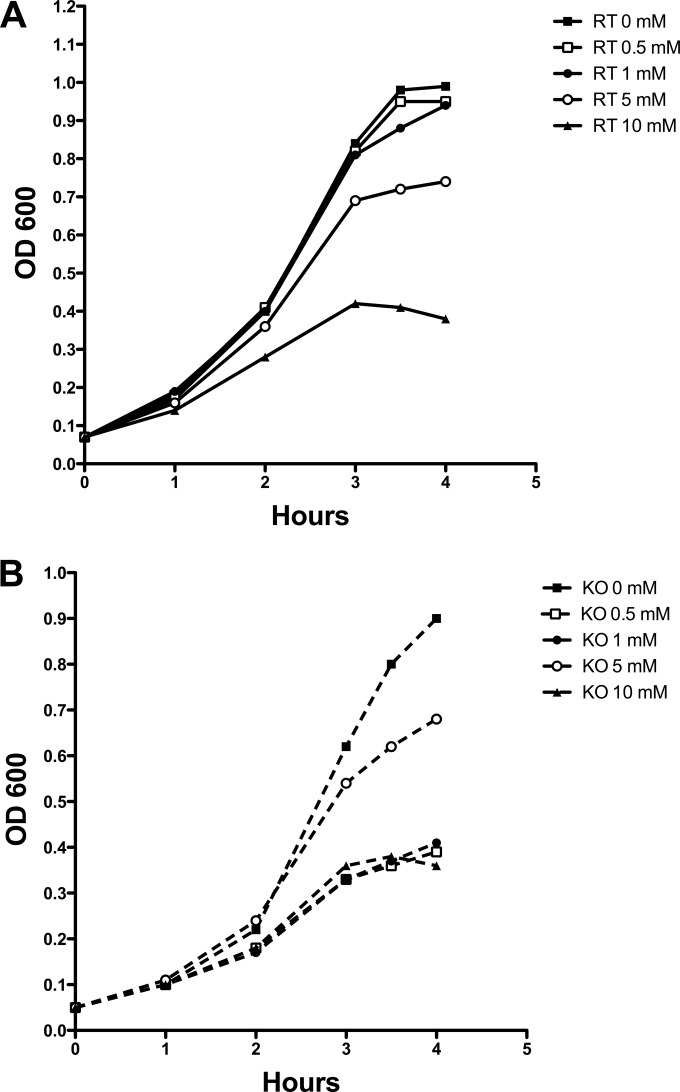
We then assessed whether excess iron in the culture medium would affect growth of the KO mutant strain. Although THY is not a chemically defined medium, the residual iron in THY does not cause growth inhibition in either strain, as shown in Fig. 3. Ferrous chloride was added to THY medium inoculated with mid-log-phase bacteria. As shown in Fig. 5, concentrations of ≥10 mM ferrous chloride were required to demonstrate inhibition of growth of the RT strain in THY. In contrast, the growth of KO strain was inhibited by the addition of as little as 0.5 mM in THY.
FIG 5.
Growth of the RT (A) and KO (B) strains in the presence of ferrous chloride. Bacteria were grown to mid-log phase and diluted in THY containing different concentrations of ferrous chloride. Background absorbance caused by the addition of ferrous chloride was subtracted.
The dpr mutant is more susceptible to macrophage killing and defective in nasopharyngeal colonization.
We examined whether the KO strain demonstrates increased susceptibility to phagocytic killing by macrophage and neutrophils. While we could not detect any differences in survival in a neutrophil surface killing assay (Fig. 6A), susceptibility to killing by macrophages was clearly increased in the KO strain. The adherence rate of KO strain to macrophages was similar to that of the RT strain (Fig. 6B); however, the KO strain was significantly more susceptible to killing by macrophages than the RT strain at 1, 2, and 3 h after gentamicin treatment to kill extracellular bacteria (P < 0.0001 by two-way ANOVA) (Fig. 6C).
FIG 6.
Compared to the RT strain, the dpr mutant is more susceptible to killing by macrophages but not neutrophils. (A) A neutrophil surface killing assay was performed using a range of cell-to-bacterium ratios. Percentages of surviving bacteria of the RT and KO strains are shown. (B) The RT and KO strains were incubated with murine macrophages at an MOI of 20 for 30 min. acrophages were washed with fresh medium, and numbers of total adherent plus intracellular bacteria were determined by plating cell lysates. (C) Surviving intracellular bacteria were counted after gentamicin treatment to kill extracellular bacteria, sequential washes, and plating of cell lysates.
Based on these results, we wished to examine whether the dpr mutant would be defective in colonization of mice, particularly since it had been shown that clearance of pneumococcal colonization is mediated by macrophages in mice that had not previously been exposed to pneumococcus (36). Thus, C57/BL6 mice were colonized with RT or KO strains and nasopharyngeal washes obtained at different time intervals. As shown in Fig. 7A, the density of colonization of the KO strain at 6 h was about 1 log lower than the density of colonization of the RT strain. The difference in densities of colonization between the two strains appears to persist for at least 2 more days, and by day 7, the KO strain was undetectable in nasal washes (P < 0.0001 compared to density of colonization by RT strain). To evaluate the early defect in colonization further, and to compare the two strains in the same animal, we performed mixed-infection competition studies, as described before (29). Ten mice were challenged with a 1:1 ratio of KO and RT strains and sampled at days 1, 3, and 7, and the ratio of KO to RT in nasal washes was determined using strain-specific primers and PCR. No KO strain was detected from nasal sampling at any of the time points (Fig. 7B), thus indicating that the KO strain was outcompeted by the RT strain as early as day 1.
FIG 7.
The ability of the dpr mutant to colonize the nasopharynx is severely impaired. (A) Groups of 5 or 10 mice were challenged intranasally with either the RT or KO strain, and their nasal colonization densities were determined at different time points. Colonization densities of the RT and KO strains at each time point were compared by the Mann-Whitney U test. (B) Competition between RT and KO strain in mice. Mice were intranasally challenged with 107 CFU of a 1:1 ratio of RT and KO strains, and samples were taken at days 1, 3, and 7. Total numbers of CFU were determined by growth on BAP. The absolute quantity of strain-specific genomic DNA in a sample was determined using strain-specific primer sets and then used to calculate the ratio of the two strains in the NP. No KO strain was detected at any time point.
Overall, these data indicate that the deletion of dpr greatly reduces the ability of the strain to colonize and renders the strain unable to compete with the RT strain in the NP.
DISCUSSION
S. pneumoniae lives in an oxygenated environment during colonization and encounters various oxidative stresses, derived presumably both from the host immune response and bacterial production of hydrogen peroxide. The precise mechanism by which pneumococcus neutralizes reactive oxygen species remains unclear, but several processes have been suggested, as reviewed in reference 37. Here we show that the S. pneumoniae Dpr binds iron and that the dpr mutant strain has significantly less total iron content. The dpr mutant strain has increased sensitivity to multiple stress conditions, including pH, temperature, iron, and oxidative stress. The dpr mutant is also significantly more sensitive to macrophage killing and highly attenuated in a nasal colonization model in mice.
dpr was first discovered by a library screening for a gene(s) responsible for resistance to peroxide or hydroperoxide toxicity in S. mutans (11). Dpr belongs to the Dps member family and has a structure similar to that of either ferritin or Dps (14, 15, 38–40). Studies have showed that dpr serves to protect Gram-positive bacteria against oxidative and other stresses (11–13). Unlike Dps, which protects DNA through direct association, Dpr does not bind to DNA (16). Instead, it has been suggested that its protection against oxidative stress is the result of chelation of intracellular free iron (17, 41). We show here that Dpr binds iron and that the dpr mutant has increased sensitivity to iron. These results are consistent with an iron-binding function to reduce the oxidative stress that results from the Fenton reaction. These results are also consistent with the findings of another study which showed that dpr expression increased in the presence of hemoglobin (42). Two iron-related regulators, RitR and IdtR, have been identified in pneumococcus (18, 43), and RitR was demonstrated to play a regulatory role in the expression of dpr, but whether IdtR is also involved in regulation of dpr expression still needs to be determined. Overall, our results are consistent with a role of dpr in conferring pneumococcal resistance to multiple stress conditions.
We also show here that dpr is in an operon with the downstream genes dhfr and clpX, suggesting that their transcriptional regulation and function may be related, as has been demonstrated in other bacterial species. In E. coli, ClpXP rapidly degrades Dps (the Dpr homolog) in stationary phase (44). However, whether Dpr concentration is similarly controlled by ClpX in pneumococcus in stationary phase is unknown.
The dpr mutant is significantly defective in resistance to macrophage killing and its ability to colonize mice. As an immunogen, Dpr has been tested in different pneumococcal animal models and shown to confer protection against pneumococcal colonization and invasive diseases (45–47). This study thus provides additional evidence for an important role of Dpr in pneumococcal pathogenesis.
ACKNOWLEDGMENTS
We thank Porter W. Anderson for his critical review of the manuscript and helpful discussions.
C.-Z.H. gratefully acknowledges support from National Science Foundation of China (grant no. 81273329). R.M. gratefully acknowledges support from the Translational Research Program at Boston Children's Hospital. This work was supported by a Boston Children's Hospital faculty development award and NIH grant R21-AI103480 from the National Institute of Allergy and Infectious Diseases (both to Y.-J.L.).
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185:6815–6825. 10.1128/JB.185.23.6815-6825.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- 3.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. 10.1126/science.2834821 [DOI] [PubMed] [Google Scholar]
- 4.Tai SS, Lee CJ, Winter RE. 1993. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun. 61:5401–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JS, Gilliland SM, Holden DW. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572–585. 10.1046/j.1365-2958.2001.02414.x [DOI] [PubMed] [Google Scholar]
- 6.Romero-Espejel ME, Gonzalez-Lopez MA, Olivares-Trejo JDJ. 2013. Streptococcus pneumoniae requires iron for its viability and expresses two membrane proteins that bind haemoglobin and haem. Metallomics 5:384–389. 10.1039/c3mt20244e [DOI] [PubMed] [Google Scholar]
- 7.Brown JS, Ogunniyi AD, Woodrow MC, Holden DW, Paton JC. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 69:6702–6706. 10.1128/IAI.69.11.6702-6706.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. 2002. Characterization of Pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70:4389–4398. 10.1128/IAI.70.8.4389-4398.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almiron M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646–2654. 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- 10.Bozzi M, Mignogna G, Stefanini S, Barra D, Longhi C, Valenti P, Chiancone E. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272:3259–3265. 10.1074/jbc.272.6.3259 [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182:3740–3747. 10.1128/JB.182.13.3740-3747.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niven DF, Ekins A. 2001. Iron content of Streptococcus suis and evidence for a dpr homologue. Can. J. Microbiol. 47:412–416. 10.1139/w01-027 [DOI] [PubMed] [Google Scholar]
- 13.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2008. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect. Immun. 76:4038–4045. 10.1128/IAI.00477-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haikarainen T, Thanassoulas A, Stavros P, Nounesis G, Haataja S, Papageorgiou AC. 2011. Structural and thermodynamic characterization of metal ion binding in Streptococcus suis Dpr. J. Mol. Biol. 405:448–460. 10.1016/j.jmb.2010.10.058 [DOI] [PubMed] [Google Scholar]
- 15.Kauko A, Haataja S, Pulliainen AT, Finne J, Papageorgiou AC. 2004. Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation. J. Mol. Biol. 338:547–558. 10.1016/j.jmb.2004.03.009 [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol. 184:2931–2939. 10.1128/JB.184.11.2931-2939.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulliainen AT, Kauko A, Haataja S, Papageorgiou AC, Finne J. 2005. Dps/Dpr ferritin-like protein: insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol. Microbiol. 57:1086–1100. 10.1111/j.1365-2958.2005.04756.x [DOI] [PubMed] [Google Scholar]
- 18.Ulijasz AT, Andes DR, Glasner JD, Weisblum B. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 186:8123–8136. 10.1128/JB.186.23.8123-8136.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffitt KL, Malley R, Lu YJ. 2012. Identification of protective pneumococcal T(H)17 antigens from the soluble fraction of a killed whole cell vaccine. PLoS One 7:e43445. 10.1371/journal.pone.0043445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung MC. 1985. A specific iron stain for iron-binding proteins in polyacrylamide gels: application to transferrin and lactoferrin. Anal. Biochem. 148:498–502. 10.1016/0003-2697(85)90258-1 [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa T, Mizunoe Y, Kawabata S, Takade A, Harada M, Wai SN, Yoshida S. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010–1017. 10.1128/JB.185.3.1010-1017.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bricker AL, Camilli A. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131–135. 10.1111/j.1574-6968.1999.tb13460.x [DOI] [PubMed] [Google Scholar]
- 23.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat. Genet. 45:656–663. 10.1038/ng.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354. 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon SB, Irving GR, Lawson RA, Lee ME, Read RC. 2000. Intracellular trafficking and killing of Streptococcus pneumoniae by human alveolar macrophages are influenced by opsonins. Infect. Immun. 68:2286–2293. 10.1128/IAI.68.4.2286-2293.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dockrell DH, Lee M, Lynch DH, Read RC. 2001. Immune-mediated phagocytosis and killing of Streptococcus pneumoniae are associated with direct and bystander macrophage apoptosis. J. Infect. Dis. 184:713–722. 10.1086/323084 [DOI] [PubMed] [Google Scholar]
- 27.Lu YJ, Gross J, Bogaert D, Finn A, Bagrade L, Zhang Q, Kolls JK, Srivastava A, Lundgren A, Forte S, Thompson CM, Harney KF, Anderson PW, Lipsitch M, Malley R. 2008. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog. 4:e1000159. 10.1371/journal.ppat.1000159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. 2001. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect. Immun. 69:4870–4873. 10.1128/IAI.69.8.4870-4873.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Gierahn T, Thompson CM, Trzcinski K, Ford CB, Croucher N, Gouveia P, Flechtner JB, Malley R, Lipsitch M. 2012. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog. 8:e1002989. 10.1371/journal.ppat.1002989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365–374 [PubMed] [Google Scholar]
- 31.Piotrowski A, Burghout P, Morrison DA. 2009. spr1630 is responsible for the lethality of clpX mutations in Streptococcus pneumoniae. J. Bacteriol. 191:4888–4895. 10.1128/JB.00285-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson GT, Ng WL, Gilmour R, Winkler ME. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 185:2961–2966. 10.1128/JB.185.9.2961-2966.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basset A, Turner KH, Boush E, Sayeed S, Dove SL, Malley R. 2012. An epigenetic switch mediates bistable expression of the type 1 pilus genes in Streptococcus pneumoniae. J. Bacteriol. 194:1088–1091. 10.1128/JB.06078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196. 10.1128/AEM.67.11.5190-5196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 189:6532–6539. 10.1128/JB.00813-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Clarke TB, Weiser JN. 2009. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J. Clin. Invest. 119:1899–1909. 10.1172/JCI36731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. 2013. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol. 21:187–195. 10.1016/j.tim.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 38.Haikarainen T, Tsou CC, Wu JJ, Papageorgiou AC. 2010. Crystal structures of Streptococcus pyogenes Dpr reveal a dodecameric iron-binding protein with a ferroxidase site. J. Biol. Inorg. Chem. 15:183–194. 10.1007/s00775-009-0582-9 [DOI] [PubMed] [Google Scholar]
- 39.Haikarainen T, Tsou CC, Wu JJ, Papageorgiou AC. 2010. Structural characterization and biological implications of di-zinc binding in the ferroxidase center of Streptococcus pyogenes Dpr. Biochem. Biophys. Res. Commun. 398:361–365. 10.1016/j.bbrc.2010.06.071 [DOI] [PubMed] [Google Scholar]
- 40.Haataja S, Penttinen A, Pulliainen AT, Tikkanen K, Finne J, Papageorgiou AC. 2002. Expression, purification and crystallization of Dpr, a ferritin-like protein from the Gram-positive meningitis-associated bacterium Streptococcus suis. Acta Crystallogr. D Biol. Crystallogr. 58:1851–1853. 10.1107/S0907444902012970 [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto Y, Fukui K, Koujin N, Ohya H, Kimura K, Kamio Y. 2004. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J. Bacteriol. 186:5997–6002. 10.1128/JB.186.18.5997-6002.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta R, Shah P, Swiatlo E. 2009. Differential gene expression in Streptococcus pneumoniae in response to various iron sources. Microb. Pathog. 47:101–109. 10.1016/j.micpath.2009.05.003 [DOI] [PubMed] [Google Scholar]
- 43.Gupta R, Bhatty M, Swiatlo E, Nanduri B. 2013. Role of an iron-dependent transcriptional regulator in the pathogenesis and host response to infection with Streptococcus pneumoniae. PLoS One 8:e55157. 10.1371/journal.pone.0055157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephani K, Weichart D, Hengge R. 2003. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol. Microbiol. 49:1605–1614. 10.1046/j.1365-2958.2003.03644.x [DOI] [PubMed] [Google Scholar]
- 45.Lu YJ, Zhang F, Sayeed S, Thompson CM, Szu S, Anderson PW, Malley R. 2012. A bivalent vaccine to protect against Streptococcus pneumoniae and Salmonella typhi. Vaccine 30:3405–3412. 10.1016/j.vaccine.2012.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vintini E, Villena J, Alvarez S, Medina M. 2010. Administration of a probiotic associated with nasal vaccination with inactivated Lactococcus lactis-PppA induces effective protection against pneumococcal infection in young mice. Clin. Exp. Immunol. 159:351–362. 10.1111/j.1365-2249.2009.04056.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green BA, Zhang Y, Masi AW, Barniak V, Wetherell M, Smith RP, Reddy MS, Zhu D. 2005. PppA, a surface-exposed protein of Streptococcus pneumoniae, elicits cross-reactive antibodies that reduce colonization in a murine intranasal immunization and challenge model. Infect. Immun. 73:981–989. 10.1128/IAI.73.2.981-989.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]