Abstract
The pneumococcus is the world's foremost respiratory pathogen, but the mechanisms allowing this pathogen to proceed from initial asymptomatic colonization to invasive disease are poorly understood. We have examined the early stages of invasive pneumococcal disease (IPD) by comparing host transcriptional responses to an invasive strain and a noninvasive strain of serotype 1 Streptococcus pneumoniae in the mouse lung. While the two strains were present in equal numbers in the lung 6 h after intranasal challenge, only the invasive strain (strain 1861) had invaded the pleural cavity at that time point; this correlated with subsequent development of bacteremia in mice challenged with strain 1861 but not the noninvasive strain (strain 1). Progression beyond the lung was associated with stronger induction of the type I interferon (IFN-I) response in the lung at 6 h. Suppression of the IFN-I response through administration of neutralizing antibody to IFNAR1 (the receptor for type I interferons) led to significantly reduced invasion of the pleural cavity by strain 1861 at 6 h postchallenge. Our data suggest that strong induction of the IFN-I response is a key factor in early progression of invasive serotype 1 strain 1861 beyond the lung during development of IPD.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is responsible for almost 1 million deaths annually in children under the age of 5 worldwide (1) and is the leading cause of debilitating diseases such as pneumonia, invasive disease (bacteremia, meningitis), and otitis media (2). However, despite the enormous burden of pneumococcal disease, the pathogen is commonly found residing asymptomatically in the nasopharynx of healthy carriers (3). Episodes of pneumococcal disease almost always follow at least transient carriage, usually of a recently acquired strain (3–5). Therefore, detailed knowledge of the mechanism that underlies the very early stages of the transition from carriage to disease is fundamental to understanding pneumococcal pathogenesis.
An intriguing feature of pneumococcal biology is the considerable strain-strain variation in invasive potential that exists, even between strains of the same serotype (6). By examining a number of serotype 1 clinical isolates of various levels of invasive capacity, we previously identified accessory genomic regions (ARs) that might be responsible for facilitating unusually severe invasive pneumococcal disease (IPD) in mice and humans (6). That work showed that while the highly virulent strains, such as strain 1861 (ST 3079), were able to survive in the lungs and rapidly invade the blood of mice following intranasal (i.n.) challenge, the noninvasive strains, such as strain 1 (ST 304), were cleared from the lungs and not detected in the blood. Interestingly, these strains were detected in equal numbers in the nasopharynx at both 48 h and 96 h postchallenge, suggesting that the key difference between the two groups of strains was the ability of the invasive strains to survive in the lungs and translocate to the blood. While strain-specific differences between other strains in the capsule and pneumolysin (Ply) have been shown to influence the immune response and alter the course of disease (7), previously studied strains 1 and 1861 produce the same amount of type 1 capsular polysaccharide and express the same amount of the same variant of Ply (6). A number of strain-specific bacterial factors with potential roles in the development of IPD have been identified and are under investigation. However, the role of the host innate immune response in the translocation of the pneumococcus from the lungs to the blood has yet to be elucidated. In addition, the role of the pleural cavity in the development of IPD is unknown, but it has recently been suggested to act as the conduit through which pneumococci pass from the lungs to the blood (8). In this study, we investigated which elements of the early immune response might influence the progression of the pneumococcus from the lung to the blood as well as potential involvement of the pleural cavity. Using PCR arrays to examine mRNA expression of various key immune genes in the lung, we have identified a role for the type I interferon (IFN-I) response in the ability of invasive strain 1861 to progress beyond the lung to cause invasive disease.
MATERIALS AND METHODS
Bacterial strains and media.
S. pneumoniae serotype 1 clinical strains 1 and 1861 used in this study have been described previously (6). Opaque-phase variants of all strains selected on Todd-Hewitt broth supplemented with 1% yeast extract (THY)-catalase plates (9) were used in all animal experiments. Before infection, the bacteria were grown in serum broth (SB) (nutrient broth [10 g/liter peptone {Oxoid}, 10 g/liter Lab Lemco powder {Oxoid}, and 5 g/liter NaCl] and 10% [vol/vol] donor horse serum) to an absorbance at 600 nm (A600) of 0.16, which approximates 1 × 108 CFU/ml.
Ethics statement.
This study was conducted in compliance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (7th edition; 2004) and the South Australian Animal Welfare Act 1985. All animal experiments were approved by the Animal Ethics Committee of the University of Adelaide.
Animal studies.
Outbred 5- to 6-week-old female CD1 (Swiss) mice were used in all animal experiments. For i.n. challenge, mice were anesthetized by intraperitoneal (i.p.) injection of sodium pentobarbital (Nembutal; Rhone-Merieux) at a dose of 66 μg per g of body weight, followed by i.n. challenge with 50 μl of bacterial suspension containing approximately 1 × 107 CFU in SB. The challenge dose was confirmed retrospectively by serial dilution and plating on blood agar. At indicated time points postchallenge, mice were euthanized by CO2 asphyxiation, alternating between groups throughout to ensure that equivalent time points were investigated for each comparison. For determining CFU counts, groups of 5 to 12 animals were used, as indicated in the figure legends. Blood was collected by syringe from the posterior vena cava. The pleural cavity was subjected to lavage with 1 ml sterile phosphate-buffered saline (PBS) containing 2 mM EDTA introduced through the diaphragm. Pulmonary vasculature was perfused by infusion of sterile PBS through the heart. Lungs for CFU counts were subsequently excised into 2-ml vials containing 1 ml sterile PBS and 2.8-mm-diameter ceramic beads. Tissues were homogenized using a Precellys 24 tissue homogenizer (Bertin Technologies) at 3 cycles of 30 s at 5,000 rpm. At each time point, a 40-μl aliquot of homogenized lung was serially diluted in SB and plated on blood agar to determine the number of CFU present in each lung sample. At 6 h postchallenge, 100-μl aliquots of undiluted pleural lavage fluid and blood samples were plated on blood agar to determine the number of CFU in each sample. At 24 h postchallenge, 20-μl aliquots of blood and pleural lavage samples were serially diluted and plated on blood agar to determine the number of CFU in these niches. Data were analyzed in GraphPad Prism using unpaired t tests or analysis of variance, as described in the figure legends.
Isolation of RNA.
Lungs from resting and challenged mice (n = 4 per group) were excised, following perfusion as described above, and transferred to 2-ml vials containing 1 ml TRIzol reagent (Life Technologies) and 2.8-mm-diameter ceramic beads for immediate homogenization as described above. RNA was isolated from TRIzol-treated samples per the manufacturer's instruction. All RNA samples were cleaned using a PureLink RNA minikit (Life Technologies) per the manufacturer's instruction, including on-column DNase treatment with a PureLink DNase Set (Life Technologies) to remove any contaminating genomic DNA (gDNA) before use in quantitative arrays. Removal of gDNA was confirmed by PCR amplification of the gene encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase) with and without reverse transcriptase.
Quantification of gene expression by PCR array.
Gene expression was analyzed by quantitative reverse transcription-PCR (qRT-PCR) on a LightCycler 480 II system (Roche), using either the mouse innate and adaptive immune response RT2 Profiler PCR array or the mouse type I interferon response RT2 Profiler PCR array with accompanying reagents (Qiagen), per the manufacturer's instruction. A detailed explanation of the control measures included in the array design can be found at the manufacturer's website. Briefly, housekeeping genes (Actb, B2m, Gapdh, Gusb, Hsp90ab1) are included in the array to normalize PCR array data. A genomic DNA control primer set is included to detect nontranscribed genomic DNA contamination. Reverse transcription controls (RTC) are included to determine cDNA synthesis efficiency, and positive PCR controls (PPC) are included to test the efficiency of the PCR itself. The latter two controls (RTC and PPC) also test for interwell and intraplate efficiency. Data were analyzed using PCR Array Data Analysis Software provided by the manufacturer.
Suppression of IFN-I response.
For IFNAR1 neutralization, mice were i.p. administered 300 μg of anti-IFNAR1 antibody (clone MAR1-5A3, low-endotoxin functional grade; Leinco Technologies) (10) or an isotype control at 48 h and 24 h prior to bacterial challenge. At 0 h, animals were i.n. administered 100 μg of antibody or isotype, concurrent with bacterial challenge. Tissues were retrieved and processed for bacterial counts as described above. Suppression of the IFN-I response due to anti-IFNAR1 treatment was confirmed by comparing the levels of expression of the IFN-stimulated Cxcl10 and Ifng genes in lungs from resting, untreated mice and 1861-infected mice treated with isotype control or anti-IFNAR1, each at 6 h postchallenge (n = 4 per group). qRT-PCR was performed using a Superscript III One-Step RT-PCR kit (Invitrogen) and primers RHCxcl10F (5′-AGT GCT GCC GTC ATT TTC TG-3′), RHCxcl10R (5′-ATT CTC ACT GGC CCG TCA T-3′), RHIFNgF (5′-CGG CAC AGT CAT TGA AAG CCT A-3′), and RHIFNgR (5′-GTT GCT GAT GGC CTG ATT GTC-3′) on a LightCycler 480 II system (Roche). Cxcl10 and Ifng expression was normalized to Gapdh expression using primers GAPDHF (5′-TCC TTG GAG GCC ATG TGG GCC AT-3′) and GAPDHR (5′-TGA TGA CAT CAA GAA GGT GGT GAA G-3′). Data are presented for each gene as fold differences relative to resting conditions.
RESULTS
Pleural cavity invasion as an early marker of invasive capacity.
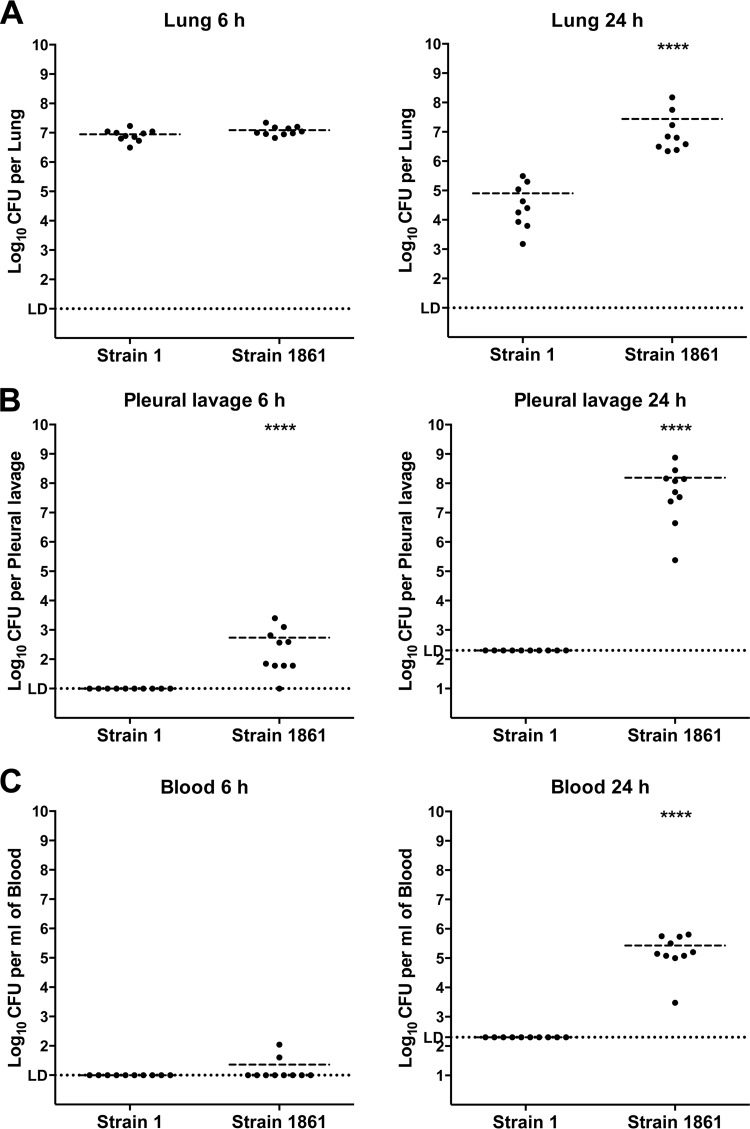
The pathogenesis of noninvasive (strain 1) and highly invasive (strain 1861) serotype 1 strains was characterized using an i.n. mouse challenge model. Following analysis of bacterial burdens in various niches at early time points (4 to 12 h postchallenge; data not shown), a time point of 6 h postchallenge (6 h) was chosen to investigate the early stages of disease, as this was the earliest time point at which bacteria could be reliably detected in the pleural cavity. Samples from 24 h postchallenge (24 h) provided confirmation of the divergent patterns of invasion reported previously for these strains (6). At 6 h, the two strains were detected in the lungs in equal numbers. However, strain 1 was significantly reduced in numbers by 24 h (Fig. 1A), consistent with its previously observed clearance from the lungs by 48 h (6). In contrast, strain 1861 persisted in the lung at 24 h (Fig. 1A). Investigation of the bacterial burden in the pleural cavity at these time points revealed a striking difference between the two strains. While strain 1 remained below the limit of detection throughout the experiment, strain 1861 was detected at low levels at 6 h in almost all mice and was present by 24 h in very high numbers (approximately 108 CFU/pleural cavity in all samples) (Fig. 1B). This pattern correlated with that seen in the blood, where strain 1 was undetectable in all samples whereas strain 1861 was detected at very low levels in some 6-h samples and was present in significant numbers in all blood samples at 24 h (Fig. 1C). These data suggest that invasion of the blood leading to bacteremia is preceded by invasion of the pleural cavity, as has been previously proposed (8), and that invasion of the normally sterile pleural cavity may be used as an early marker of invasion. The data also show that whereas pleural cavity invasion by strain 1861 had commenced at 6 h, this was not due to a difference in bacterial numbers in the lung from those determined for strain 1 but was likely due to a difference in the interactions between the host and the two bacterial strains within the lung. Therefore, the host immune response in the lung at this time point was investigated to identify factors that might explain the different pathogenic profiles of these strains.
FIG 1.
Pathogenesis of serotype 1 isolates in mice following intranasal challenge. Groups of 9 to 10 mice were challenged i.n. with 107 CFU of either strain 1 or strain 1861. The total number of CFU recovered from lungs (A), the pleural cavity (B), or blood (C) was determined at 6 h or 24 h postchallenge, as indicated. The horizontal broken lines in each strain group indicate the geometric mean number of CFU that were recovered. Statistical differences were analyzed by a two-tailed unpaired t test using log-transformed values (****, P < 0.0001). The single horizontal spotted line indicates the limit of detection (LD). At 6 h, this equates to 102 CFU/lung, 10 CFU/pleural cavity, and 10 CFU/ml blood. At 24 h, it equates to 102 CFU/lung, 2 × 102 CFU/pleural cavity, and 2 × 102 CFU/ml blood.
Proinflammatory response to challenge with serotype 1 strains in the lung.
The immune response in the lungs was characterized broadly at the level of transcription, using real-time RT-PCR to perform quantitative expression analysis of 84 key immune response genes. Total RNA was isolated from lung samples collected 6 h after challenge with strain 1 or strain 1861 as well as from lung samples from unchallenged “resting” control mice. Compared with resting lung samples, 21 of 84 genes were found to be differentially regulated (fold change ≥ 2, P < 0.05, n = 4) in a strain-independent manner (Table 1; see also Table S1 in the supplemental material). Of these, 20 showed increased expression compared to resting lung samples following challenge with either strain. Upregulated genes included those encoding proinflammatory cytokines such as interleukin-1 beta (IL-1β), IL-6, IL-23a, and tumor necrosis factor alpha (TNF-α) as well as the inflammasome component NLRP3 and a range of other immune modulatory factors such as the chemokine receptor CCR5, CD14, costimulatory molecules CD80 and CD86, granulocyte-macrophage colony-stimulating factor (GM-CSF; Csf2), the integrin CD11b (Itgam), and the associated adhesion molecule ICAM1. A number of pattern recognition receptors, both intra- and extracellular, including Toll-like receptor 4 (TLR4), TLR5, TLR8, and Nod2, were also differentially regulated. Of all the genes differentially regulated in a strain-independent manner, only Tlr5 was downregulated in comparison to resting controls. These data show that the two strains are recognized by the murine immune system and induce broadly similar, generally proinflammatory responses. Data for the complete array of 84 genes analyzed can be found in Table S1.
TABLE 1.
Strain-independent differential regulation of immune response genes in lungs at 6 h postchallengea
| Gene symbol | Strain 1 vs resting |
Strain 1861 vs resting |
||
|---|---|---|---|---|
| Fold change | Significance | Fold change | Significance | |
| Casp1 | 2.55 | * | 4.42 | ** |
| Ccr5 | 3.44 | * | 3.18 | * |
| Cd14 | 36.69 | * | 28.55 | *** |
| Cd80 | 13.00 | * | 5.97 | **** |
| Cd86 | 2.51 | * | 4.49 | ** |
| Csf2 | 14.25 | *** | 9.42 | **** |
| Icam1 | 4.37 | ** | 4.05 | ** |
| Il1a | 12.51 | *** | 20.08 | **** |
| Il1b | 56.20 | ** | 63.47 | **** |
| Il1r | 3.62 | ** | 3.31 | *** |
| Il23a | 36.57 | ** | 51.20 | ** |
| Il6 | 146.02 | *** | 161.79 | * |
| Itgam | 6.55 | ** | 12.58 | ** |
| Jak2 | 2.86 | * | 3.85 | * |
| Nlrp3 | 38.19 | ** | 54.21 | ** |
| Nod2 | 9.66 | * | 19.07 | * |
| Slc11a1 | 3.47 | * | 2.62 | * |
| Tlr4 | 2.70 | * | 5.32 | * |
| Tlr8 | 2.57 | ** | 2.40 | * |
| Tnf | 71.26 | * | 55.16 | * |
| Tlr5 | −2.26 | ** | −3.69 | *** |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse innate and adaptive immune response RT2 Profiler PCR arrays (cutoff, fold change ≥ 2). Data represent changes in gene expression in lungs from challenged animals relative to resting controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Differential regulation of immune response genes by invasive and noninvasive strains.
Analysis of genes that showed strain-dependent differential expression revealed a significant increase in expression of 8 genes in the lungs by 6 h postchallenge with strain 1861 compared to strain 1 (Table 2). Of these genes, 7 are associated with an IFN-I response, while expression of Ifng, encoding type II gamma interferon (IFN-γ), was also increased in a strain-dependent manner. Of the 7 IFN-I-related genes identified, expression levels of 3 were found to be significantly elevated, with a greater than 2-fold change in expression, in lungs from mice challenged with either strain compared to resting mice, as was the expression level of Ifng (Table 3). Expression levels of the remaining 4 genes were significantly upregulated compared to the levels seen with resting samples only after challenge with strain 1861. These data suggest that, although the two strains induce broadly similar responses, the IFN-I response induced by strain 1861 is stronger than that induced by strain 1.
TABLE 2.
Strain-dependent induction of immune response genes in lungs at 6 h postchallenge with strain 1861 versus strain 1 suggests involvement of the IFN-I responsea
| Gene symbol | Strain 1861 vs strain 1 |
|
|---|---|---|
| Fold change | Significance | |
| Crp | 6.91 | * |
| Cxcl10 | 5.11 | * |
| Ifna2 | 3.69 | * |
| Ifng | 6.22 | ** |
| Il10 | 2.22 | * |
| Irf7 | 2.73 | * |
| Mx1 | 3.39 | * |
| Stat1 | 3.50 | * |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse innate and adaptive immune response RT2 Profiler PCR arrays (cutoff used, fold change ≥ 2). Data represent changes in gene expression in lungs from animals challenged with strain 1861 relative to lungs from animals challenged with strain 1. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (P ≥ 0.05).
TABLE 3.
Strain-dependent induction of immune response genes in lungs at 6 h postchallenge with strain 1861 and strain 1 versus resting conditions suggests involvement of the IFN-I responsea
| Gene symbol | Strain 1 vs resting |
Strain 1861 vs resting |
||
|---|---|---|---|---|
| Fold change | Significance | Fold change | Significance | |
| Crp | 1.19 | ns | 8.20 | * |
| Cxcl10 | 662.83 | ** | 3,386.31 | ** |
| Ifna2 | 2.64 | ns | 9.77 | * |
| Ifng | 55.52 | ** | 345.61 | ** |
| Il10 | 5.50 | * | 12.21 | *** |
| Irf7 | 7.29 | ns | 19.88 | ** |
| Mx1 | 16.97 | * | 57.60 | *** |
| Stat1 | 1.99 | * | 6.94 | * |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse innate and adaptive immune response RT2 Profiler PCR arrays (cutoff used, fold change ≥ 2). Data represent changes in gene expression in lungs from challenged animals relative to resting controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (P ≥ 0.05). Italicized values indicate fold changes that do not meet both the cutoff and P value requirements for significance.
The IFN-I response is exacerbated in lungs from animals challenged with strain 1861.
To verify the response indicated by the data reported above, a more comprehensive set of genes associated with the IFN-I response was analyzed by a real-time RT-PCR array. This confirmed that both strains induced an IFN-I response (see Table S2 in the supplemental material), with 61 of 84 genes differentially regulated by at least one of the two bacterial strains compared to resting conditions, including some identified in the previous array. Of these, 40 genes were induced by both strains compared to resting controls. Strain-independent increases in expression were observed in 16 of these genes as listed in Table 4. The remaining 24 genes were found to be more strongly induced in lungs from animals challenged with strain 1861 than in lungs from those challenged with strain 1 (Table 5). An additional 5 genes were found to be specifically altered in expression by strain 1861 compared to strain 1 (Table 6). Of these 5 genes, 4 were found to be significantly increased in expression following challenge with the invasive strain. The apparent downregulation of the fifth gene, Traf3, is likely due to a slight (<2-fold) decrease in expression seen with strain 1861 compared to resting conditions coupled with a slight but not statistically significant increase in expression of the same gene following challenge with strain 1 (see Table S2 in the supplemental material). Together, these data further suggest that the ability of strain 1861 to invade involves an amplification of the IFN-I response. Data for the complete array of IFN-I response genes analyzed are shown in Table S2.
TABLE 4.
Strain-independent induction of genes involved in the IFN-I response in lungs at 6 h postchallengea
| Gene symbol | Strain 1 vs resting |
Strain 1861 vs resting |
||
|---|---|---|---|---|
| Fold change | Significance | Fold change | Significance | |
| Adar | 2.09 | ** | 3.6 | * |
| Ccl5 | 2.75 | * | 2.88 | * |
| Cd69 | 4.69 | ** | 16.85 | * |
| Cd80 | 21.59 | * | 24.29 | *** |
| Cd86 | 4.07 | * | 5.98 | ** |
| Ifitm3 | 2.65 | *** | 4.68 | **** |
| Ifna2 | 5.99 | * | 17.26 | * |
| Ifnb1 | 66 | * | 106.12 | * |
| Il6 | 229.84 | * | 256.8 | **** |
| Irf9 | 2.91 | ** | 5.28 | *** |
| Jak2 | 3.07 | ** | 5.21 | **** |
| Oas2 | 4.16 | **** | 7.9 | ** |
| Stat3 | 2.8 | ** | 4.01 | **** |
| Tap1 | 5.37 | *** | 8.48 | *** |
| Timp1 | 25.32 | ** | 63.21 | * |
| Tlr8 | 2.01 | * | 2.29 | * |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse type I interferon response RT2 Profiler PCR arrays (cutoff used, fold change ≥ 2). Data represent changes in gene expression in lungs from challenged animals relative to resting controls. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TABLE 5.
Strain 1861 induces stronger upregulation of genes involved in the IFN-I response than strain 1 compared to resting conditionsa
| Gene symbol | Strain 1 vs restingb |
Strain 1861 vs restingb |
Strain 1861 vs strain 1c |
|||
|---|---|---|---|---|---|---|
| Fold change | Significance | Fold change | Significance | Fold change | Significance | |
| Ccl2 | 102.86 | **** | 421.53 | ** | 4.1 | ** |
| Ccl4 | 205.36 | ** | 817.17 | **** | 3.98 | *** |
| Cxcl10 | 1,490.93 | * | 5,770.58 | ** | 3.87 | * |
| Ddx58 | 3.1 | *** | 6.21 | *** | 2 | ** |
| Eif2ak2 | 3.94 | *** | 16.62 | **** | 4.22 | *** |
| Gbp1 | 8.95 | **** | 29.33 | **** | 3.28 | *** |
| Ifi204 | 7.78 | * | 18.82 | *** | 2.42 | * |
| Ifih1 | 4.69 | *** | 20.39 | ** | 4.35 | * |
| Ifit1 | 24.37 | *** | 77.82 | ** | 3.19 | * |
| Ifit2 | 21.25 | *** | 105.02 | *** | 4.94 | *** |
| Ifit3 | 3.09 | *** | 9.22 | ** | 2.98 | ** |
| Ifitm1 | 4.21 | *** | 8.56 | ** | 2.03 | * |
| Il10 | 7.23 | *** | 14.85 | *** | 2.05 | * |
| Il15 | 2.7 | ** | 9.95 | **** | 3.69 | *** |
| Irf1 | 5.33 | **** | 11.59 | *** | 2.17 | ** |
| Irf7 | 7.32 | ** | 19.83 | *** | 2.71 | ** |
| Isg15 | 26.3 | ** | 110.24 | ** | 4.19 | ** |
| Isg20 | 3.21 | **** | 7.31 | *** | 2.27 | ** |
| Mx1 | 24.08 | * | 67.74 | **** | 2.81 | * |
| Mx2 | 14.12 | *** | 34.05 | ** | 2.41 | * |
| Oas1a | 5.59 | *** | 18.76 | **** | 3.36 | *** |
| Psme2 | 2.26 | * | 5.88 | ** | 2.6 | * |
| Stat1 | 3.1 | * | 10.02 | *** | 3.23 | ** |
| Stat2 | 3.38 | * | 8.98 | **** | 2.66 | ** |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse type I interferon response RT2 Profiler PCR arrays (cutoff used, fold change ≥ 2). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data represent changes in gene expression in lungs from challenged animals relative to resting controls.
Data represent changes in gene expression in lungs from animals challenged with strain 1861 relative to lungs from animals challenged with strain 1.
TABLE 6.
Strain 1861 specifically differentially regulates a subset of genes involved in the IFN-I responsea
| Gene symbol | Strain 1861 vs strain 1 |
|
|---|---|---|
| Fold change | Significance | |
| Bst2 | 7.89 | * |
| Crp | 4.73 | * |
| H2-k1 | 2.55 | ** |
| Nos2 | 2.76 | * |
| Traf3 | −2.13 | * |
Animals (n = 4 per group) were challenged with 107 CFU of strain 1 or strain 1861 or left unchallenged (“resting”). Lungs were harvested for RNA isolation at 6 h postchallenge, and immune response gene expression was analyzed using mouse type I interferon response RT2 Profiler PCR arrays (cutoff used, fold change ≥ 2). Data represent changes in gene expression in lungs from animals challenged with strain 1861 relative to lungs from animals challenged with strain 1. *, P < 0.05; **, P < 0.01.
Suppression of the IFN-I response inhibits progression of strain 1861 to the pleural cavity.
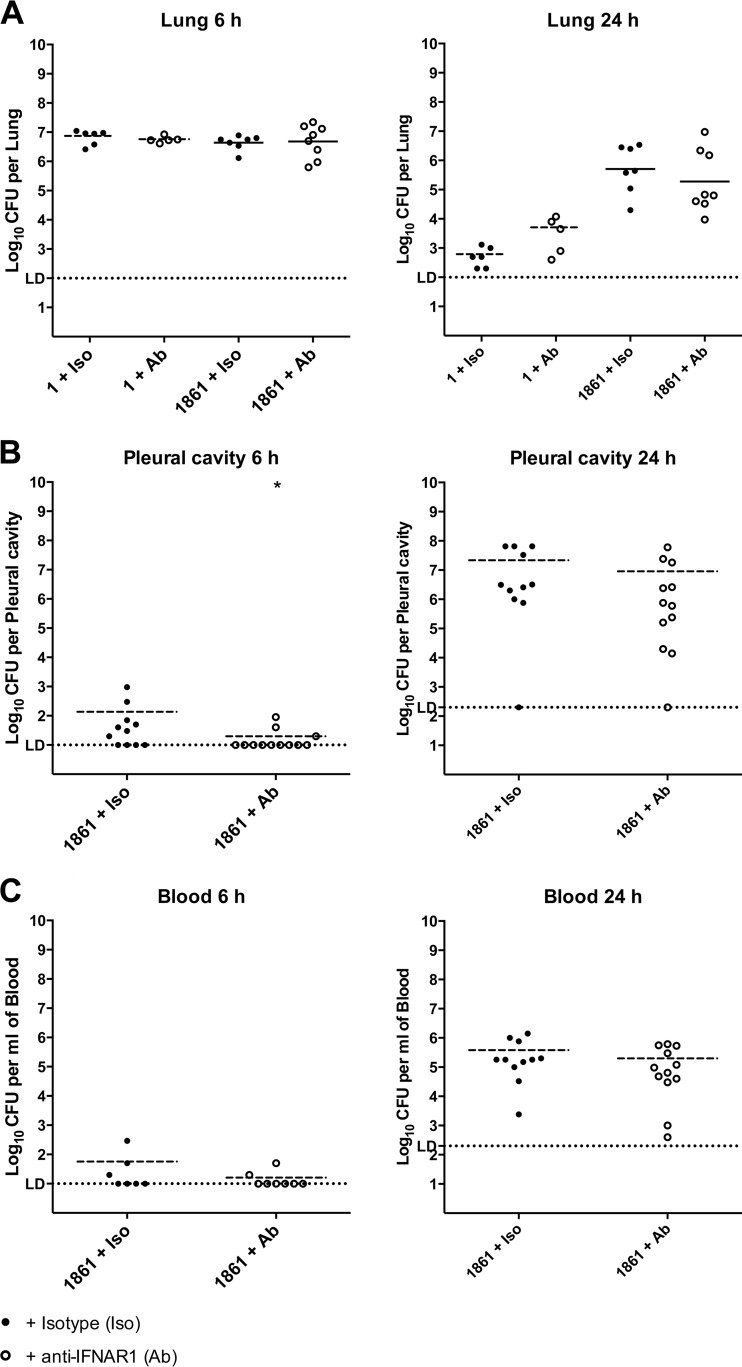
To confirm the importance of the IFN-I response in the progression to invasive disease, groups of mice were treated with either anti-IFNAR1 antibody, which blocks the IFN-I response (10, 11), or an isotype control and were challenged with either strain 1 or strain 1861. Activity of the antibody was confirmed by determining relative expression levels of the IFN-I-stimulated Cxcl10 and Ifng genes for both resting mice and mice treated with either isotype control or anti-IFNAR1 antibody and challenged with strain 1861, with samples taken at 6 h postchallenge (see Fig. S1 in the supplemental material). Numbers of pneumococci in the lungs, pleural cavity, and blood were quantified at both 6 h and 24 h postchallenge. Numbers in the lungs were unaffected by antibody treatment for either strain at both time points (Fig. 2A), suggesting that the IFN-I response is not responsible for the difference between strains 1 and 1861 in the survival rates in the lungs. However, at 6 h postchallenge with strain 1861, we observed a significant decrease in bacterial invasion of the pleural cavity following treatment with anti-IFNAR1 antibody compared to treatment with the isotype control (Fig. 2B). Not only was the mean number of CFU per pleural cavity lower in the antibody-treated group than in the isotype-treated group (one-tailed t test, Welch corrected, P = 0.035), there were also significantly fewer animals in the antibody-treated group with detectable pneumococci in the pleural cavity than in the isotype-treated group, as determined by Fisher's exact (one-tailed) test (P = 0.0296). By 24 h, this difference was no longer apparent (Fig. 2B). At 6 h, only a small number of animals challenged with strain 1861 had detectable CFU in the blood, with or without antibody treatment, while all animals developed bacteremia by 24 h following challenge with strain 1861 (Fig. 2C). Strain 1 was not detectable in the pleural cavity or blood at any time point with either treatment. These data suggest that suppression of the IFN-I response hinders progression of strain 1861 from the lungs to the pleural cavity but does not impede proliferation of the strain in the pleural cavity.
FIG 2.
Suppression of the IFN-I response delays invasion of the pleural cavity by strain 1861. Mice were treated with anti-IFNAR1 neutralizing antibody (Ab; open circles) or an isotype control (Iso; closed circles) and challenged i.n. with 107 CFU of either strain 1 or strain 1861. The total number of CFU recovered from lungs (A), the pleural cavity (B), or blood (C) was determined at 6 h or 24 h postchallenge, as indicated. The horizontal broken lines in each strain group indicate the geometric mean number of CFU that were recovered. In panel B, statistical differences were analyzed by a one-tailed unpaired t test using log-transformed values (*, P < 0.05). The single horizontal spotted line indicates the limit of detection (LD). At 6 h, this equates to 102 CFU/lung, 10 CFU/pleural cavity, and 10 CFU/ml blood. At 24 h, it equates to 102 CFU/lung, 2 × 102 CFU/pleural cavity, and 2 × 102 CFU/ml blood. Group sizes were as follows: n = 5 to 6 for strain 1 challenge groups; n = 7 to 8 for blood and lung samples from 6-h strain 1861 challenge groups; n = 7 to 8 for lung samples from 24-h 1861 challenge groups; n = 11 to 12 for all other groups.
DISCUSSION
The mechanisms that underlie the progression of S. pneumoniae from asymptomatic carriage to invasive disease are fundamental to pneumococcal pathogenesis. While many studies investigating the development of IPD have focused on the properties of the pathogen, it is clear that the host immune response is a critical element in the invasive process. In this study, we demonstrated that differences in the early immune response to two closely related serotype 1 strains play a key role in determining their ability to progress beyond the lung and cause IPD. Strong induction of an IFN-I response by strain 1861 leads to rapid invasion of the pleural cavity with subsequent development of severe bacteremia that is absent from mice challenged with noninvasive strain 1. Suppression of this response in animals challenged with strain 1861 delays the initial invasion of the pleural cavity.
The focus in this study on the earliest stages of IPD enabled the elucidation of the mechanisms involved in the transition from the lung to IPD rather than of the events occurring after bacteremia has been established. We have demonstrated that for invasive strain 1861, invasion of the pleural cavity as early as 6 h postchallenge consistently precedes detection of bacteria in the bloodstream. This supports the notion that the pleural cavity may act as a conduit for bacteria to travel from the lung to the blood (8) and highlights this niche as an important site for investigation of host-pathogen interaction in pneumococcal disease.
As the two strains were detected in equal numbers in the lungs at the 6-h time point, it is clear that the difference in invasive capacity is not simply a dose-dependent case of an overwhelming bacterial burden in or a failure to clear bacteria from this niche. Instead, when we look more closely at the immune response in the lung at this early stage, a more complex interplay between host and pathogen becomes apparent. Both strains induce a broadly proinflammatory response, causing upregulation of genes encoding various proinflammatory cytokines, pattern recognition receptors (PRRs), costimulatory molecules, and other immune modulators. However, some clear indications of divergence were observed. Although both strains induced an IFN-I response, a subset of genes indicative of an IFN-I response was more strongly upregulated by invasive strain 1861 than by noninvasive strain 1 compared to resting levels. Strain 1861 also uniquely upregulated several other IFN-I-related genes whose expression remained unchanged in the presence of strain 1. This, coupled with the demonstration that inhibition of the IFN-I response using anti-IFNAR1 led to decreased invasion into the pleural cavity at 6 h for strain 1861, indicates that the relative scale of the IFN-I response, which is independent of the bacterial numbers present in the lungs, is a critical factor in progression to IPD from the lungs for these strains. In previous work, 8 accessory genomic regions (ARs) >1 kb in size were found to be present in the genome of the highly virulent strains, including strain 1861, but absent from the less virulent strains, which included strain 1 (6). In particular, the version of the variable region of pneumococcal pathogenicity island 1 present in strain 1861 (PPI-1v1861) was responsible for a greater competitive advantage in the lungs and blood of infected mice than the equivalent region present in strain 1 (PPI-1v1) when both were expressed in the D39 background. Therefore, it is tempting to speculate that PPI-1v1861 might be responsible for a stronger IFN-I response in the lungs of mice than PPI-1v1. However, given that there is little known about the proteins encoded in either PPI-1v or the remaining 7 ARs, it is possible that any one region or combination of these regions might be responsible for the differences in the initial immune response to strains 1 and 1861.
IFN-Is are widely known to be induced in response to viruses, as well as during some intracellular bacterial infections (12). In recent years, it has become clear that IFN-Is are also involved in the response to the pneumococcus, as shown for a variety of serotypes. Previous studies have suggested a specific involvement of the IFN-I response in nasopharyngeal colonization for both a serotype 2 strain (13, 14) and a serotype 23F strain (15). For the latter strain, increasing the IFN-I response either artificially or through coinfection with influenza virus led to increased levels of pneumococcal colonization of the nasopharynx. This was attributed to an IFN-I-dependent inhibition of macrophage recruitment to the site of colonization (15). Recent work has also highlighted the importance of the IFN-I response in susceptibility to development of IPD (16, 17). Using serotype 3 strain ATCC 6303, Shahangian and colleagues reported that prior infection with influenza virus rendered mice more sensitive to secondary pneumococcal infection, an effect abrogated either by administration of neutralizing antibody against IFNAR1 or through use of an IFNAR1-null mouse (16). The IFN-I-dependent effect was linked to impaired CXC chemokine production, required for neutrophil recruitment. Similarly, artificial induction of an IFN-I response led to decreased survival rates and increased bacterial burden in both the lungs and the blood of mice with strain ATCC 6303 that persisted for up to 4 days after challenge (17). In contrast, others have indicated that the induction of an IFN-I response to the pneumococcus can be beneficial to the host, at least in the case of challenge with derivatives of commonly used serotype 2 strain D39. Parker and colleagues found that, in contrast to studies using other serotypes, induction of IFN-Is led to improved clearance of D39 from the nasopharynx (14), while a more recent study demonstrated that variants of the serotype 2 strain were less invasive in the presence of a robust IFN-I response (18), a finding linked to reduced lung permeability following induction of IFN-I and a corresponding reduction in platelet-activating factor (PAF) receptor expression.
These studies indicate that the serotype and strain of S. pneumoniae used in each study are critical to the outcome. Our data show that, for the clinically relevant serotype 1 strains tested in this study, the strength of the IFN-I response induced during primary infection is a key factor in determining the ability to invade. These data are in parallel with those from a study published earlier this year which indicated that the strength of induction of a IFN-I response is a determining factor in the pathogenicity of strains of Staphylococcus aureus, another important human pathogen, in a pneumonia model (19). As described for the strains investigated in our work, the strength of the response to S. aureus was found to correlate with increased virulence in a murine model. The report showed that, while both S. aureus strains used induced an IFN-I response in vitro, the response induced by the more virulent strain was much stronger, and the ability of this strain to cause invasive disease was significantly reduced in an IFNAR-null mouse. In comparisons of lungs from animals infected i.n., those lacking the IFN-I response had less-severe pulmonary pathology, with IFN-I signaling linked to increased tissue damage in the infected lung (19). This contrasts with the results of a study by LeMessurier and colleagues, which attributed decreased lung permeability to an increase in IFN-I levels. These studies, as well as the work presented here, indicate that the effect of IFN-I induction in response to infection is dependent on the strength of the response induced, which varies in a strain-dependent manner.
The ability to progress beyond the lung and invade normally sterile compartments of the host is a critical factor in the pathogenicity of the pneumococcus. By examining the very early stages of pneumococcal disease progression, we have shown that pleural cavity invasion by serotype 1 strain 1861 precedes detectable bacteremia, bolstering confidence in the suggestion that the pleural cavity can act as a gateway to dissemination throughout the host. Crucially, we have also demonstrated for the first time that S. pneumoniae can induce a strong IFN-I response early in primary pneumococcal infection that promotes progression of the pneumococcus from the lung to the pleural cavity, with subsequent development of IPD.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Program Grant 565526 from the National Health and Medical Research Council (NHMRC) of Australia (to J.C.P.) and Discovery Project Grant DP120101432 from the Australian Research Council (to J.C.P.). J.C.P. is a NHMRC Senior Principal Research Fellow.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02067-14.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib Pneumococcal Global Burden of Disease Study Team 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2008. 23-Valent pneumococcal polysaccharide vaccine. WHO position paper. Wkly. Epidemiol. Rec. 83:373–384 [PubMed] [Google Scholar]
- 3.Gray BM, Converse GM, III, Dillon HC., Jr 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142:923–933. 10.1093/infdis/142.6.923 [DOI] [PubMed] [Google Scholar]
- 4.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. Tonawanda/Williamsville Pediatrics. J. Infect. Dis. 175:1440–1445. 10.1086/516477 [DOI] [PubMed] [Google Scholar]
- 5.Syrjänen RK, Auranen KJ, Leino TM, Kilpi TM, Mäkelä PH. 2005. Pneumococcal acute otitis media in relation to pneumococcal nasopharyngeal carriage. Pediatr. Infect. Dis. J. 24:801–806. 10.1097/01.inf.0000178072.83531.4f [DOI] [PubMed] [Google Scholar]
- 6.Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, Paton JC. 2011. A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6:e19650. 10.1371/journal.pone.0019650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell AM, Mitchell TJ. 2010. Streptococcus pneumoniae: virulence factors and variation. Clin. Microbiol. Infect. 16:411–418. 10.1111/j.1469-0691.2010.03183.x [DOI] [PubMed] [Google Scholar]
- 8.Wilkosz S, Edwards LA, Bielsa S, Hyams C, Taylor A, Davies RJ, Laurent GJ, Chambers RC, Brown JS, Lee YC. 2012. Characterization of a new mouse model of empyema and the mechanisms of pleural invasion by Streptococcus pneumoniae. Am. J. Respir. Cell Mol. Biol. 46:180–187. 10.1165/rcmb.2011-0182OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, Schreiber RD. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26:804–819. 10.1089/jir.2006.26.804 [DOI] [PubMed] [Google Scholar]
- 11.Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, Schreiber RD. 2005. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 6:722–729. 10.1038/ni1213 [DOI] [PubMed] [Google Scholar]
- 12.Carrero JA. 2013. Confounding roles for type I interferons during bacterial and viral pathogenesis. Int. Immunol. 25:663–669. 10.1093/intimm/dxt050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce EA, Popper SJ, Falkow S. 2009. Streptococcus pneumoniae nasopharyngeal colonization induces type I interferons and interferon-induced gene expression. BMC Genomics 10:404. 10.1186/1471-2164-10-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, Schindler C, Prince A. 2011. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. mBio 2:e00016-11. 10.1128/mBio.00016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, Davis KM, Weiser JN. 2011. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121:3657–3665. 10.1172/JCI57762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. 2009. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119:1910–1920. 10.1172/JCI35412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian X, Xu F, Lung WY, Meyerson C, Ghaffari AA, Cheng G, Deng JC. 2012. Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria. PLoS One 7:e41879. 10.1371/journal.pone.0041879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeMessurier KS, Hacker H, Chi L, Tuomanen E, Redecke V. 2013. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 9:e1003727. 10.1371/journal.ppat.1003727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker D, Planet PJ, Soong G, Narechania A, Prince A. 2014. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. PLoS Pathog. 10:e1003951. 10.1371/journal.ppat.1003951 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.