Abstract
Coxiella burnetii, the causative agent of Q fever, is a human intracellular pathogen that utilizes the Icm/Dot type IVB secretion system to translocate effector proteins into host cells. To identify novel C. burnetii effectors, we applied a machine-learning approach to predict C. burnetii effectors, and examination of 20 such proteins resulted in the identification of 13 novel effectors. To determine whether these effectors, as well as several previously identified effectors, modulate conserved eukaryotic pathways, they were expressed in Saccharomyces cerevisiae. The effects on yeast growth were examined under regular growth conditions and in the presence of caffeine, a known modulator of the yeast cell wall integrity (CWI) mitogen-activated protein (MAP) kinase pathway. In the presence of caffeine, expression of the effectors CBU0885 and CBU1676 caused an enhanced inhibition of yeast growth, and the growth inhibition of CBU0388 was suppressed. Furthermore, analysis of synthetic lethality effects and examination of the activity of the CWI MAP kinase transcription factor Rlm1 indicated that CBU0388 enhances the activation of this MAP kinase pathway in yeast, while CBU0885 and CBU1676 abolish this activation. Additionally, coexpression of CBU1676 and CBU0388 resulted in mutual suppression of their inhibition of yeast growth. These results strongly indicate that these three effectors modulate the CWI MAP kinase pathway in yeast. Moreover, both CBU1676 and CBU0885 were found to contain a conserved haloacid dehalogenase (HAD) domain, which was found to be required for their activity. Collectively, our results demonstrate that MAP kinase pathways are most likely targeted by C. burnetii Icm/Dot effectors.
INTRODUCTION
Coxiella burnetii is a Gram-negative intracellular pathogen and the causative agent of Q fever, a zoonotic disease with flu-like symptoms that may develop into a severe chronic state (1). Typically, humans are infected with C. burnetii by inhalation of contaminated aerosols containing the bacteria that are shed by domestic ruminants (2). Due to its low infectious dose and environmental stability, wild-type C. burnetii has been classified as a category B select agent by the U.S. Centers for Disease Control and Prevention (3, 4).
C. burnetii utilizes an Icm/Dot type IVB secretion system, which has been shown to be required for its intracellular growth in human macrophages (5, 6). The Icm/Dot secretion system was first identified in the evolutionary closely related intracellular pathogen Legionella pneumophila, the causative agent of Legionnaires' disease, where it was found to be essential for all aspects of its pathogenesis (reviewed in references 7 and 8). The Icm/Dot secretion systems of C. burnetii and L. pneumophila are very similar in their gene content and organization (9, 10), and several of the Icm/Dot components of these two pathogens were shown to be functionally similar (11, 12). Both the C. burnetii and L. pneumophila Icm/Dot secretion systems were shown to translocate proteins, termed effectors, into host cells, but their arsenals of effectors were found to be different. This difference probably leads to their different intracellular lifestyles: C. burnetii resides within a phagosome that traffics through the endocytic pathway and matures into an acidic phagolysosome-like compartment (reviewed in references 13 and 14), whereas L. pneumophila inhibits phagosome-lysosome fusion, and its phagosome matures into an endoplasmic reticulum-like compartment (reviewed in references 7 and 8). This different intracellular habitat most likely leads to another difference in the functionality of the Icm/Dot secretion system: C. burnetii effectors were found to translocate into host cells only 8 h postinfection (after the acidification of its vacuole), in contrast to L. pneumophila effectors, which were shown to be translocated minutes postinfection (6, 15).
Extensive study of the Icm/Dot secretion system of L. pneumophila has led to the identification of about 300 effector proteins, which represent about 10% of its proteome (reviewed in references 16 and 17). To date, about 100 effector proteins have been identified in C. burnetii, which constitute about 5% of its proteome. The C. burnetii effector proteins were identified thus far based on different criteria, including (i) existence of eukaryotic motifs such as ankyrin domains (18, 19), (ii) presence of a PmrA binding site in their upstream regulatory region (20, 21), (iii) identification of their C-terminal translocation signal (17), and (iv) location on plasmids (22, 23). Most intriguingly, most of the effectors identified are unique proteins not present in any other sequenced organism (24, 25).
Although the functions of most of the C. burnetii effectors are currently unknown, the functions of a few effectors have been determined. Three effectors (AnkG, CaeA, and CaeB) were shown to inhibit apoptosis in macrophages (26, 27), and an additional effector, CvpA, was found to subvert clathrin-mediated vesicular trafficking (28). Important clues for possible additional functions that might be performed by Icm/Dot effectors came from several studies that investigated the host pathways and factors required for C. burnetii intracellular growth. In one study, a genome-wide screen was performed using gene silencing by small interfering RNA. This screen identified several host factors which affect C. burnetii intracellular growth, including components of the retromer complex, small GTPases Rab5A and Rab7A, subunits of the vacuolar-type H ATPase, and syntaxin-17, a member of the SNARE protein family (29). Another screen examined the role of eukaryotic kinase signaling in the establishment of the C. burnetii phagosome. Inhibition of 11 kinases, including protein kinase C (PKC) and protein kinase A (PKA), altered phagosome formation and prevented pathogen growth (30). In addition, another study determined the phosphorylation state of multiple host proteins during C. burnetii infection. This study identified increased levels of three proteins during C. burnetii infection: (i) phosphorylated extracellular signal-regulated kinase 1/2 (Erk1/2), which is part of a mitogen-activated protein (MAP) kinase pathway; (ii) P38 (which is also part of a MAP kinase pathway); and (iii) Akt (31).
The study of effectors in many bacterial pathogens is challenging due to functional redundancy among effectors, as well as due to the lack of genetic systems in some pathogens. One way to overcome these difficulties is the use of the yeast Saccharomyces cerevisiae as a eukaryotic model system to study effector proteins (32–34). The yeast system has been very useful in the study of effectors that manipulate phosphorylation cascades in eukaryotic cells, such as the MAP kinase pathways (32). Effectors from several bacterial pathogens, including Shigella flexneri (IpgB2 and OspF) (35, 36), Yersinia species (YopJ) (37), Vibrio parahaemolyticus (VopA) (38), and Pseudomonas syringae (HopX1) (39), target MAP kinase signaling pathways, and their functions were discovered using genetic analyses in yeast.
The goal of this study was to identify novel C. burnetii effectors by using the powerful machine-learning approach that was used previously to identify L. pneumophila effectors (40) and then to functionally characterize the newly identified effectors using yeast genetics. We identified 13 novel C. burnetii effectors, and functional characterization in yeast demonstrated that three C. burnetii effectors are involved in modulating a MAP kinase pathway.
MATERIALS AND METHODS
Data sets used for machine learning.
For each of the three learning phases (see Results), the open reading frames (ORFs) of C. burnetii Nine Mile phase II RSA493 were divided into three sets: (i) validated effectors, which in the first learning phase included effectors that were validated in previous studies and in the subsequent learning phases included additionally newly identified effectors from this study and from studies published in the course of this research (see Data Set S1 in the supplemental material); (ii) noneffectors, a manually curated set of 750 ORFs whose annotation suggests that they are highly unlikely to be effectors (see Data Set S1 in the supplemental material); and (iii) unknowns, the rest of the ORFs in the genome. The genomic sequences used for the bioinformatic analyses were C. burnetii Nine Mile phase II RSA493 (accession numbers NC_002971 and NC_004704), Homo sapiens NCBI Build 36.3, Bos taurus NCBI Build 4.1, and L. pneumophila (accession number NC_002942).
Features used for machine learning.
For each ORF, 62 features measuring different characteristics were measured (see Data Set S2 in the supplemental material). These included protein sequence similarity to known C. burnetii and Legionella effectors; sequence similarity to any C. burnetii or L. pneumophila ORF excluding effectors; sequence similarity to host proteins (human and cow); analysis of the Icm/Dot C-terminal translocation signal (17); protein length; protein topology, including transmembrane regions, coiled-coil domains, and myristoylation motif; GC content; regulatory elements; and amino acid composition, including single amino acids, different combinations of amino acids, and similarity of the profile of amino acids to the amino acid profile of C. burnetii effectors. Table 1 presents the groups of features, and Data Set S2 in the supplemental material details each one of the features.
TABLE 1.
Features used in the machine-learning scheme
| Feature groupa | No. of features | Importance in learning phaseb |
||
|---|---|---|---|---|
| 1st | 2nd | 3rd | ||
| Similarity to known C. burnetii effectors | 2 | 3 | 4 | |
| Similarity to known Legionella effectors | 2 | 4 | 3 | 4 |
| Similarity to any C. burnetii ORF | 2 | |||
| Similarity to any L. pneumophila ORF excluding effectors | 2 | 2 | 1 | 1 |
| Similarity to host proteins | 4 | |||
| C-terminal translocation signal | 1 | 1 | 3 | |
| Amino acid composition | 28 | 5 | 2 | 2 |
| Regulatory elements | 1 | 5 | 5 | |
| Protein topology | 11 | |||
| Genome organization | 7 | |||
| Protein length | 1 | |||
| GC content | 1 | |||
Short description of the different feature groups. Similarity to host proteins includes homology to proteins present in the human and cow genomes. The C-terminal signal was calculated according to the HSMM Icm/Dot translocation signal score (17). Amino acid composition includes content of single amino acids and groups of amino acids along the entire protein length. Regulatory elements score for the PmrA binding site. Protein topology includes predictions for transmembrane and coiled-coil domains. Genome organization takes into account the distance between effector-encoding genes.
The numbers indicate the ranks of the top five feature groups that contributed the most to each of the three learning phases.
Feature computation for machine learning.
For sequence similarity, two measures were used: (i) the BLAST hit score of the most similar hit and (ii) the number of BLAST hits with E values lower than 0.01. For the regulatory element score, we used the PmrA position-specific scoring matrix (PSSM) from reference 20. The PmrA score was the highest log PSSM score within the region from 150 nucleotides (nt) upstream to 50 nt downstream of the ORF start codon. Amino acid composition was calculated as the percentage of the protein that consists of the relevant amino acid or group of amino acids. For each amino acid, we computed a score by taking the log of the likelihood ratio between its frequency in effectors and that in noneffectors. The similarity of a given ORF in terms of amino acid composition to effectors was calculated as the average of this score over all amino acids within that ORF. Protein topology-related features were calculated as follows. The myristoylation motif was predicted based on the existence of CAAX in the amino acid sequences. Coiled-coil regions were computed using COILS (41), and transmembrane regions were computed using HMMTOP (42). The Icm/Dot translocation signal was computed using the hidden semi-Markov model (HSMM) described in reference 17 with the C. burnetii proteome as background.
Machine-learning scheme.
The features of the training set comprising the known effectors and the noneffectors were used to train four classifiers: (i) naive Bayes (43, 44); (ii) Bayesian networks (45), using the Tree Augmented Bayes Network (TAN) search algorithm (46) to search for network structure; (iii) support vector machine (SVM) (47, 48), with a radial basis function (RBF) kernel; and (iv) random forest (49). The “Wrapper” procedure for feature selection (50) was carried out for each classifier, excluding random forest, which performs feature selection internally. The performance of each classifier was estimated using the mean area under the precision-recall curve (AUPRC) over 10-fold cross-validation. The final classification score of an ORF was a weighted mean of its scores from the four classifiers trained on the full training set. The performance estimate of each classifier was used as the classifier's weight. For random forest, we used the randomForest R package (51), based on the original Fortran implementation by Breiman (49). For the other classifiers, the WEKA Java library was used with default parameters unless otherwise stated. Feature importance was estimated by random forest (49). The AUPRC values were calculated using AUCCalculator 0.2 (52).
Bacterial and yeast strains, plasmids, and primers.
The C. burnetii strain used in this study was Nine Mile phase II RSA493 (53). The L. pneumophila wild-type strain used in this study was JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (54); also used was a mutant strain derived from JR32, which contains a kanamycin (Km) cassette instead of the icmT gene (GS3011) (12). The E. coli strain used was MC1022 (55). The S. cerevisiae wild-type strain used in this study was BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ), and we also used mutant strains derived from BY4741 (56), which contain a G418 cassette instead of the bck1, mkk1, mkk2, mpk1, hog1, kdx1, kss1, fus3, or smk1 gene (kindly provided by Martin Kupiec, Tel Aviv University). Plasmids and primers used in this work are listed in Tables S1 and S2 in the supplemental material, respectively.
Construction of CyaA and 13× myc fusions.
In order to construct CyaA fusions, the pMMB-cyaA-C (20) vector, which contains a polylinker under Ptac control at the end of the cyaA gene, was used. The pGREG523 or pGREG525 vector was used for the overexpression of 13× myc-tagged effectors in yeast (57). These vectors contain a polylinker under the yeast GAL1 promoter at the end of a 13× myc tag. The C. burnetii genes examined were amplified by PCR using a pair of primers containing suitable restriction sites (see Table S2 in the supplemental material). The PCR products were subsequently digested with the relevant enzymes and cloned into pUC-18 to generate the plasmids listed in Table S1 in the supplemental material. The plasmid inserts were sequenced to verify that no mutations were introduced during the PCR. The inserts were then digested with the same enzymes and cloned into the suitable plasmids described above to generate the CyaA and 13× myc fusions (see Table S1 in the supplemental material).
Site-directed mutagenesis.
Site-specific mutants in the putative haloacid dehalogenase (HAD) domain of CBU0885 and CBU1676 were constructed by the PCR overlap-extension approach (58) as described in reference 20. The primers CBU0885M-F and CBU0885M-R (see Table S2 in the supplemental material) were used to generate pZB-CBU0885-GREG-M, and the primers CBU1676M-F and CBU1676M-R (see Table S2 in the supplemental material) were used to generate pZB-CBU1676-GREG-M.
CyaA translocation assay.
Differentiated HL-60-derived human macrophages plated in 24-well tissue culture dishes at a concentration of 2.5 × 106 cells/well were used for the assay. Bacteria were grown on charcoal yeast extract (CYE) agar plates containing chloramphenicol for 48 h. The bacteria were scraped off the plates and suspended in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) medium, the optical density at 600 nm (OD600) was adjusted to 0.2 in AYE medium containing chloramphenicol, and the resulting cultures were grown on a roller drum for 17 to 18 h until they reached an OD600 of about 4 (stationary phase). The bacteria were then diluted in fresh AYE medium to obtain an OD600 of 0.2 and grown for 2 h. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cultures were grown for an additional 2 h. Cells were infected with bacteria harboring the appropriate plasmids at a multiplicity of infection (MOI) of approximately 4, and the plates were centrifuged at 180 × g for 5 min, followed by incubation at 37°C under CO2 (5%) for 2 h. Cells were then washed twice with ice-cold phosphate-buffered saline (PBS) (1.4 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4) and lysed with 200 μl of lysis buffer (50 mM HCl, 0.1% Triton X-100) at 4°C for 30 min. Lysed samples were then boiled for 5 min and centrifuged for 10 min. The samples were neutralized with NaOH, and 110 μl of each sample was then transferred to a new tube followed by an addition of 220 μl cold ethanol (95%). Samples were then centrifuged for 5 min at 4°C, and the supernatant was transferred to a new tube and stored at −20°C until the next step was performed. The samples were dried in a Speed-Vac and suspended in 110 μl of sterile double-distilled water (DDW). Samples were incubated at 42°C for 5 min, followed by 5 min of incubation at room temperature. The levels of cyclic AMP (cAMP) were determined using the cAMP Biotrak enzyme immunoassay system (Amersham Biosciences) according to the manufacturer's instructions.
C. burnetii reverse transcription-quantitative PCR (RT-qPCR).
C. burnetii Nine Mile phase II RSA493 was grown in ACCM-2 axenic medium (59, 60) at 37°C, with 5% CO2 and 2.5% O2. Samples were taken at 3 days (exponential phase) or 6 days (post-exponential phase), and bacteria were collected by filtration. Filters were flash frozen and stored at −80°C. For intracellular samples, HEK293T cells (ATCC CRL-11268) were infected with ACCM-2-grown C. burnetii at an MOI of 10 as described in reference 61. After 14 h, the cells were washed with PBS to remove extracellular bacteria. The infected cells were collected 3 days (exponential phase) and 7 days (post-exponential phase) after infection. Medium was replaced with RNALater (Ambion), cells were lysed in 1% (wt/vol) saponin (Sigma), and the pellet was flash frozen and stored at −80°C. Bacterial RNA was isolated with TRIzol (Invitrogen), and cDNA was produced using the iScript Select kit (Bio-Rad) according to the manufacturer's instructions. The qPCR was performed using SYBR green reagents and the StepOne Plus system (Applied Biosystems). Primers for qPCR were designed using Primer3 and are listed in Table S2 in the supplemental material. The threshold cycle (ΔCT) was calculated for each RT-PCR product with the corresponding 16S value. The starting quantity was calculated by raising 2 to the power of the CT. Biological triplicates were averaged for each growth condition. Using Cluster 3.0, the data were log2 transformed and the gene expression values were centered around the mean. Distances between genes were calculated using the Pearson correlation coefficient (correlation uncentered), and hierarchical clustering was done using average linkage. The heat map was generated using Java Treeview (62).
Yeast growth assay.
C. burnetii effector-encoding genes were cloned under the GAL1 promoter in the pGERG523 and pGREG525 yeast overexpression vectors as described above. Plasmids were transformed into yeast cells using a standard lithium acetate protocol (63), and transformants were selected for histidine prototrophy on minimal synthetic defined (SD) dropout plates. Resulting transformants were then grown overnight in liquid SD culture medium at 30°C, cell number was adjusted, and a series of 10-fold dilutions was made. The cultures were then spotted onto the respective SD dropout plates containing 2% glucose or galactose. When indicated, the plates were supplemented with 1 M NaCl (Merck) or 2 mM caffeine (Sigma).
Yeast β-galactosidase assays.
Yeast strains containing the 2× Rlm1-lacZ reporter plasmid (64) (kindly provided by Guido Sessa, Tel Aviv University) were cotransformed with the plasmids expressing the effectors or with a vector control. Cells were grown overnight in SD medium containing 2% glucose and then diluted in fresh SD medium containing 2% glucose and grown for an additional 4 h to reach an OD600 of 0.8 to 1. Cultures were then washed and divided into two tubes containing 1 ml of fresh SD medium containing 2% galactose. One culture was transferred to a preheated SD medium at 39°C (heat shock), and the second was kept at 30°C. The cultures were incubated for 1 h at the indicated temperatures, and their OD600 levels were determined. The cells were then subjected to a β-galactosidase assay. The cultures were centrifuged and resuspended in 1 ml of Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol, pH 7) followed by the addition of 5 μl of 0.1% sodium dodecyl sulfate (SDS) and 3 drops of chloroform. The cultures were immediately vortexed for 10 s and placed in a 28°C water bath for 5 min. The β-galactosidase activity was determined as described in reference 65.
Western blot analysis.
For all protein fusions described above, the formation of a fusion protein with a proper size was validated by Western blotting using anti-CyaA antibody 3D1 (Santa Cruz Biotechnology, Inc.) for CyaA fusions or the anti-myc antibody 9E10 (Santa Cruz Biotechnology, Inc.) for the 13× myc fusions. Anti-actin antibody C14 (MP Biomedicals, Inc.) was used as a loading control for the 13× myc fusions expressed in yeast.
RESULTS
Machine-learning scheme.
We predicted novel C. burnetii Nine Mile phase II RSA493 Icm/Dot effectors using a machine-learning approach similar to the one that we previously used to predict effectors in L. pneumophila (40). We improved our machine-learning scheme by including an additional powerful classifier, random forest (49), and by weighting the vote of each classifier based on its estimated performance (see Materials and Methods). Additionally, we adapted and significantly enlarged the set of features taken into account for effector prediction. The features used in this study included sequence similarity to known Coxiella and Legionella ORFs and effectors, similarity to host proteins, Icm/Dot translocation signal, GC content, amino acid composition, genome organization, protein topology, gene regulation, and more. Table 1 summarizes the feature groups used, and a detailed description of the features is presented in Data Set S2 in the supplemental material. In our analyses, we excluded proteins shorter than 35 amino acids, since these are unlikely to encode genuine effectors. We also excluded ORFs considered pseudogenes according to other C. burnetii isolates, since we wanted to investigate effectors with similar functions across the different C. burnetii isolates (25). To examine effector translocation, we used the well-established L. pneumophila model of Icm/Dot translocation that was shown to correlate entirely with effector translocation by C. burnetii (21, 24, 25). We conducted two learning and validation phases and a third final learning phase to predict additional putative effectors.
Learning and validation phase 1.
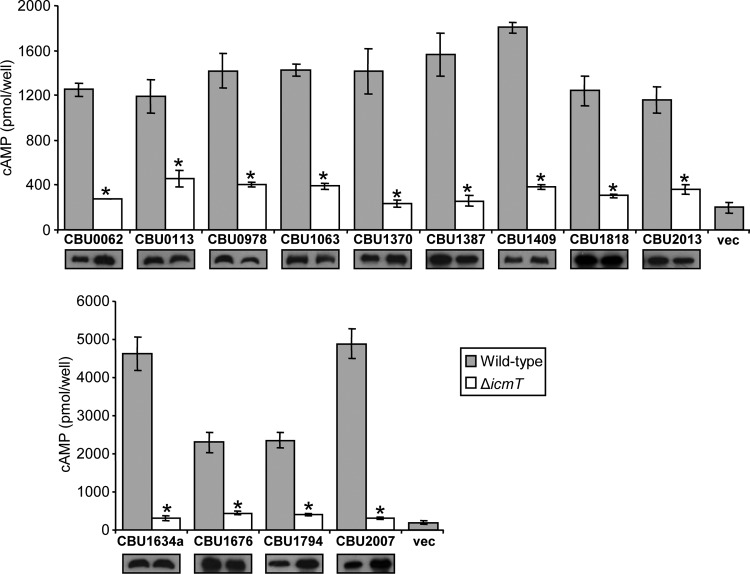
The training set of the first learning phase included 65 known effectors (the effectors used in each phase are listed in Data Set S1 in the supplemental material). The machine-learning analysis resulted with a list of predictions ranked by their score, and we decided to include in our analysis the 67 ORFs that obtained a score of 0.7 or higher. These 67 genes included 50 already known effectors and three genes that were previously shown not to translocate into host cells (17, 25), as well as 14 newly predicted effectors (see Data Set S3 in the supplemental material). We cloned these 14 genes and were able to obtain fusion proteins of proper size for 13 of them (see Data Set S3 in the supplemental material). Next, these 13 fusion proteins were tested for translocation using the CyaA reporter system; all of them were expressed at similar levels in the wild-type strain JR32 and in the icmT deletion mutant, and 10 of them (CBU0113, CBU0978, CBU1063, CBU1387, CBU1409, CBU1634a, CBU1676, CBU1794, CBU1818, and CBU2013) exhibited IcmT-dependent translocation (Fig. 1 and Table 2).
FIG 1.
Icm/Dot-dependent translocation of C. burnetii novel effectors identified by the machine-learning approach. The L. pneumophila wild-type strain JR32 (gray bars) and the icmT deletion mutant GS3011 (white bars) harboring the CyaA fusion proteins (indicated below each bar) were used to infect HL-60-derived human macrophages, and the cAMP levels of the infected cells were determined as described in Materials and Methods. Vector control is indicated as “vec.” The bar heights represent the mean amounts of cAMP per well obtained in at least three independent experiments; error bars indicate standard deviations. The effectors were divided according to the levels of the cAMP obtained in the wild-type L. pneumophila strain. The cAMP levels of each fusion were found to be significantly different (*, P < 0.001, paired Student's t test) between the wild-type strain and the icmT deletion mutant. The C. burnetii novel effectors were examined by Western analysis for their expression in the wild-type strain JR32 (left) and the icmT deletion mutant GS3011 (right) using an anti-CyaA antibody.
TABLE 2.
C. burnetii effectors identified in this study
| Locusa | Name | Size (aa) | Predicted domainb | Homolog(s) in: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| CbuD | CbuG | RSA331 | RSA334 | A35 | CbuK | ||||
| CBU0062 | Cem1 | 501 | J domain | 2044 | A0133 | B0120 | |||
| CBU0113 | Cem2 | 70 | 1994 | 1899 | A0202 | 1971 | |||
| CBU0978 | Cem3 | 155 | 1069 | 1027 | A0960 | 1005 | A0892 | 0861 | |
| CBU1063 | Cem4 | 468 | 1164 | 0941 | A0871 | 1085 | |||
| CBU1370 | Cem5 | 328 | 1457 | 0643 | A1526 | 1420 | |||
| CBU1387 | Cem6 | 216 | 0607 | 0624 | A1546 | 0581 | A1424 | 1458 | |
| CBU1409 | Cem7 | 278 | 0587/0588 | 1480/1482 | |||||
| CBU1634a | Cem8 | 49 | 0363 | 0388 | A1819 | 0323 | A1677 | 1857 | |
| CBU1676 | Cem9 | 361 | HAD | 0325 | 0345 | ||||
| CBU1794 | Cem10 | 272 | 0020 | 0172 | A1991 | A1939 | 0087 | ||
| CBU1818 | Cem11 | 481 | 0047 | 0191 | A2017 | 2106 | A1966 | 0114 | |
| CBU2007 | Cem12 | 395 | 2108 | 2016 | A0078 | 2172 | A2130 | 2058 | |
| CBU2013 | Cem13 | 365 | 2113 | 2022 | A0073 | 2177 | A2135 | 2064 | |
CBU1409 has two paralogs in strains CbuD and CbuK; CBU1676 has a paralog (CBU0885) in C. burnetii RSA493. The effectors CBU0113, CBU2007, and CBU2013 were also identified by others during the course of this study (21).
J-domain, domain of the HSP70 interaction site; HAD, haloacid dehalogenase-like superfamily that includes many types of phosphatases.
Learning and validation phase 2.
The 10 newly validated effectors from the first learning phase were added to the training set of the second learning phase as well as two additional effectors reported during our work (22) (see Data Set S1 in the supplemental material), generating a list of 77 effectors. The machine-learning analysis scored all the genes in the genome, and this time, we analyzed the 71 ORFs that obtained a score of 0.76 or higher. These 71 genes included 62 already known effectors, two ORFs that were found not to translocate in the first learning phase, and the single ORF that we were unable to clone for the first learning phase, as well as six newly predicted effectors (see Data Set S3 in the supplemental material). We cloned these six genes and were able to obtain CyaA fusion proteins of proper size for all of them. Next, these six fusion proteins were tested for translocation using the CyaA reporter system, and three of them (CBU0062, CBU1370, and CBU2007) also exhibited IcmT-dependent translocation (Fig. 1 and Table 2).
Learning phase 3.
In total, we discovered 13 novel C. burnetii effectors and these novel effectors were termed Cem for Coxiella effector identified by machine learning. These effectors are listed in Table 2, which includes information regarding homologs in other C. burnetii strains, paralogs within C. burnetii, and protein sequence motifs. The number of validated C. burnetii effectors at the end of the second validation phase was 80. At the same time that this work was being carried out, a study that identified 43 C. burnetii effectors was published (21). Three of these 43 effectors were also identified in our study (CBU0113, CBU2007, and CBU2013). Due to this significant increase in the number of C. burnetii effectors, we decided to perform a third learning phase for which no experimental validation was performed. This learning phase included 120 validated C. burnetii effectors (see Data Set S1 in the supplemental material). In this machine-learning phase, 86 ORFs obtained a score of 0.8 and higher. These 86 ORFs included 70 validated effectors, two ORFs that were found not to translocate in the first learning phase, and the single ORF that we were unable to clone in the first learning phase as well as 13 newly predicted effectors (see Data Set S4 in the supplemental material). The relatively good accuracy of our predictions (68%) suggests that more than half of these 13 newly predicted effectors (listed in Data Set S4 in the supplemental material) probably encode effector proteins.
Feature groups that contributed the most to effector identification.
The features used as input to the machine-learning classifiers described above were divided into 12 groups (Table 1), which include 62 features (see Data Set S2 in the supplemental material). Examination of the features' importance indicated that features that belong to six of these groups were the ones that contributed the most to the learning. These were similarity to known C. burnetii effectors, similarity to known Legionella effectors, similarity to any L. pneumophila ORF excluding effectors, the C-terminal signal, amino acid composition, and regulatory elements. The feature group of similarity to any L. pneumophila ORF excluding effectors includes two features, (i) BLAST score of best hit against a database of all L. pneumophila proteins excluding effectors and (ii) number of BLAST hits with E values of <0.01 against a database of all L. pneumophila proteins excluding effectors, and both were found to be strong negative features. This result indicates that if a C. burnetii ORF has a homologous protein in L. pneumophila that is not an effector, the C. burnetii ORF is probably not an effector protein as well. Four other feature groups—similarity to known C. burnetii effectors, similarity to known Legionella effectors, C-terminal signal, and regulatory elements—were all found to be strong positive features. Another feature group, amino acid composition, contains 28 features, only three of which were found to have a significant contribution to the classification: (i) similarity to the amino acid composition of known C. burnetii effectors; (ii) combined content of serine, asparagine, glutamic acid, and lysine; and (iii) combined content of valine, alanine, glycine, and isoleucine. The first two features were found to be positive, and the third one was found to be negative. The first feature in this group compares the overall amino acid profile of an ORF to the one of effectors. The two other features were generated since we observed that the amino acids serine, asparagine, glutamic acid, and lysine are more abundant in effectors than in noneffectors and that valine, alanine, glycine, and isoleucine are the least abundant in effectors. The reason for this difference in amino acid profile between effectors and noneffectors is currently not known, but a possible explanation is that the requirement of effectors to unfold during translocation by the Icm/Dot transporter (66) and to refold with their C-terminal ends exposed generated evolutionary constraints that affect the amino acid composition of the effector proteins.
The newly identified C. burnetii effectors are expressed during infection.
In order to determine whether the 13 newly validated effectors (listed in Table 2) are expressed in C. burnetii, the levels of their mRNA were measured using RT-qPCR with C. burnetii Nine Mile phase II grown in ACCM-2 axenic medium as well as in HEK293T cells (see Materials and Methods). The levels of expression of these effector-encoding genes were compared to those of two known C. burnetii effector-encoding genes (CBU1751-coxDFB5 and CBU0635) (24, 25) (Fig. 2). Two main patterns of expression were obtained (Fig. 2). Three effectors (CBU1063, CBU1634a, and CBU1794) were found to have a higher level of expression at the post-exponential growth phase when examined in vivo in HEK293T cells. Several of the other effectors had a pattern of expression similar to those of two previously identified C. burnetii effectors (Fig. 2). These results indicate that all the newly validated effector-encoding genes are expressed in C. burnetii during growth in host cells as well as during growth in axenic medium.
FIG 2.
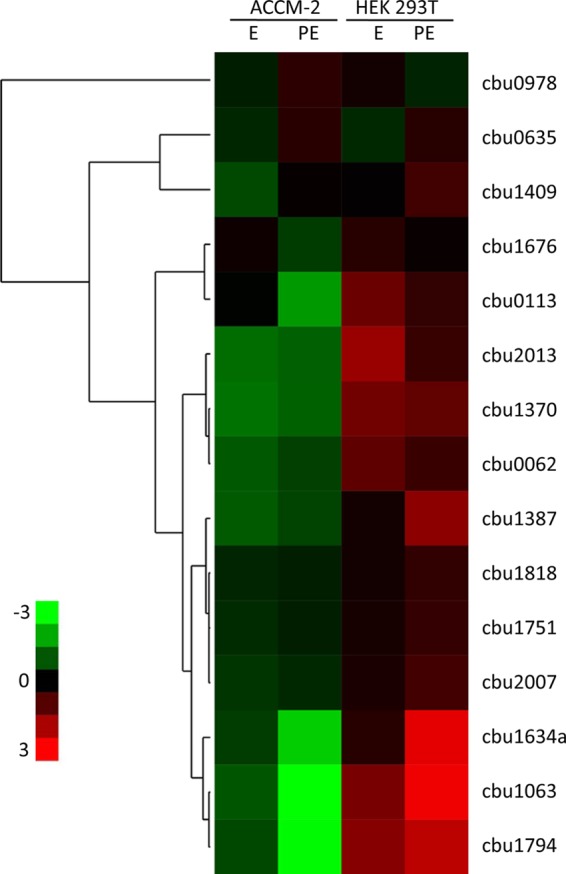
Gene expression levels of the C. burnetii effectors identified. Gene expression levels of C. burnetii effectors were determined during growth in ACCM-2 axenic medium and in HEK293T cells. The heat map shows the logarithm of expression values of the 13 effectors validated in this study as well as two previously identified effectors (CBU1751 and CBU0635). The values are the RNA levels normalized to 16S RNA levels during exponential (E) and post-exponential (PE) phases as described in Materials and Methods.
One of the C. burnetii effectors inhibits yeast cell growth.
With the aim of obtaining a first indication for the function performed by the newly identified C. burnetii effectors (listed in Table 2), they were cloned under the control of the galactose-inducible promoter (GAL1 promoter) and expressed in the yeast S. cerevisiae. It was previously shown, using effectors from various pathogenic bacteria, including C. burnetii and L. pneumophila, that ectopic expression of effectors sometimes results in inhibition of yeast growth (21, 25, 32, 33, 67, 68). Yeast growth inhibition suggests that a conserved and essential eukaryotic process that is modulated by the effector in the host cell was also modulated in the yeast cell, resulting in inhibition of yeast cell growth (33, 34, 67, 69). Examination of the 13 newly identified C. burnetii effectors (CBU0062, CBU0113, CBU0978, CBU1063, CBU1370, CBU1387, CBU1409, CBU1634a, CBU1676, CBU1794, CBU1818, CBU2007, and CBU2013) in yeast revealed that all of them were expressed (Fig. 3B and data not shown), and one effector (CBU1818-Cem11) inhibited yeast growth (Fig. 3A; see also Fig. S1 in the supplemental material). This result indicates that this effector affects a conserved eukaryotic process that is growth limiting under regular yeast growth conditions.
FIG 3.
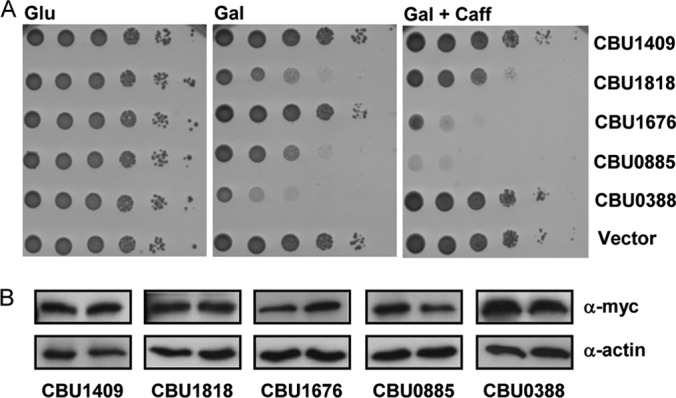
The growth inhibition effect of three C. burnetii effectors was altered in the presence of caffeine. (A) The C. burnetii effectors (indicated on the right) were cloned under the GAL1 promoter and grown on plates containing glucose (Glu), galactose (Gal, inducing conditions), or galactose supplemented with 2 mM caffeine (Gal + Caff) in the wild-type S. cerevisiae BY4741. pGREG523 (Vector) was used as a negative control. (B) The protein levels of the C. burnetii effectors, indicated below each pair, on a galactose-containing plate (left) and a galactose- and caffeine-containing plate (right) were determined by Western analysis using anti-myc antibody. Actin was used as a loading control.
Three C. burnetii effectors had an altered effect on yeast growth in the presence of caffeine.
It was previously shown that effectors can target a pathway that is conserved but is not normally rate limiting for yeast growth (32, 33). To examine whether this situation also occurs with the C. burnetii effectors that we identified, yeast strains expressing the 13 newly identified effectors (listed in Table 2) were grown in the presence of two well-characterized yeast stressors: caffeine and NaCl. These two stressors were chosen since they were previously shown to affect MAP kinase pathways in yeast, and these pathways are important for C. burnetii infection (see the introduction). Caffeine has pleiotropic effects on yeast: it activates the cell wall integrity (CWI) MAP kinase pathway, and growth sensitivity to caffeine is often associated with defects in components of this pathway (64, 70). NaCl is an osmotic stressor that activates the high-osmolarity glycerol (HOG) MAP kinase pathway and is highly toxic to mutants of this pathway (71, 72). The addition of NaCl to the medium did not result in clear changes in the growth of yeast expressing the effectors examined (see Fig. S1 in the supplemental material). However, the addition of caffeine resulted in a marked change in the phenotype of one effector (CBU1676-Cem9) (Fig. 3A). CBU1676 did not inhibit yeast growth under standard growth conditions, but in the presence of caffeine, its expression severely inhibited yeast growth (Fig. 3A). As indicated in Table 2, CBU1676 has a homologous effector in C. burnetii (CBU0885-CetCb4). The results obtained with CBU1676 directed us to examine the effect of CBU0885 on yeast growth under regular growth conditions and after the addition of caffeine (Fig. 3A). Interestingly, the expression of CBU0885 under regular growth conditions moderately inhibited yeast growth, an effect that was markedly enhanced with the addition of caffeine (Fig. 3A). Examination of the levels of expression of these effectors on plates containing galactose with and without the addition of caffeine indicated that the effectors were expressed at similar levels under the two conditions (Fig. 3B). These results indicate that both the CBU1676 and CBU0885 effectors interfere with the ability of the yeast cells to respond to the addition of caffeine and that therefore their expression results in inhibition of yeast growth under these conditions.
In previous work from our group, several C. burnetii effectors were expressed in yeast cells and were examined for their inhibition of yeast growth under regular and stress conditions (unpublished results). In this screen, we identified a C. burnetii effector (CBU0388-CetCb2) that strongly inhibits yeast growth under regular growth conditions (Fig. 3A). Remarkably, its effect on yeast growth was completely suppressed in the presence of caffeine (Fig. 3A). This effect was not a result of changes in the level of expression of this effector under the different conditions since similar levels of expression were observed with and without caffeine (Fig. 3B). This intriguing result led us to include CBU0388 in the forthcoming analyses since it might have an opposite function from the one mediated by the effectors CBU1676 and CBU0885 described above.
Three C. burnetii effectors modulate a MAP kinase pathway in yeast.
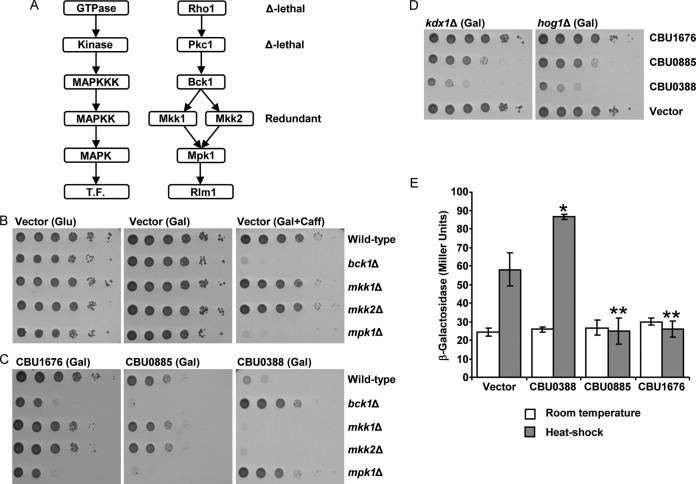
Yeast cells treated with caffeine require the CWI MAP kinase pathway, which activates an adaptation response by inducing the expression of proteins that allow the yeast to grow in the presence of caffeine (64, 70). This pathway consists of a cascade of phosphorylation reactions initiated with the GTPase Rho1, which activates Pkc1, which in turn activates a basic three-protein kinase module. This module includes the MAP kinase-kinase-kinase Bck1 that phosphorylates and thus activates two redundant MAP kinase-kinases (MAPKKs) (Mkk1 and Mkk2) that then phosphorylate and thus activate the MAP kinase Mpk1 (70) (Fig. 4A). Deletion of either mpk1 or bck1 results in yeast strains that are hypersensitive to caffeine (73) (Fig. 4B). Therefore, we decided to test for possible relationships between these three C. burnetii effectors and the CWI MAP kinase pathway.
FIG 4.
Three C. burnetii effectors affect the CWI MAP kinase pathway in yeast. (A) Diagram of the yeast Rho1-activated Pkc1 CWI MAP kinase pathway. The function of each protein is indicated on the left. T.F., transcription factor. (B) Yeast deletion mutants in the CWI MAP kinase pathway are sensitive to caffeine. The deletion mutant strains indicated on the right were examined on plates containing glucose (Glu), galactose (Gal), or galactose and caffeine (Gal+Caff). (C) Examination of the inhibition of yeast growth mediated by the three C. burnetii effectors CBU1676, CBU0885, and CBU0388 in deletion mutants of the CWI MAP kinase pathway. The C. burnetii effectors (indicated above) were overexpressed in the wild-type S. cerevisiae BY4741 (Wild-type) and in the bck1, mkk1, mkk2, and mpk1 deletion mutants (bck1Δ, mkk1Δ, mkk2Δ, and mpk1Δ, respectively). pGREG523 (Vector) was used as a control. (D) The yeast growth inhibition of the C. burnetii effectors CBU1676, CBU0885, and CBU0388 was also examined in two other yeast MAP kinase deletion mutants, the hog1 and kdx1 strains (hog1Δ and kdx1Δ, respectively). The glucose control plates for panels C and D are presented in Fig. S1 in the supplemental material. (E) Three C. burnetii effectors modulate the activity of the Rlm1 transcription factor. The activity of the Rlm1-regulated lacZ reporter was examined following incubation of S. cerevisiae BY4741 containing a vector or expressing the C. burnetii effector CBU1676, CBU0885, or CBU0388 under normal conditions (white bars) and after heat shock (gray bars). β-Galactosidase activity was measured as described in Materials and Methods. Data are the means ± standard deviations (error bars) of three independent transformations. The assay was repeated three times, and similar results were obtained. The levels of expression of the Rlm1 reporter after heat shock of yeast containing each of the effectors were found to be significantly different (*, P < 0.05; **, P < 0.001, paired Student's t test) in comparison to the vector control.
To test whether CBU1676 and CBU0885 modulate the CWI MAP kinase pathway, these effectors were transformed into the bck1Δ, mkk1Δ, mkk2Δ, and mpk1Δ mutants and assayed for their effect on yeast growth (Fig. 4C). The bck1Δ and mpk1Δ mutants expressing these effectors showed increased inhibition of yeast growth in comparison to the wild-type yeast strain when examined on medium containing galactose (Fig. 4C). Growth of the mkk1Δ and mkk2Δ deletion mutants, which are functionally redundant (70), was comparable to that of the wild-type strain. Our analysis suggests functional relationships between yeast genes and the C. burnetii CBU1676 and CBU0885 effectors by identifying yeast deletion mutants that aggravate the CBU1676 and CBU0885 inhibition of yeast growth. The yeast deletion mutants (bck1Δ and mpk1Δ) that are hypersensitive to the expression of CBU1676 and CBU0885 effectively define synthetic lethal interactions with these effectors. Thus, these results indicate possible functional relationships between the yeast genes encoding the CWI MAP kinase pathway and CBU1676 and CBU0885 (see Discussion).
To examine for possible relationships between CBU0388 and the CWI MAP kinase pathway, we introduced CBU0388 into the four deletion mutants indicated above and determined its inhibition on yeast growth on medium containing galactose (Fig. 4C). Since expression of CBU0388 inhibits wild-type yeast growth, any deletion mutant that will grow when CBU0388 is expressed should identify the corresponding gene as a potential target for CBU0388-induced inhibition of yeast growth. Out of the four strains tested, the bck1 and mpk1 deletion mutants resulted in complete suppression of the CBU0388 inhibitory effect. We were unable to examine a deletion in the gene encoding Pkc1 or Rho1 (the two upstream components of the CWI MAP kinase cascade [Fig. 4A]) because these are inviable. In addition, we did not observe suppression of the growth phenotype of CBU0388 with either mkk1 or mkk2 deletions because these genes are functionally redundant. While yeast strains carrying deletions in bck1 and mpk1 grew well when CBU0388 was expressed (Fig. 4C), deletion of genes encoding components of other MAP kinases (hog1, kdx1, fus3, kss1, and smk1) did not affect its inhibition of yeast growth (Fig. 4D and data not shown). Thus, our results are consistent with CBU0388, CBU0885, and CBU1676 specifically targeting the CWI MAP kinase pathway in yeast.
Three C. burnetii effectors affect the activity of the Rlm1 transcription factor in yeast.
The results presented above (Fig. 3 and 4B and C) led us to examine whether the effects observed with the CBU0388, CBU0885, and CBU1676 effectors were a direct result of improper activation of the CWI MAP kinase pathway. To this end, we monitored the effects of these three effectors on the Rlm1 transcription factor, which is activated by Mpk1 (Fig. 4A), using an Rlm1 transcriptional reporter. This well-characterized reporter contains two Rlm1 binding sites fused to the minimal CYC1 promoter driving lacZ (64). The expression of this reporter gene responds appropriately to perturbations that activate the CWI MAP kinase pathway, such as heat shock conditions, and it was previously used to examine the effect of bacterial effectors on this pathway (36, 38). As can be seen in Fig. 4E, expression of CBU0388 resulted with stronger activation after heat shock, and the expression of CBU0885 and CBU1676 abolished the heat shock activation of the Rlm1 reporter. These results fit perfectly with the results presented above (Fig. 3 and 4B and C) and strongly indicate that these three C. burnetii effectors modulate the CWI MAP kinase pathway in yeast.
CBU1676 and CBU0388 suppress the effect of each other on yeast growth.
The “mirror image” results obtained with CBU0388 in comparison to CBU0885 and CBU1676 strongly suggest that CBU1676 and CBU0885 perform a function that antagonizes that of CBU0388. It was previously shown in L. pneumophila that effectors performing opposite functions could be studied in yeast. SidM/DrrA was shown to function as a Rab1 GEF (guanine exchange faction), and it also AMPylates Rab1 (74–77). In addition, this effector was also shown to inhibit yeast growth (78). A screen that was conducted in yeast identified SidD as a deAMPylator of Rab1 that counteracts the activity of SidM and thus suppresses its inhibition of yeast growth (78). A similar observation was obtained with the effectors AnkX and Lem3 (79).
In order to examine whether CBU0388 performs the opposite function of CBU1676 and CBU0885, these effectors were coexpressed in yeast (Fig. 5). When CBU1676 and CBU0388 were examined together on galactose-containing plates, a clear suppression of the CBU0388 inhibition of yeast growth was observed (Fig. 5A). When the yeast strains expressing CBU1676 and CBU0388 were examined on plates containing galactose and caffeine, CBU0388 did not inhibit yeast growth and CBU1676 inhibited yeast growth (Fig. 3A and 5A). Under these conditions, when the two effectors were coexpressed, CBU0388 suppressed the inhibition of yeast growth mediated by CBU1676. These results strongly indicate that CBU0388 and CBU1676 modulate the same pathway in yeast, but in an opposite manner. When a similar analysis was performed using CBU0885 and CBU0388, no suppression of the yeast growth inhibition mediated by either of these effectors was observed (Fig. 5B). The difference between the suppression with CBU1676 and that with CBU0885 is probably a result of the strong inhibition of yeast growth mediated by CBU0885 in comparison to the one mediated by CBU1676. An alternative explanation is that CBU1676 and CBU0388 affect the same target in an opposite manner, leading to suppression of each other's effects, while CBU0388 and CBU0885 affect the same pathway in an opposite manner but do not affect the exact same target; therefore, no suppression of each other's inhibition of yeast growth was observed.
FIG 5.
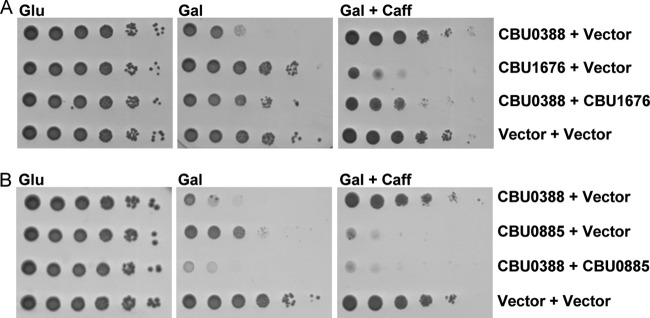
CBU1676 suppresses the yeast growth inhibition of CBU0388 and vice versa. (A) CBU1676, CBU0388, or the two effectors together were overexpressed in wild-type S. cerevisiae BY4741. pGREG523 (Vector) and pGREG525 (Vector) were used as negative controls for CBU0388 and CBU1676, respectively. (B) The same analysis was also performed with CBU0388 and CBU0885. The effectors were examined on glucose (Glu)-, galactose (Gal)-, and galactose- and caffeine (Gal + Caff)-containing plates.
CBU1676 and CBU0885 harbor a functional HAD domain.
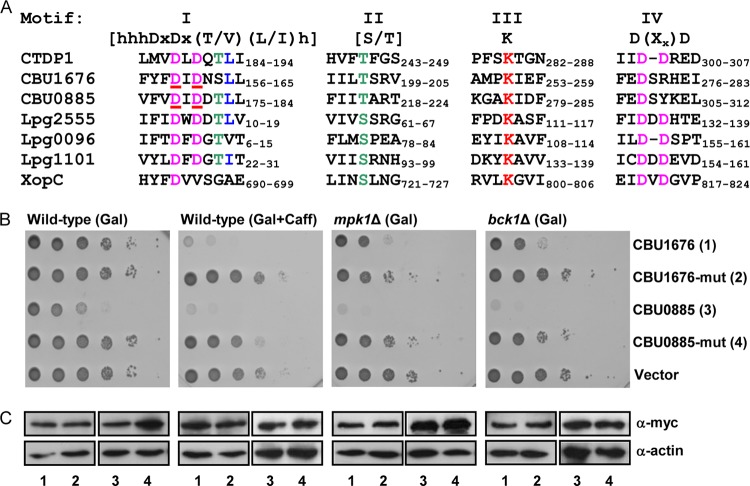
The CBU1676 and CBU0885 effectors are members of a family of proteins that contain a haloacid dehalogenase (HAD) domain. The HAD superfamily is represented in the proteomes of organisms from all three domains of life and performs numerous biological functions, including those of phosphoesterases, ATPases, phosphatases, and dehalogenases, among others (80, 81). Crystallographic studies of prokaryotic and eukaryotic HAD phosphatases revealed that all members of the HAD phosphatase superfamily share the same structural arrangement of the active core (80, 81). The catalytic core residues are highly conserved throughout the HAD phosphatase family and cluster into four signature motifs in the primary amino acid sequence (Fig. 6A). The alignment of CBU1676 and CBU0885 with other HAD family members demonstrated that the critical residues forming the catalytic core were present in these C. burnetii effectors (Fig. 6A). We examined whether mutations in the predicted catalytic sites would affect the ability of CBU1676 and CBU0885 to inhibit yeast cell growth in the presence of caffeine: the DID motifs in CBU0885 at positions 178 to 180 and in CBU1676 at positions 159 to 161 were changed to AIA (these mutated residues are underlined in the first motif of the HAD domain in Fig. 6A). We found that both mutated proteins did not inhibit yeast growth under these conditions (Fig. 6B). In addition, when these mutated effectors were expressed in deletion mutants of the CWI MAP kinase pathway, they did not exhibit the growth inhibition observed with the corresponding wild-type effectors (Fig. 6B). The reduction in the growth inhibition of the mutated effectors was not a result of changes in their level of expression (Fig. 6C). Together, these results demonstrate that the HAD domain present in CBU1676 and CBU0885 is required for their activity on the CWI MAP kinase pathway.
FIG 6.
The HAD domain of CBU1676 and CBU0885 is required for their function. (A) Amino acid sequence alignments of HAD motifs I to IV of the human CTDP1 phosphatase; the C. burnetii effectors CBU1676 and CBU0885; the L. pneumophila effectors Lpg2555, Lpg0096, and Lpg1101; and the Xanthomonas campestris pv. vesicatoria effector XopC. The highlighted catalytic core residues were previously identified by crystal structure (81). The amino acids that were mutated are underlined in motif I. (B) The yeast growth inhibition of CBU1676 and CBU0885 was eliminated by mutations in their HAD domain. The C. burnetii wild-type and mutated (DID to AIA in residues 178 to 180 of CBU0885 and residues 159 to 161 of CBU1676) effectors were grown on plates containing galactose (Gal) or galactose with caffeine (Gal+Caff) in the wild-type S. cerevisiae BY4741 (Wild-type) and in the mpk1 and bck1 deletion mutants (mpk1Δ and bck1Δ) on plates containing galactose (Gal). pGREG523 (Vector) was used as a negative control. The glucose control plates are presented in Fig. S3 in the supplemental material. (C) The protein levels of the wild-type and mutated myc-tagged effectors were determined by Western blotting using anti-myc antibody. The lanes correspond to those indicated in panel B. Actin was used as a loading control.
DISCUSSION
MAP kinase-signaling cascades are ubiquitous eukaryotic cellular processes that often serve as targets for bacterial effector proteins. In mammals as well as in yeast, these signaling pathways regulate a variety of cellular activities. Although the environmental stimuli and therefore the cellular responses mediated by these signaling cascades differ between yeast and mammals, the cascade components themselves are highly conserved and are characterized by a phosphorelay of three kinases (71).
Numerous effectors from several bacterial pathogens were found to modulate components of MAP kinase pathways, and many of them were studied in yeast. For example, both the Yersinia YopJ and the Vibrio VopA effectors function as MAPKK acetyltransferases (37, 38) and the Shigella IpaH9.8 functions as a MAPKK ubiquitin E3 ligase (82). Two additional Shigella effectors, IpgB2 and OspF, were found to modulate a MAP kinase cascade (35, 36), and the characterization of these two effectors, using yeast genetics, resulted in phenotypes similar to the ones presented in our study. Overexpression of the Shigella effector IpgB2 resulted in the inhibition of wild-type yeast growth. To identify the possible target of this effector, it was screened on an ordered array of 5,000 viable yeast deletion mutants to identify deletion mutants in which its growth inhibition will be suppressed. Out of the 5,000 deletion mutants tested, only deletions in three genes, bck1, mpk1, and rlm1, resulted in suppression of the IpgB2 inhibition of yeast growth. These results fit perfectly with the results obtained with the C. burnetii effector CBU0388. This effector, when overexpressed, inhibited wild-type yeast growth, and its effect was suppressed in both the bck1Δ and mpk1Δ mutants (Fig. 4C). Further study of the Shigella IpgB2 revealed that it stimulates cellular responses analogous to those to GTP-active RhoA. This factor is homologous to the yeast Rho1 GTPase, which is an upstream component of the CWI MAP kinase cascade (Fig. 4A). Our results using the Rlm1 reporter system (Fig. 4E) also indicated that the activation of the CWI MAP kinase pathway was stronger when CBU0388 was expressed. Another Shigella effector, OspF, was also studied using yeast genetics (33, 36). The overexpression of OspF in yeast did not inhibit yeast growth, but it mediated a strong growth inhibition in the presence of caffeine (33). This effector was also examined in the yeast deletion mutant collection with the aim of identifying yeast genes whose deletion will result in a synthetic lethal effect. This analysis yielded the identification of many yeast deletion mutants, most of which were related to the CWI signaling pathway, including bck1 and mpk1 (36). These results fit perfectly with the results presented for CBU1676 and CBU0885 for which the growth inhibition was stronger in the presence of caffeine and in the bck1Δ and mpk1Δ mutants (Fig. 4C). Further study of the Shigella OspF effector revealed that it inhibits the MAP kinase pathway by functioning as a MAP kinase phosphothreonine lyase (83). Our results using the Rlm1 reporter system (Fig. 4E) also demonstrated that the activation of the CWI MAP kinase pathway was abolished when either CBU0885 or CBU1676 was expressed. It is important to note that the three C. burnetii effectors examined share no sequence homology with the Shigella effectors described. The similar results that were obtained with the three C. burnetii effectors described in our study and the two Shigella effectors described above, as well as the result showing that CBU1676 and CBU0388 suppress each other's growth inhibition (Fig. 5), strongly suggest that, similarly to the Shigella effectors, these C. burnetii effectors also modulate a MAP kinase pathway during infection of host cells.
In addition to identifying the MAP kinase pathway as the likely pathway modulated by CBU1676 and CBU0885, we also determined the importance of the HAD domain present in these effectors (Fig. 6). The HAD domain superfamily includes phosphoesterases, ATPases, phosphatases, and dehalogenases, which act on a diverse set of substrates. Phosphatases of the HAD domain superfamily are a very large class of enzymes that were shown to dephosphorylate a wide range of low- and high-molecular-weight substrates (80, 81). Using site-directed mutagenesis of conserved residues of the HAD domain of CBU1676 and CBU0885, we showed that this domain is critical for the function of these effectors in yeast (Fig. 6B). Similar to our results, it was shown previously that the Xanthomonas campestris effector XopC does not inhibit yeast growth, but when it was examined in the presence of caffeine, it caused a strong growth inhibition (84). Notably, this effector was found to contain a HAD domain (Fig. 6A), and mutagenesis of conserved amino acids of this domain abolished the growth inhibition by XopC on yeast growth (84). The results showing the importance of the HAD domain for the activity of the C. burnetii CBU1676 and CBU0885 effectors and the analysis in yeast which indicates that these two effectors modulate a MAP kinase pathway suggest that these two effectors modulate the phosphorylation state of one or more components in the MAP kinase pathway.
Both C. burnetii and L. pneumophila utilize very similar Icm/Dot secretion systems for intracellular growth and pathogenesis, and yet their effector repertoires are different. However, two phenomena related to the effector repertoires of these two pathogens seem to be similar. First, many L. pneumophila effectors were found to be functionally redundant: they function on the same host pathway, and even on the same host factor (85, 86). For example, seven L. pneumophila effectors (SidM/DrrA, SidD, AnkX, Lem3, LidA, SidC, and LepB) were shown to target the host factor Rab1 (74, 76–79, 87–89). The few effectors that were studied thus far in C. burnetii already suggest a similar functional redundancy between effectors. Three effectors (AnkG, CaeA, and CaeB) were found to modulate apoptosis in host cells (26, 27), and in this study, we found three effectors (CBU0388, CBU1676, and CBU0885) that modulate a MAP kinase pathway. A second phenomenon that was observed in L. pneumophila effectors was that pairs of effectors can mediate opposite functions: SidM/DrrA was shown to AMPylate Rab1, while SidD functions as a Rab1 deAMPylator (75, 78). In addition, AnkX was shown to function as a phosphocholinator of Rab1, and Lem3 counteracts its activity and functions as a dephosphocholinator of Rab1 (79, 90). The opposite functions mediated by these pairs of effectors were determined using analyses of inhibition of yeast growth (78, 79, 90). In a similar manner, we found in this study that CBU1676 counteracts the yeast growth inhibition mediated by CBU0388 and vice versa, suggesting that C. burnetii effectors also mediate opposite functions during infection. Further studies are required in order to uncover the degree of redundancy that exists among the C. burnetii effectors and the specific functions mediated by the C. burnetii effectors on host MAP kinase pathways.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by grant 2009070 from the United States-Israel Binational Science Foundation (to G.S., T.P., and H.A.S.) and in part by grant no. 3-8178 from the Chief Scientist Office of the Ministry of Science, Israel (to G.S. and T.P.). D.B. was a fellow of the Converging Technologies Program of the Israeli Council for Higher Education. K.S. and H.A.S. acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Research Consortium (National Institutes of Health award 1-U54-AI-057153).
Footnotes
Published ahead of print 23 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01729-14.
REFERENCES
- 1.Fenollar F, Fournier PE, Carrieri MP, Habib G, Messana T, Raoult D. 2001. Risks factors and prevention of Q fever endocarditis. Clin. Infect. Dis. 33:312–316. 10.1086/321889 [DOI] [PubMed] [Google Scholar]
- 2.Maurin M, Raoult D. 1999. Q fever. Clin. Microbiol. Rev. 12:518–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad AF. 2007. Pathogenic rickettsiae as bioterrorism agents. Clin. Infect. Dis. 45(Suppl 1):S52–S55. 10.1086/518147 [DOI] [PubMed] [Google Scholar]
- 4.Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. 2003. Q fever: a biological weapon in your backyard. Lancet Infect. Dis. 3:709–721. 10.1016/S1473-3099(03)00804-1 [DOI] [PubMed] [Google Scholar]
- 5.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2(4):e00175-11. 10.1128/mBio.00175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. 10.1371/journal.pone.0054566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13–24. 10.1038/nrmicro1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26:261–283. 10.1146/annurev-cellbio-100109-104034 [DOI] [PubMed] [Google Scholar]
- 9.McDonough JA, Newton HJ, Roy CR. 2012. Coxiella burnetii secretion systems. Adv. Exp. Med. Biol. 984:171–197. 10.1007/978-94-007-4315-1_9 [DOI] [PubMed] [Google Scholar]
- 10.Segal G, Feldman M, Zusman T. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29:65–81. 10.1016/j.femsre.2004.07.001 [DOI] [PubMed] [Google Scholar]
- 11.Zamboni DS, McGrath S, Rabinovitch M, Roy CR. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965–976. 10.1046/j.1365-2958.2003.03626.x [DOI] [PubMed] [Google Scholar]
- 12.Zusman T, Yerushalmi G, Segal G. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71:3714–3723. 10.1128/IAI.71.7.3714-3723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton HJ, Roy CR. 2011. The Coxiella burnetii Dot/Icm system creates a comfortable home through lysosomal renovation. mBio 2(5):e00226-11. 10.1128/mBio.00226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. 2013. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 11:561–573. 10.1038/nrmicro3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826–831. 10.1073/pnas.0406239101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. 2011. Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2:208. 10.3389/fmicb.2011.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U. S. A. 110:E707–E715. 10.1073/pnas.1215278110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. 10.1126/science.1158160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191:4232–4242. 10.1128/JB.01656-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 63:1508–1523. 10.1111/j.1365-2958.2007.05604.x [DOI] [PubMed] [Google Scholar]
- 21.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J. Bacteriol. 195:3914–3924. 10.1128/JB.00071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maturana P, Graham JG, Sharma UM, Voth DE. 2013. Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii. J. Bacteriol. 195:3269–3276. 10.1128/JB.00180-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J. Bacteriol. 193:1493–1503. 10.1128/JB.01359-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 107:21755–21760. 10.1073/pnas.1010485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 7:e1002056. 10.1371/journal.ppat.1002056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. 2013. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell. Microbiol. 15:675–687. 10.1111/cmi.12066 [DOI] [PubMed] [Google Scholar]
- 27.Luhrmann A, Nogueira CV, Carey KL, Roy CR. 2010. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc. Natl. Acad. Sci. U. S. A. 107:18997–19001. 10.1073/pnas.1004380107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc. Natl. Acad. Sci. U. S. A. 110:E4770–E4779. 10.1073/pnas.1309195110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. 2013. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. mBio 4(1):e00606-12. 10.1128/mBio.00606-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain SK, Broederdorf LJ, Sharma UM, Voth DE. 2010. Host kinase activity is required for Coxiella burnetii parasitophorous vacuole formation. Front. Microbiol. 1:137. 10.3389/fmicb.2010.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voth DE, Heinzen RA. 2009. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect. Immun. 77:205–213. 10.1128/IAI.01124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siggers KA, Lesser CF. 2008. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4:8–15. 10.1016/j.chom.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. 2008. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 4:e9. 10.1371/journal.ppat.0040009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curak J, Rohde J, Stagljar I. 2009. Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 12:18–23. 10.1016/j.mib.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 35.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE. 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124:133–145. 10.1016/j.cell.2005.10.031 [DOI] [PubMed] [Google Scholar]
- 36.Kramer RW, Slagowski NL, Eze NA, Giddings KS, Morrison MF, Siggers KA, Starnbach MN, Lesser CF. 2007. Yeast functional genomic screens lead to identification of a role for a bacterial effector in innate immunity regulation. PLoS Pathog. 3:e21. 10.1371/journal.ppat.0030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon S, Liu Z, Eyobo Y, Orth K. 2003. Yersinia effector YopJ inhibits yeast MAPK signaling pathways by an evolutionarily conserved mechanism. J. Biol. Chem. 278:2131–2135. 10.1074/jbc.M209905200 [DOI] [PubMed] [Google Scholar]
- 38.Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, Siegel RM, Orth K. 2004. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J. Biol. Chem. 279:51953–51937. 10.1074/jbc.M407001200 [DOI] [PubMed] [Google Scholar]
- 39.Salomon D, Bosis E, Dar D, Nachman I, Sessa G. 2012. Expression of Pseudomonas syringae type III effectors in yeast under stress conditions reveals that HopX1 attenuates activation of the high osmolarity glycerol MAP kinase pathway. Microbiology 158:2859–2869. 10.1099/mic.0.062513-0 [DOI] [PubMed] [Google Scholar]
- 40.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5:e1000508. 10.1371/journal.ppat.1000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- 42.Tusnady GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- 43.Langley P, Iba W, Thompson K. 1992. An analysis of Bayesian classifiers, p 223–228 In Swartout E. (ed), Proceedings of the 10th National Conference on Artificial Intelligence. AAAI Press/MIT Press, San Jose, CA [Google Scholar]
- 44.Morrison DF. 1990. Multivariate statistical methods.. McGraw-Hill, New York, NY [Google Scholar]
- 45.Heckerman D, Geiger D, Chickering D. 1995. Learning Bayesian networks: the combination of knowledge and statistical data. Mach. Learn. 20:197–243. 10.1007/BF00994016 [DOI] [Google Scholar]
- 46.Friedman N, Geiger D, Goldszmidt M. 1997. Bayesian network classifiers. Mach. Learn. 29:131–163. 10.1023/A:1007465528199 [DOI] [Google Scholar]
- 47.Vapnik V. 1999. The nature of statistical learning theory.. Springer, New York, NY [Google Scholar]
- 48.Burges CJC. 1998. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2:121–167. 10.1023/A:1009715923555 [DOI] [Google Scholar]
- 49.Breiman L. 2001. Random forests. Mach. Learn. 45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 50.Kohavi R, John GH. 1997. Wrappers for feature subset selection. Artif. Intell. 97:273–324. 10.1016/S0004-3702(97)00043-X [DOI] [Google Scholar]
- 51.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22 [Google Scholar]
- 52.Davis J, Goadrich M. 2006. The relationship between Precision-Recall and ROC curves, p 233–240 In Proceedings of the 23rd International Conference on Machine Learning. Association for Computing Machinery, New York, NY [Google Scholar]
- 53.Moos A, Hackstadt T. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect. Immun. 55:1144–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207. 10.1016/0022-2836(80)90283-1 [DOI] [PubMed] [Google Scholar]
- 56.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 57.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. 2014. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J. Bacteriol. 196:681–692. 10.1128/JB.01175-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 59.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl. Environ. Microbiol. 77:3720–3725. 10.1128/AEM.02826-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 106:4430–4434. 10.1073/pnas.0812074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect. Immun. 78:3465–3474. 10.1128/IAI.00406-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 63.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96. 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 64.Jung US, Sobering AK, Romeo MJ, Levin DE. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781–789. 10.1046/j.1365-2958.2002.03198.x [DOI] [PubMed] [Google Scholar]
- 65.Miller JH. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 66.Amyot WM, deJesus D, Isberg RR. 2013. Poison domains block transit of translocated substrates via the Legionella pneumophila Icm/Dot system. Infect. Immun. 81:3239–3252. 10.1128/IAI.00552-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heidtman M, Chen EJ, Moy MY, Isberg RR. 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell. Microbiol. 11:230–248. 10.1111/j.1462-5822.2008.01249.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viner R, Chetrit D, Ehrlich M, Segal G. 2012. Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis. PLoS Pathog. 8:e1002988. 10.1371/journal.ppat.1002988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campodonico EM, Chesnel L, Roy CR. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56:918–933. 10.1111/j.1365-2958.2005.04595.x [DOI] [PubMed] [Google Scholar]
- 70.Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175. 10.1534/genetics.111.128264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen RE, Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311–1340. 10.1016/j.bbamcr.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hohmann S. 2009. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett. 583:4025–4029. 10.1016/j.febslet.2009.10.069 [DOI] [PubMed] [Google Scholar]
- 73.Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J. 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 61:1147–1166. 10.1111/j.1365-2958.2006.05300.x [DOI] [PubMed] [Google Scholar]
- 74.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949. 10.1126/science.1192276 [DOI] [PubMed] [Google Scholar]
- 75.Neunuebel MR, Chen Y, Gaspar AH, Backlund PS, Jr, Yergey A, Machner MP. 2011. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333:453–456. 10.1126/science.1207193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8:971–977. 10.1038/ncb1463 [DOI] [PubMed] [Google Scholar]
- 77.Machner MP, Isberg RR. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318:974–977. 10.1126/science.1149121 [DOI] [PubMed] [Google Scholar]
- 78.Tan Y, Luo ZQ. 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475:506–509. 10.1038/nature10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan Y, Arnold RJ, Luo ZQ. 2011. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U. S. A. 108:21212–21217. 10.1073/pnas.1114023109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. 2006. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361:1003–1034. 10.1016/j.jmb.2006.06.049 [DOI] [PubMed] [Google Scholar]
- 81.Seifried A, Schultz J, Gohla A. 2013. Human HAD phosphatases: structure, mechanism, and roles in health and disease. FEBS J. 280:549–571. 10.1111/j.1742-4658.2012.08633.x [DOI] [PubMed] [Google Scholar]
- 82.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1:77–83. 10.1016/j.chom.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 83.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science 315:1000–1003. 10.1126/science.1138960 [DOI] [PubMed] [Google Scholar]
- 84.Salomon D, Dar D, Sreeramulu S, Sessa G. 2011. Expression of Xanthomonas campestris pv. vesicatoria type III effectors in yeast affects cell growth and viability. Mol. Plant Microbe Interact. 24:305–314. 10.1094/MPMI-09-10-0196 [DOI] [PubMed] [Google Scholar]
- 85.O'Connor TJ, Boyd D, Dorer MS, Isberg RR. 2012. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338:1440–1444. 10.1126/science.1229556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. U. S. A. 108:14733–14740. 10.1073/pnas.1111678108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. 10.1038/nature10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. 10.1038/nature06336 [DOI] [PubMed] [Google Scholar]
- 89.Machner MP, Isberg RR. 2006. Targeting of host rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11:47–56. 10.1016/j.devcel.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 90.Goody PR, Heller K, Oesterlin LK, Muller MP, Itzen A, Goody RS. 2012. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 31:1774–1784. 10.1038/emboj.2012.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.