Abstract
A heterogeneous subset of extraintestinal pathogenic Escherichia coli (ExPEC) strains, referred to as uropathogenic E. coli (UPEC), causes most uncomplicated urinary tract infections. However, no core set of virulence factors exists among UPEC strains. Instead, the focus of the analysis of urovirulence has shifted to studying broad classes of virulence factors and the interactions between them. For example, the RTX nonfimbrial adhesin TosA mediates adherence to host cells derived from the upper urinary tract. The associated tos operon is well expressed in vivo but poorly expressed in vitro and encodes TosCBD, a predicted type 1 secretion system. TosR and TosEF are PapB and LuxR family transcription factors, respectively; however, no role has been assigned to these potential regulators. Thus, the focus of this study was to determine how TosR and TosEF regulate tosA and affect the reciprocal expression of adhesins and flagella. Among a collection of sequenced UPEC strains, 32% (101/317) were found to encode TosA, and nearly all strains (91% [92/101]) simultaneously carried the putative regulatory genes. Deletion of tosR alleviates tosA repression. The tos promoter was localized upstream of tosR using transcriptional fusions of putative promoter regions with lacZ. TosR binds to this region, affecting a gel shift. A 100-bp fragment 220 to 319 bp upstream of tosR inhibits binding, suggesting localization of the TosR binding site. TosEF, on the other hand, downmodulate motility when overexpressed by preventing the expression of fliC, encoding flagellin. Deletion of tosEF increased motility. Thus, we present an additional example of the reciprocal control of adherence and motility.
INTRODUCTION
Urinary tract infections (UTIs) are the second most common bacterial infection in humans (1). UTIs can be classified as complicated or uncomplicated infections. Uncomplicated UTIs, occurring in otherwise healthy individuals, are self-limited infections of the bladder, referred to as cystitis (2–4). However, upon bacterial ascension into the kidney, a more serious infection referred to as pyelonephritis can develop (2, 4). Pyelonephritis, in turn, can lead to the development of bacteremia and sometimes fatal urosepsis (5, 6).
UTIs normally occur when uropathogens that colonize the intestine alongside commensal organisms gain access to the periurethral area and then ascend to the urinary bladder (7, 8). A heterogeneous subset of extraintestinal pathogenic Escherichia coli (ExPEC) strains, referred to as uropathogenic E. coli (UPEC), causes the overwhelming majority of uncomplicated UTIs (4). UPEC strains carry a battery of virulence factors, including adhesins, toxins, and iron acquisition systems, which promote uropathogenesis (9, 10). However, no core set of virulence factors has been identified. Instead, any given UPEC strain appears to use various virulence factors from these three classes of virulence determinants to colonize the urinary tract (11, 12). This thesis requires that we consider established, newly discovered, and putative virulence factors, as well as the interactions among them, to better understand urovirulence.
Adhesins represent one broad class of virulence determinants. Fimbrial adhesins assembled via the chaperone-usher pathway are the most extensively studied adherence factors (13, 14). Indeed, the genes necessary to synthesize two chaperone-usher fimbriae, type 1 and P fimbriae (pyelonephritis-associated pili), were among the first cloned virulence factor genes (15, 16) and are important during experimental and human UTIs, respectively (17–23). In addition, the genes for seven other putative chaperone-usher fimbriae are carried by prototype UPEC strain CFT073 alone (24). On the other hand, nonfimbrial adhesins have garnered less attention than chaperone-usher adhesins. These adhesins, nevertheless, can also contribute to uropathogenesis (25–30), underscoring the importance of continued study of this adhesin class.
In addition to adhesins, flagellum-mediated motility also contributes to the development of infection ascending to the upper urinary tract (31–33). It is now recognized, however, that adherence genes and flagellar genes can be reciprocally coordinated (34–39). In this network, it is logical that an adherent bacterium should not be motile and a motile bacterium should not be adherent. That is, when fimbrial genes are expressed, flagellar genes should be repressed, and vice versa.
With respect to nonfimbrial adhesins, we have previously described that UPEC strain CFT073 contains within its aspV pathogenicity island (PAI) an RTX (repeats-in-toxin) nonfimbrial adhesin, referred to as TosA (or type one secretion A; originally annotated UpxA) (24, 26). RTX proteins are typically thought of as toxins that are secreted through a type 1 secretion system and diffuse away from the bacterium to mediate effects on the host. This is exemplified by the family prototype α-hemolysin (40–46). However, adhesins that are secreted in the same manner but that remain associated with the bacterial cell surface are a growing group of RTX proteins composed of at least six other well-characterized members (47). We presume that TosA contributes to uropathogenesis by binding to receptors on the surface of host epithelial cells derived from the upper urinary tract. Indeed, deletion of tosA creates a fitness and virulence defect for E. coli CFT073 during an experimental transurethral cochallenge of mice with the parental wild-type strain (26) or independent challenge (48). The same mutant also shows a fitness defect in the spleens and livers during bacteremia, suggesting a function for TosA during urosepsis (27). The tosA gene was previously found in an estimated one-fourth of UPEC strains (11, 26).
An intriguing feature of the tos operon is its strong in vivo expression but poor in vitro expression (27). Indeed, TosA was discovered in an in vivo-induced antigen technology (IVIAT) screen that identified gene products preferentially expressed in vivo (48). The mechanism that explains tight regulation is not understood. Therefore, the focus of this study was to identify regulatory elements associated with tosA expression and the consequences of this regulation as it relates to the reciprocal regulation of motility and adherence. We found that TosR, a PapB family member, represses expression of tosA, while TosE and TosF, two members of the LuxR family, mediate the repression of motility. This work furthers our understanding of how adhesins are regulated and helps to describe the underlying network governing the interplay between adherence and motility.
MATERIALS AND METHODS
Strain construction.
E. coli CFT073 ΔtosR, ΔtosE ΔtosF, and ΔtosR ΔtosE ΔtosF deletion mutants were generated and screened via PCR in an unmarked ΔlacZ background using the primers described in Table 1 and the bacteriophage lambda red recombineering method previously described (49). The original ΔlacZ construct was selected for on lysogeny broth (LB) agar (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl, 15 g/liter agar) containing chloramphenicol (20 μg/ml) and unmarked as previously described (49). All other mutants were selected for on LB plates containing kanamycin (25 μg/ml). In the case of the ΔtosR aph+ strain, the deletion mutation was unmarked as described above to produce the ΔtosR strain. The tosR mutation was also moved into a clean background of wild-type E. coli CFT073 by transduction using phage ΦEB49 (50), with the following modification: phage lysate and an overnight culture were incubated together at room temperature for 20 min at a ratio of 1:5. These constructs were verified by PCR using the primers listed in Table 1. The ΔtosR mutation was also verified by DNA sequencing.
TABLE 1.
Oligonucleotide primers used in the study

F, forward, R, reverse.
TosR-His6 was constructed by cloning tosR into the NcoI and HindIII sites of pBAD-myc-HisA (Invitrogen). A tosEF overexpression construct was generated by cloning tosEF into the PstI and XhoI sites of pBAD-myc-HisA. All constructs were verified by PCR, and the pBAD-tosR-His6 construct was verified by DNA sequencing. Plasmids were maintained in LB containing ampicillin (100 μg/ml). The primers used for generating and screening the plasmid constructs are described in Table 1.
lacZ transcriptional fusions of intergenic regions within the tos operon were generated by cloning the 600-, 233-, and 198-bp regions upstream of tosR, tosC, and tosA, respectively, into the EcoRI and BamHI sites of pRS551 (36), generating pRS551-(PR, PC, PA)-lacZ. The constructs were verified by PCR and DNA sequencing in the case of pRS551-PC-lacZ and pRS551-PA-lacZ. Plasmids were maintained in LB containing kanamycin (25 μg/ml). The primers used to generate and screen these transcriptional fusions are listed in Table 1.
Bioinformatics.
A structural prediction of the TosA 335-amino-acid tandem repeats (27) was constructed by entering this sequence into the Phyre2 server (51) under the normal modeling mode option. The highest-scoring predicted structure was selected as the putative structure of the TosA repeat amino acid sequence.
To construct a GC sliding window plot of the tos operon, a sequence of 12,200 bp, including the entire tos operon and adjacent nucleotide sequences, was entered into the SeqinR R environment (52). We modified a sliding window plot of the GC content program (http://a-little-book-of-r-for-bioinformatics.readthedocs.org/en/latest/src/chapter2.html) to construct a 200-bp sliding window GC content plot of the tos operon in R (version 3.0.1), which was fit to a representation of the tos locus. The average GC content for each gene and the total E. coli CFT073 chromosome was estimated using the sequence statistics feature of the SeqBuilder program (DNAStar).
The prevalence of the genes encoding predicted regulators TosR, TosE, and TosF was estimated by entering the first 100 amino acids of TosA from E. coli CFT073 into the BLAST query tool available on the Broad Institute's UTI Bacteremia Initiative website (https://olive.broadinstitute.org/comparisons/ecoli_uti_bacteremia.3) (E. coli UTI Bacteremia Initiative, Broad Institute [broadinstitute.org], unpublished data). A search was performed against the genomes present in this database with an arbitrary E value cutoff of 1 × 10−20. The results from this search represent strains carrying the tosA gene. The E. coli CFT073 amino acid sequences for TosR, TosE, and TosF were then subjected to the same BLAST search. The resulting hits from these searches were correlated with tosA prevalence. The predicted amino acid sequences of the TosR, TosE, and TosF variants were aligned with the MegAlign (DNAStar) and Clustal V programs. In addition, within the SeqinR (52), Biostrings (53), and gdata (54) R environments, we analyzed the GC content of the tos genes from the UPEC strains described above using several algorithms that we developed.
Deletion mutant and overexpression construct experimental culture conditions.
The tos operon deletion constructs were cultured at 37°C in LB containing kanamycin (25 μg/ml) to mid-log phase (A600 ≈ 0.5). Bacteria were harvested by centrifugation at 6,000 × g for 10 min. The cell pellet was resuspended in 10 mM HEPES, pH 8.3 to 8.9, and centrifuged again. The bacterial cell pellet was again resuspended in 10 mM HEPES, pH 8.3 to 8.9. The cell suspension was stored at −30°C prior to quantification with a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) and Western blotting.
The pBAD-tosR-His6 construct was induced in the unmarked ΔtosR background with 0.05, 0.2, 0.6, 3, and 10 mM l-arabinose in LB containing ampicillin (100 μg/ml) until the culture reached mid-log phase (A600 ≈ 0.5). Whole-cell proteins were collected, stored, and quantified as described above.
To assay fliC expression, the pBAD-tosEF construct was induced in E. coli CFT073 with no or 30 mM l-arabinose in tryptone broth (10 g/liter tryptone, 5 g/liter NaCl) containing ampicillin (100 μg/ml) for 2.5 h. Material was harvested as described above, with the exception that the culture was centrifuged only once at 1,100 × g for 10 min prior to resuspension in 10 mM HEPES, pH 8.3 to 8.9, and the suspension was stored at −30°C.
Western blotting of deletion mutants and overexpression constructs.
To detect TosA, total protein from the tos deletion mutants, E. coli CFT073, or E. coli CFT073 carrying the pBAD overexpression constructs was collected and Western blotting was performed. Briefly, equal amounts of total proteins, determined using a Pierce BCA protein assay kit (Thermo Scientific), from specific constructs were resolved, transferred, and blotted with polyclonal anti-TosA antibodies (27), polyclonal anti-FliC antibodies (37), or an anti-His6 antibody (Invitrogen).
β-Galactosidase assay.
Miller assays were performed as previously described (55), with the exception that bacteria harboring the lacZ transcriptional fusions described above were cultured to mid-log phase (A600 ≈ 0.6 to 0.8) in LB containing kanamycin (25 μg/ml) and, after resting on ice and centrifugation, were resuspended in Z buffer (pH 7.0; 60 mM Na2HPO4·7H2O, 40 mM NaH2PO4, 1 M KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol).
TosR-His6 purification.
TosR-His6 protein was isolated by incubating 50-ml cultures of CFT073 harboring pBAD-tosR-His6 to mid-log phase (A600 ≈ 0.5) in LB containing ampicillin (100 μg/ml) and subsequently inducing expression with 10 mM arabinose for 2.5 h. Bacteria from this culture were pelleted at 2,700 × g and stored at −30°C. TosR-His6 was extracted using a modified QIAexpressionist protocol (Qiagen). Briefly, the cell pellet was resuspended in lysis buffer (pH 8.0; 50 mM NaH2PO4, 300 mM NaCl, 40 mM imidazole) and passed three times through a French pressure cell at 1,200 lb/in2. Cellular debris was cleared by centrifugation at 10,000 × g. The Ni-nitrilotriacetic acid (NTA) agarose (Invitrogen) was equilibrated as described in the QIAexpressionist protocol (Qiagen), with the exception that half of the volumes of Ni-NTA and lysis buffer were used in this step. Cleared lysate was incubated with Ni-NTA agarose at room temperature for 30 min and subsequently run through a column. The column bed was washed three times with washing buffer (pH 8.0; 50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole), and bound proteins were eluted with elution buffer (pH 8.0; 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole). Eluted proteins were concentrated with 10-kDa-cutoff Amicon ultracentrifugal filters (Millipore) and quantified using a 2-D Quant kit (GE Healthcare); this concentrate was dissolved in 10 mM HEPES, pH 8.3 to 8.9. The purity of the TosR-His6 concentrate was assessed on a 12% SDS-polyacrylamide gel, and the gel was stained with SimplyBlue SafeStain (Life Technologies). The presence of TosR-His6 was confirmed by Western blotting, as described above.
Electrophoretic mobility shift assay of the tos operon promoter.
PCR was performed to amplify PtosR and lacZ DNA probes using the gel shift primers described in Table 1 and Easy-A high-fidelity enzyme (Agilent). These probes were terminally labeled with digoxigenin-11-ddUTP (DIG-ddUTP) using a 2nd-generation DIG gel shift kit (Roche Applied Science). Assessment of probe labeling efficiency, TosR-His6 DNA binding reactions, and resolution and detection of shifted DNA probes were all performed as described in the same kit protocol, with the exception that the 25-min binding reaction mixtures contained only binding buffer [pH 7.6; 100 mM HEPES, 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (vol/vol) Tween, 150 mM KCl], in addition to proteins and labeled or unlabeled DNA probes. The concentrations of gel running and transfer buffers were increased to 1× TBE (89.0 mM Tris, 89.0 mM boric acid, 2.0 mM EDTA, pH 8.0), and the anti-DIG-alkaline phosphatase detection antibody dilution used was decreased to 1:1,000 from 1:10,000.
Unlabeled ∼100-bp PtosR fragments were generated using the primers described in Table 1. An electrophoretic mobility shift assay was again performed as described above on DIG-ddUTP-labeled PtosR using the aforementioned PCR products as unlabeled competitors. However, 5% of the amount of the labeled probe described above was used in these competition electrophoretic mobility shift competition assays.
RNA extraction and RT-PCR.
cDNAs were synthesized from equal amounts of RNAs extracted from wild-type E. coli CFT073 and the ΔtosR constructs in exponential phase (A600 ≈ 0.4 to 0.5) that had been cultured in LB as previously described (48). An exception to this extraction protocol was that half the amount of cellular material previously described was stopped with half the amount of stopping solution (5% phenol in ethanol). Reverse transcriptase (RT) PCR was performed on equal amounts of the above-described cDNAs using primers, described in Table 1, directed against the tosAE intergenic region or the tosEF intergenic region. Equal volumes of each PCR mixture were loaded into the sample lanes, and DNA amplicons were resolved on a 1% (wt/vol) agarose gel.
Motility assays.
Overnight cultures of E. coli CFT073 harboring pBAD-myc-HisA, pBAD-tosR-His6, and pBAD-tosEF were normalized to an A600 of 1.0 in 10 mM HEPES, pH 8.3 to 8.9, and stabbed into soft agar (10 g/liter tryptone, 5 g/liter NaCl, 2.5 g/liter agar) containing ampicillin (100 μg/ml) with 33.3 mM l-arabinose. After 17 h of incubation at 30°C, the diameter of the zone of swimming was measured. The ΔtosR, ΔtosE ΔtosF, ΔtosR ΔtosE ΔtosF, and wild-type constructs were also assayed as described above, with the exception that the soft agar did not contain antibiotics or l-arabinose.
Growth curve generation.
Overnight cultures of E. coli CFT073 harboring pBAD-myc-HisA, pBAD-tosR-His6, and pBAD-tosEF were diluted 1:100 into tryptone broth (10 g/liter tryptone, 5 g/liter NaCl) containing ampicillin (100 μg/ml) and 30 mM l-arabinose. Constructs were cultured at 30°C for 24 h in a Bioscreen C automated growth curve system, with A600 readings being recorded every 15 min. This procedure was the same for the ΔtosR, ΔtosE ΔtosF, ΔtosR ΔtosE ΔtosF, and wild-type constructs, with the exception that the tryptone broth did not contain antibiotics or l-arabinose.
Statistical analysis.
The statistical significance of all single comparisons was determined using an unpaired Student's t test. Multiple comparisons were made using an unpaired analysis of variance (ANOVA) followed by Tukey's multiple-comparisons test. With the exception of the tos operon GC content plot graphed in R (version 3.0.1), all other graphs and statistical testing were performed with GraphPad Prism software (version 6.0).
RESULTS
The tos operon of E. coli CFT073 encodes the TosA adhesin, the type 1 secretion system, and three putative regulators.
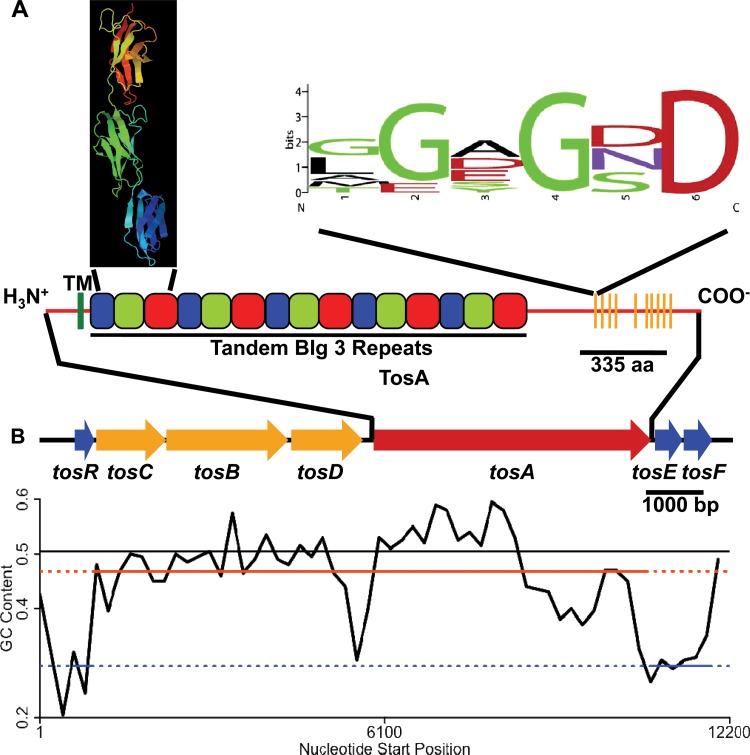
In E. coli CFT073, the tos operon encodes the high-molecular-weight RTX nonfimbrial adhesin, TosA (Fig. 1A and B). In addition, the tos operon contains genes for a putative type 1 secretion system, tosCBD. TosC is predicted to form the outer membrane pore through which TosA is released from the secretion system, TosB is the predicted ATPase/TosA recognition factor, and TosD forms the predicted periplasmic channel through which TosA passes (27, 56). Three open reading frames of previously unknown function, now annotated tosR, tosE, and tosF, are also located within the tos locus. TosR is a homolog of the PapB family of adhesin regulators, and TosE and TosF align with LuxR family members (27).
FIG 1.
The tos operon encodes the genes for the RTX nonfimbrial adhesin TosA, a secretion system, and putative regulators but does not have a uniform GC content. (A) The entire predicted 2,516-amino-acid (aa) sequence of TosA is represented by a red horizontal line. Near the amino terminus, a predicted transmembrane domain is designated TM (vertical green line). Tandem blue, green, and red boxes represent the predicted bacterial immunoglobulin-like family 3 (BIg 3) folds. The corresponding predicted Ig fold structures (modeled using Phyre2 against SiiE from S. enterica with 98.5% confidence) are represented in the black box inset. Near the carboxyl terminus, the positions of 10 tandem RTX repeats are denoted with orange vertical lines. The sequence logo of the RTX repeats is noted (27). (B) Within the tos locus, blue arrows represent genes (tosR and tosEF) encoding predicted DNA binding proteins, orange arrows represent genes (tosCBD) encoding a predicted type 1 secretion system, and a red arrow represents the gene (tosA) encoding the RTX nonfimbrial adhesin. The entire tos locus is fit to a 200-bp sliding window plot of the GC content. Black line, the average GC content of 80 UPEC genomes; orange-red and blue lines, the average GC content of tosCBDA and of tosR and tosEF in the same 80 genomes, respectively.
To assess whether TosA may possess structural features found in other nonfimbrial adhesins (47), a bioinformatics approach was taken to predict TosA features. We previously identified that TosA contains five tandem 335-amino-acid sequence repeats (27). A Phyre2 model (Fig. 1A) (51) predicts that the structure of these repeats is similar to that of the bacterial immunoglobulin-like domain group 3 (BIg 3) repeats found in the Salmonella enterica nonfimbrial adhesin SiiE (57). Like other RTX nonfimbrial adhesins (47), TosA also contains 10 RTX repeats near its carboxyl terminus (27) and a putative transmembrane domain near its amino terminus.
tos operon genes are broadly conserved among UPEC strains.
As the GC content of genes within an operon tends to be similar (58), to determine whether this was also the case for the tos operon, we tracked the tos operon GC content using a 200-bp sliding window (data not shown). The GC content of the structural genes (tosCBDA), 48.5%, was similar to that of the E. coli CFT073 backbone (average, 50.5%). However, the GC content of the putative regulatory genes, tosR, tosE, and tosF, was 29.1%, significantly lower than that of the chromosome, in general, and that of tosCBDA, in particular. Thus, these data reveal that the putative tos operon regulatory genes have a distinct GC content.
Given the differences in GC content among tosR and tosEF in E. coli CFT073, to determine whether the same regulatory genes are conserved in tos operons of other UPEC strains, we analyzed 317 sequenced genomes found in the Broad Institute's UTI Bacteremia Initiative database (E. coli UTI Bacteremia Initiative, Broad Institute [broadinstitute.org]). Among the UPEC strains associated with this study, tosA was found in 32% (101 of 317) of isolates, a prevalence slightly higher than that previously estimated by us in a PCR-based survey of our UPEC strain collection (11). Of the strains with tosA, the overwhelming majority (92 of 101 [91%]) also contained tosR and tosEF. Among strains with single copies of tosA and at least one gene among the tosR and tosEF genes (96/101), our GC content analysis program was able to determine the GC content for 83.3% (80/96) of these strains. The GC content disparities among tos structural and regulatory genes described above held among these strains with the tos genes (Fig. 1B). Thus, while the GC content of tosR and tosEF (29.5%) is considerably lower than that of tosCBDA (46.8%), these genes are conserved and linked to each other within UPEC strains. In agreement with this, we did not find these putative regulators in a UPEC background without tosA. However, 4.0% (4/101) and 7.9% (8/101) of strains with tosA did not contain tosR and tosEF, respectively. Therefore, the tosR and tosEF regulators are conserved and linked among the overwhelming majority of sequenced UPEC strains with tosA.
The TosR, TosE, and TosF amino acid sequences of the tos operon show a clonal nature.
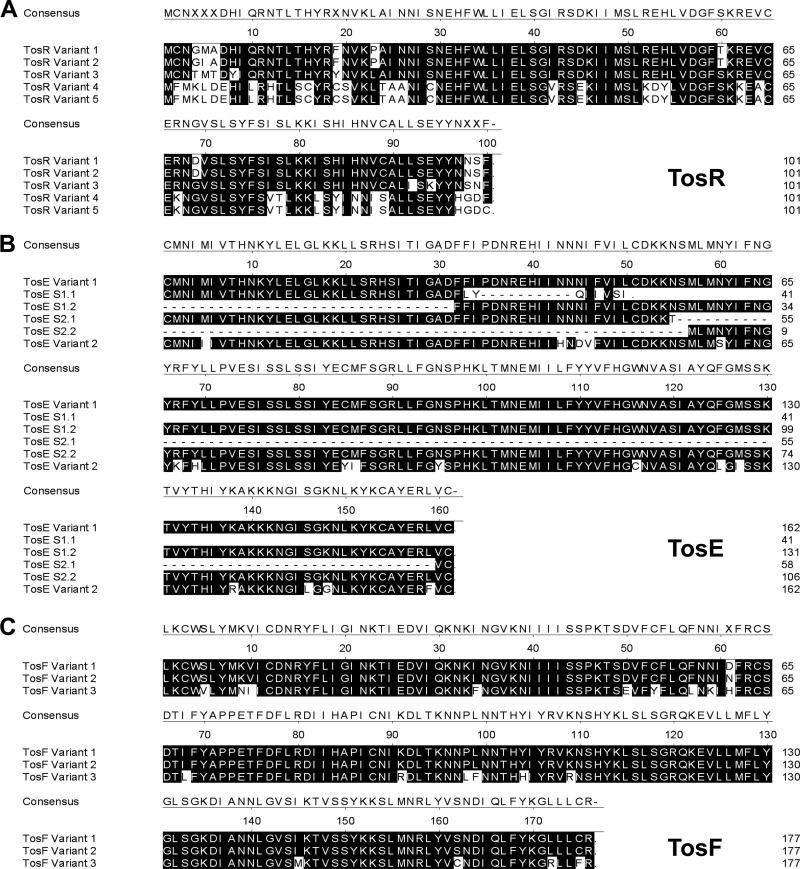
To determine whether the TosR amino acid sequence is conserved among UPEC strains, the predicted TosR amino acid sequence was aligned against the sequences of UPEC strains with genes encoding TosR in the Broad Institute's UTI Bacteremia Initiative database (Fig. 2A). Based on the unique TosR amino acid sequences, we assigned each TosR sequence to one of five sequence variant groups. All variants had an overall predicted sequence identity of 59.0% among each other. Variant 1 accounted for 84.5% of the TosR sequences and was the variant found in E. coli CFT073. Variants 2, 3, 4, and 5 were less prevalent, accounting for 3.1%, 7.2%, 3.1%, and 2.1% of TosR sequences, respectively. It is interesting to note that residues L35, L36, L55, V56, Y74, F75, and S76 were completely conserved among all TosR sequence variants. These residues have previously been shown to be involved in PapB oligomerization (59). In addition, E53 and H54 of TosR sequence variants 1 to 3 and D53 and Y54 of TosR sequence variants 4 and 5 have properties identical or similar to those of D53 and Y54 of PapB, which are again involved in PapB oligomerization (59). Further conservation was observed at putative residue C65, which was previously found to be important for PapB DNA binding (59). Likewise, K61 is completely conserved among all TosR sequence variants and has properties similar to those of R61 of PapB, which was also shown to be involved in DNA binding (59). Thus, from these observations, it can be suggested that strong selective pressure drives sequence conservation among TosR sequence variants.
FIG 2.
Multiple-sequence alignment of TosR (A), TosE (B), and TosF (C) amino acid sequence variants. (A) Shaded residues denote conservation with the predicted TosR consensus sequence where at least three residues at a given position are conserved. (B) Shaded residues denote conservation with the predicted TosE consensus sequence where at least four residues at a given position are conserved. The sequences denoted with an S prefix are predicted to result from a TosE frameshift mutation. (C) Shaded residues match the TosF consensus sequence where at least two residues at a given position are conserved.
TosE and TosF were also conserved among UPEC isolates. As with TosR, TosE and TosF were grouped on the basis of their unique amino acid sequences. For TosE, there were four variants, including two frameshift mutants that are predicted to disrupt TosE function (Fig. 2B). Variant 1 accounted for 79.6% of TosE sequences and was the variant found in E. coli CFT073. Frameshift mutants 1 and 2 and variant 2 accounted for 7.5%, 5.4%, and 7.5% of the TosE sequences, respectively. TosF had three sequence variants, albeit two were virtually identical (Fig. 2C). Variant 1 accounted for 86.0% of sequences and was also the variant found in E. coli CFT073. TosF variants 2 and 3 accounted for 7.5% and 6.5% of amino acid sequences, respectively. A strain harboring TosR variant 1 contained TosE variant 1, frameshift mutant 1 or frameshift mutant 2, and TosF variant 1 or 2. If a strain harbored TosR variant 2, it always contained TosE frameshift mutant 2 and TosF variant 1. All strains harboring TosR variant 3 contained TosE variant 2 and TosF variant 3. However, little consistent homology with nonhypothetical LuxR family members makes functional characterization of the conserved residues in TosEF variants difficult, unlike for TosR, as described above.
TosR is a negative regulator of tosA and a PapB family homolog.
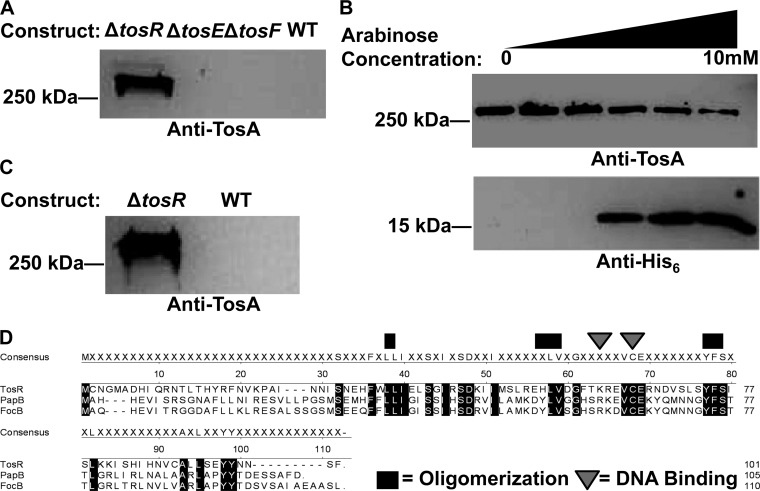
To test the hypothesis that one or more of the identified putative regulators associated with the tos operon exert a regulatory function on tosA expression, deletion mutations of these putative regulatory genes were constructed in E. coli CFT073. Deletion of tosR resulted in a substantial increase in TosA production, as assessed by Western blotting of whole-cell protein using anti-TosA serum (Fig. 3A). However, no change in TosA production was observed after the deletion of tosE and tosF. Additionally, overexpression of tosE and tosF did not result in altered TosA levels (data not shown). Overexpression of TosR-His6 from an arabinose-inducible construct partially complemented the tosR deletion in the unmarked mutant background, repressing tosA expression (Fig. 3B), which is consistent with TosR being a negative regulator of tosA and which indicates that TosR-His6 is biologically activity.
FIG 3.
tosA is negatively regulated by the PapB family member TosR. (A) A Western blot with polyclonal anti-TosA serum reveals that TosA (∼250-kDa) is overexpressed in the ΔtosR aph+ construct but is poorly expressed in both the ΔtosE ΔtosF construct and wild-type (WT; ΔlacZ) E. coli CFT073. (B) A trans-complementation assay in a ΔtosR background using TosR-His6 induced from a plasmid (pBAD-tosR-HisA) with arabinose concentrations ranging from 0 to 10 mM shows that TosA levels (detected as described above) are inversely related to TosR-His6 (∼15 kDa) levels (detected on a Western blot using a His6 antibody). (C) TosA levels (detected as described in the legends to panels A and B) remain high in a ΔtosR aph+ phage-transduced construct compared to those in the wild-type control. All lanes in a respective Western blot were loaded with equal amounts of whole-cell protein, as determined using a Pierce BCA protein assay kit (Thermo Scientific). (D) Alignment of TosR (variant 1), FocB, and PapB reveals that all three share amino acid sequence identify at domains previously shown to be important for oligomerization and DNA binding. The consensus sequence represents residues conserved between the PapB family members shown.
To rule out the possibility that increased tosA expression was due to a secondary mutation, we transduced the tosR deletion mutation into a clean E. coli CFT073 background. This ΔtosR mutation transductant still overproduced TosA compared to the level of TosA production by the wild-type E. coli CFT073 parental strain (Fig. 3C). Thus, we confirmed that the TosA overproduction phenotype associated with ΔtosR is not due to a secondary mutation.
The predicted TosR amino acid sequence of E. coli strain CFT073, when subjected to BLAST analysis, identified PapB family regulators as potential homologs. Using sequence alignment against other PapB family regulators found in E. coli strain CFT073 (Fig. 3D), TosR was found to share 27.7% amino acid sequence identity with PapB and 26.7% identity with FocB. All three proteins share significant sequence similarity with each other, and the predicted TosR protein structure is nearly identical to the structure of FocB (60) (see Fig. S1 in the supplemental material). TosR, PapB, and FocB also carry conserved residues previously shown to contribute to oligomerization and DNA binding (59).
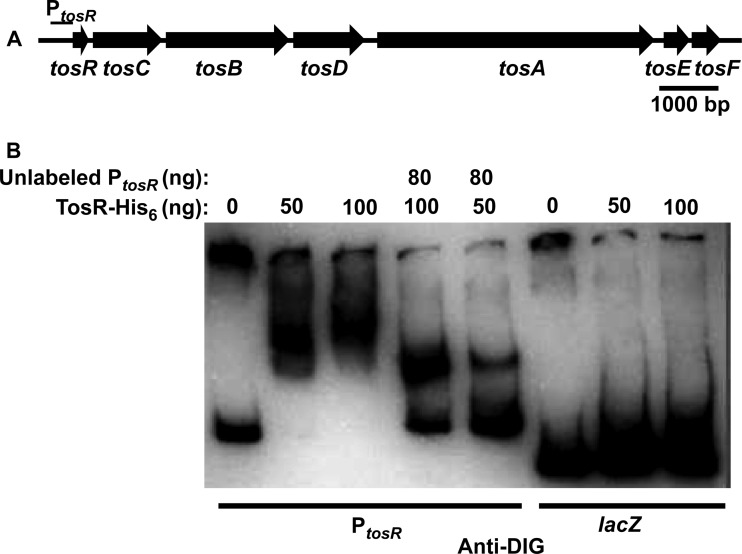
TosR-His6 binds to PtosR, which contains the tos operon promoter.
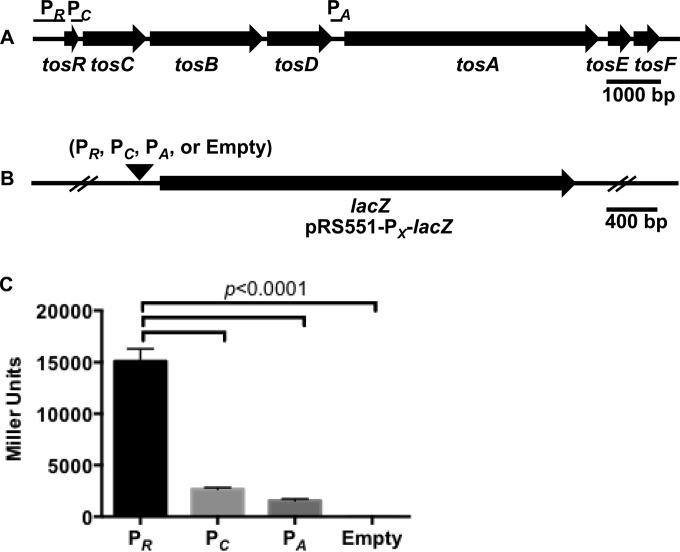
To determine whether the promoter driving tosA expression could be identified, we generated lacZ transcriptional fusions of tos intergenic regions (Fig. 4A), which, from their location upstream of tosA or its cognate secretion system, were predicted to be the most probable location of Ptos (Fig. 4B). These included the 600 bp upstream of tosR (with the addition of the first 30 bp of tosR), the 233 bp upstream of tosC (including the final 181 bp of tosR), and the 199 bp between tosD and tosA. Using Western blotting, we attempted to find an optimal condition for TosA synthesis. Conditions included culturing of wild-type E. coli CFT073 to stationary phase, culturing during exponential phase, static culture, and exposure to different osmotic stresses, human urine, low iron, and different carbon sources. However, none of the aforementioned conditions resulted in reproducible high levels of TosA synthesis compared with the levels achieved in the strain with the ΔtosR mutation cultured to exponential phase (data not shown). Thus, the transcriptional fusions were all assayed using the Miller assay in a strain with the ΔtosR background cultured to exponential phase, which is an optimal condition for tos operon expression. All three putative constructs had elevated transcriptional activity compared to that for the empty vector control (Fig. 4C). However, the construct designated PR, which contains PtosR, was significantly upregulated among all constructs (P < 0.0001). In full agreement with our previous findings regarding tos operon structure (27), we concluded that PtosR contains Ptos, the tos operon promoter.
FIG 4.
The region upstream of tosR exhibits transcriptional activity. (A) Within a representation of the tos operon, sequences harboring possible sites for tos promoters are denoted PR, PC, and PA. (B) A black arrowhead indicates the position where the PR, PC, and PA sequences were inserted into the BamHI and EcoRI sites upstream of lacZ in pRS551 to produce the indicated transcriptional fusions. The empty construct was native pRS551 plasmid. (C) The activity of each transcriptional fusion was assayed by the Miller assay. The transcriptional activity, determined indirectly through measurement of β-galactosidase activity (in Miller units), associated with the PR construct is significantly higher than that associated with the PC and PA constructs (P < 0.0001). Black and gray bars indicate the average values of Miller units for each construct (n = 6). Significance was determined by using Tukey's multiple-comparisons test following ANOVA (P < 0.0001). Error bars indicate the SD about the mean.
PapB family members usually mediate a regulatory function through binding DNA upstream of a PapB family member gene. To determine whether this is the case for TosR, we performed an electrophoretic mobility shift assay on the DNA sequence upstream of tosR, PtosR (Fig. 5A). TosR-His6 shifted labeled PtosR DNA, and competition with unlabeled PtosR in the same binding reaction inhibited labeled probe shifting (Fig. 5B). TosR-His6 did not bind to an unrelated labeled lacZ sequence, demonstrating that TosR-His6 binds specifically to PtosR DNA.
FIG 5.
TosR-His6 binds DNA derived from the region upstream of tosR. (A) Within a representation of the tos operon, the location of the 399-bp PtosR sequence used for the DNA binding assay whose results are shown in panel B is indicated. (B) DIG terminally labeled PtosR or a lacZ fragment was treated with the indicated amounts of TosR-His6 and with or without excess unlabeled PtosR, as indicated. Shifted and unshifted probes were detected with an anti-DIG antibody.
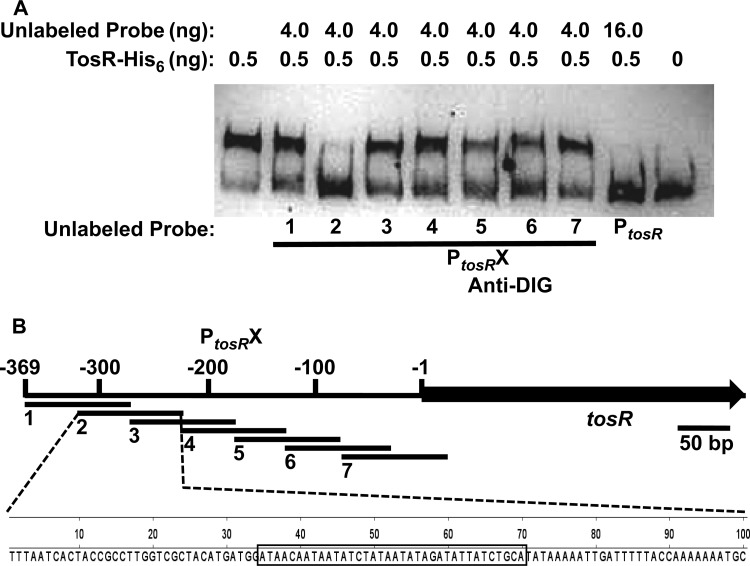
The sequence immediately upstream of a gene encoding a PapB family member is often AT rich (61, 62), and tandem repeated AT-rich nonomers often demarcate PapB family member binding sites (61, 63). Indeed, the region immediately upstream of tosR is AT rich (Fig. 1B). Therefore, to identify the putative TosR binding site, we PCR amplified seven ∼100-bp fragments of PtosR, each of which overlapped by 50 bp. An electrophoretic mobility shift assay was performed using these PtosR fragments as unlabeled competitors (Fig. 6A). Unlabeled PtosR2 (220 to 319 bp upstream of tosR) unshifted labeled PtosR, while none of the adjacent fragments did. Plotting of the positions of the seven PtosR fragments revealed that an AT-rich repetitive sequence was complete only in PtosR2 and was only partially present in the two adjacent fragments, PtosR1 and PtosR3 (Fig. 6B). Thus, we predict that the TosR binding site is centered within this sequence.
FIG 6.
The TosR binding site is centered on PtosR2. (A) The DIG terminally labeled PtosR probe is unshifted only by addition of excess unlabeled PtosR2 or full-length unlabeled PtosR. (B) PtosR fragments 1 to 7 are indicated on a schematic of the PtosR region. The complete sequence of PtosR2 is indicated, with the boxed region indicating the predicted TosR binding site. The tosR open reading frame is indicated.
TosE and TosF contribute to the reciprocal regulation of adherence and motility.
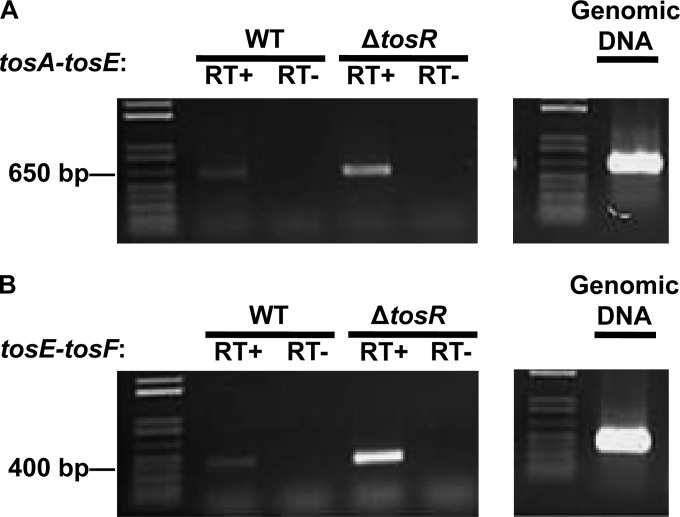
It has previously been observed that adhesin operons regulated by PapB family members harbor genes that encode proteins that suppress motility. These genes are located at the 3′ end of adhesin operons (34–36, 64). To first confirm that tosEF are indeed part of the tos transcript, we performed RT-PCR using primers directed against the junctions between tosA and tosE and tosE and tosF. We found that tosEF were part of the tos transcript, as a strain that overexpresses tosA (the ΔtosR strain) had a corresponding larger amount of transcript with the tosAE (Fig. 7A) and tosEF junctions (Fig. 7B).
FIG 7.
tosEF are part of the tos operon. RT-PCR assays with primers directed against the intergenic region between tosAE (A) and tosEF (B) were performed in the indicated backgrounds. The expected fragment sizes of 619 bp (A) and 402 bp (B) were observed only with reaction mixtures containing cDNAs (RT+) and not with reaction mixtures containing only input RNA (RT−). Equal amounts of input RNAs were used to synthesize all cDNAs, and equal amounts of all cDNAs were used as inputs in the PCRs whose results are shown here. Likewise, equal volumes of all PCR mixtures were loaded in each lane.
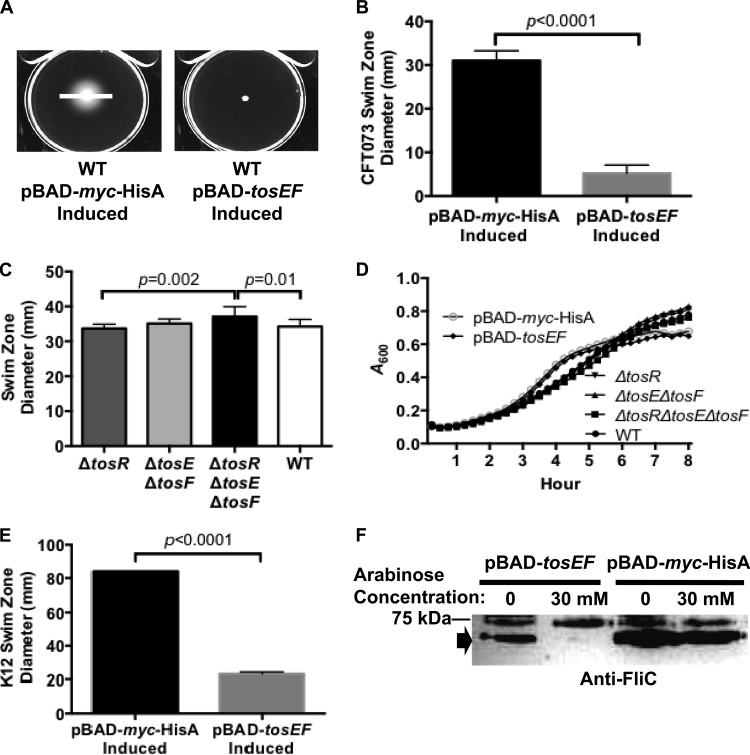
To determine whether TosEF affect motility, we performed motility assays in the presence and absence of tosE and tosF expression. TosEF overproduction resulted in a substantial decrease in swimming motility when this construct was stabbed into soft agar and incubated for 17 h compared to the swimming motility for an empty vector control (P < 0.0001) (Fig. 8A and B). Likewise, compared to the motility of strains with the ΔtosR and wild-type backgrounds, a ΔtosR ΔtosE ΔtosF mutant was statistically significantly, but modestly, more motile in soft agar (P ≤ 0.01) (Fig. 8C). The differences in swimming motility in soft agar between CFT073 ΔtosR, CFT073 ΔtosE ΔtosF, and the CFT073 wild type were not significant. It remains unclear why such a disparity exists between the results described above for the TosEF overproduction construct and the tos deletion constructs. However, no differences in growth rates between constructs could account for the differential motility observed (Fig. 8D). Intriguingly, we found that overexpression of tosEF in E. coli K-12 MG1655 (a non-UPEC strain) also inhibited motility (Fig. 8E), suggesting that TosEF suppress motility through a broadly conserved mechanism. Indeed, we found that overexpression of tosEF in E. coli CFT073 cultured in tryptone broth results in the reduced production of FliC (Fig. 8F). Thus, this shows that TosEF, coincident with tosA expression, mediate motility repression by reducing flagellin expression.
FIG 8.
TosEF downmodulate motility. (A) E. coli CFT073 harboring either pBAD-tosEF or pBAD-myc-HisA was stabbed into soft agar and incubated for 17 h with 33.3 mM l-arabinose. White lines, swimming zone diameter for each induced construct. (B) Average swimming zone diameters are represented by black or gray bars (n = 9). Error bars represent the SD about the mean, and significance between the differences in diameters was determined using Student's t test (P values are indicated). (C) Average (n = 9) swimming zone diameters of strains with the indicated deletion backgrounds or wild-type CFT073, measured after 17 h, are represented by black, gray, or white bars (n = 9). Significance, indicated by the associated P values, was determined by using Tukey's multiple-comparisons test following ANOVA (p = 0.002). Error bars indicate the SD about the mean. (D) Growth curves of the indicated constructs were traced over a period of 8 h. Each point represents an average A600 reading at a given time point (n = 12). (E) E. coli K-12 MG1655 harboring either pBAD-myc-HisA or pBAD-tosEF was stabbed into soft agar and incubated for 17 h with 33.3 mM l-arabinose. Average (n = 9) swimming zone diameters of each overexpression construct are represented by a black or gray bar. Error bars represent the SD about the mean, and the significance between the differences in the diameters was determined using Student's t test (the P value is indicated). (F) Western blotting with polyclonal anti-FliC serum reveals that FliC (∼65 kDa) (black arrow) levels are reduced in wild-type CFT073 harboring pBAD-tosEF induced with l-arabinose at the indicated concentrations compared to the level in a strain of the same background harboring pBAD-myc-HisA. All lanes of this Western blot were loaded with equal amounts of whole-cell protein, as determined by using a Pierce BCA protein assay kit (Thermo Scientific).
DISCUSSION
UTIs are common human infections. While most uncomplicated UTIs are caused by UPEC, no core set of virulence factors has been identified. Thus, a comprehensive understanding of uropathogenesis demands an understanding of broad virulence factor classes and the underlying networks connecting them. We previously identified a novel E. coli adhesin, referred to as TosA, which was expressed only in vivo during experimental infection (27, 48). At that time, it was unclear how the tos operon was regulated. Here, we have shed light on tosA regulation and its function in the reciprocal regulation between adherence and motility. TosR negatively regulates expression of the tos operon, while TosEF downregulate motility when the TosA adhesin is expressed.
We previously found that TosA contains an internal repetitive region of about 1,675 amino acids comprised of five repeats of 335 amino acids each. Protein structure prediction revealed that these internal repeats may have a structure similar to that of the bacterial immunoglobulin-like domain group 3 (BIg 3) repeats found in another nonfimbrial adhesin of S. enterica, SiiE (57). These immunoglobulin folds mediate protein ligand interactions, which endow SiiE with adhesive properties (65). In addition, these immunoglobulin folds, coupled with Ca2+ binding, could also promote SiiE length extension (57), bringing it into proximity with its cognate receptor on the host cell. However, whether the BIg 3 repeats found in TosA mediate adherence, extend TosA such that an element in the carboxyl terminus can mediate adherence, or some combination of these two is still unknown.
tosR, tosE, and tosF, in addition to tosCBDA, are part of the tos locus and are well conserved among UPEC strains harboring tosA. As differences in GC content often demarcate operon boundaries (58), the GC content differences between the tos regulatory and structural genes suggest that tosCBDA might be modular, where the respective regulator genes might be deleted from or inserted into the tos operon. However, we do not preclude the possibility that these GC content differences also reflect a poorly understood tos operon regulatory mechanism, such as differential nucleoid structuring or differential RNA stability. Nonetheless, all three regulator genes are broadly conserved among UPEC strains also carrying tosA, and the typical operon structure is represented by tosRCBDAEF.
The tos operon is conserved among UPEC strains. The vast majority of these tos operons carried by UPEC strains fit tightly into one of five closely related variants, on the basis of the predicted TosR amino acid sequence encoded by the respective operon. Further support for this clonal nature of the tos operon comes from the fact that TosR variants are associated with specific TosE and TosF variants. We conclude that the tos operon present in E. coli CFT073 represents the archetype of the tos operon, as its TosR and TosEF sequence variants are the most prevalent among UPEC strains. However, the origin of the tos operon in its present form is still a matter of conjecture. Nevertheless, its presence on the E. coli CFT073 aspV PAI makes acquisition by horizontal gene transfer likely. PAIs, such as the aspV PAI, are often acquired in such a manner.
The gene encoding TosR is located immediately upstream of tosCBDA. Deletion of tosR results in robust tosA expression, which leads us to conclude that TosR is a negative regulator of tosA expression. Phage transduction of the original ΔtosR mutation into a clean background resulted in high levels of expression of tosA, suggesting that this phenotype is not the result of an unknown secondary mutation. In addition, a His-tagged version of TosR complements the tosR deletion mutation, demonstrating that TosR itself mediates this negative regulation.
Members of the PapB family regulate adhesin operons by binding in the DNA minor groove (60, 61). Minor groove binding proteins often take advantage of the inherent DNA structure of AT-rich regions to mediate target recognition and binding (66–69), which is likely the case for PapB family members (61–63). Indeed, the region upstream of the tos operon, which contains the tos promoter, is an AT-rich sequence. A biologically active His-tagged TosR specifically binds an AT-rich sequence within PtosR. Therefore, we propose that TosR mediates its negative regulatory effect by binding in the minor groove of an AT-rich sequence within the tos operon promoter.
Based on the findings from the present study, we can begin to speculate about how tos regulation fits into other genetic networks within uropathogenic E. coli. While focusing on these underlying networks, such as motility, provides a more complete picture of tosA regulation, determination of this broad mechanism was beyond the scope of the present study. Nevertheless, TosA is an adhesin, and the reciprocal regulation that exists between adhesins and flagellar motility (34–36, 38, 39) represents a starting point for understanding the relationship between tosA and the complete E. coli virulence network. Immediately downstream of tosRCBDA are the two genes encoding TosE and TosF, whose predicted amino acid sequences qualify them to be members of the LuxR helix-turn-helix family of transcriptional regulators (27). Simultaneous deletion or overexpression of tosEF does not affect tosA expression. However, genes encoding factors that downmodulate motility are often found downstream of adhesin operons (34–36). When a respective adhesin operon is induced, these motility-downmodulating genes are also expressed. This, in turn, leads to the downregulation of motility. Indeed, consistent with this prediction, TosE and TosF overproduction results in the downmodulation of motility. Motility repression has also been observed among other LuxR family members, such as CsgD, FimZ, RcsB, and RcsA (70–72).
It was previously unknown, however, whether tosE and tosF were transcriptionally coupled to tosRCBDA under any conditions; it was only known that their expression, like that of tosRCBDA, was poor in vitro (27). We predict that expression of tosRCBDA and tosEF is coordinated to support the reciprocal regulation of adherence and motility. For example, it is possible that the loss of tosR results in increased expression of tosCBDA and tosEF, consistent with the tos transcript structure described above. The subsequent loss of tosE in the ΔtosR background results in enhanced swimming motility by virtue of the loss of tosEF. This increased expression and possible cotranscription are also supported by the fact that a mutant with the ΔtosE ΔtosF mutations alone does not replicate the phenotypes of a ΔtosR ΔtosE ΔtosF mutant.
The reason for the modest differences in motility observed between the tos operon deletion mutants and the tosEF overexpression construct remains unclear. Nevertheless, one intriguing possibility might be that tosEF expression in tos operon mutants is not as uniformly high as that in an overexpression vector construct. In this hypothesis, only certain cell subpopulations might express sufficiently high levels of tosEF to suppress motility, and others expressing lower levels of tosEF might not fully suppress motility. Thus, the composite of these two phenotypes is a swim zone with a reduced, but not a severely reduced, diameter. It is also possible that the more modest expression of tosEF from the chromosome is insufficient to override the dependency for an additional, yet undefined, signal to mediate motility suppression. Therefore, the effect that TosEF has on motility in this scenario would be more modest.
We postulate that, in the case of TosEF, motility repression is an event that occurs upstream of fliC expression, as evidenced by the fact that TosEF overproduction results in reduced FliC synthesis. However, fliC is expressed as a class III gene late in the flagellar assembly gene network (73). Thus, while the tos operon and its expression appear to be part of the network underlying reciprocal regulation between adherence and motility, more work is required to elucidate the precise mechanism of TosEF regulation of the flagellar assembly gene network.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by Public Health Service grant AI059722 from the National Institutes of Health.
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01608-14.
REFERENCES
- 1.Schappert SM, Rechtsteiner EA. 2008. Ambulatory medical care utilization estimates for 2006. Natl. Health Stat. Report 6:1–29 http://www.cdc.gov/nchs/data/nhsr/nhsr008.pdf [PubMed] [Google Scholar]
- 2.Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113:5–13. 10.1016/S0002-9343(02)01054-9 [DOI] [PubMed] [Google Scholar]
- 3.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 52:e103–e120. 10.1093/cid/ciq257 [DOI] [PubMed] [Google Scholar]
- 4.Foxman B. 2010. The epidemiology of urinary tract infection. Nat. Rev. Urol. 7:653–660. 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- 5.Kalra OP, Raizada A. 2009. Approach to a patient with urosepsis. J. Glob. Infect. Dis. 1:57–63. 10.4103/0974-777X.52984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikaheimo R, Siitonen A, Karkkainen U, Mustonen J, Heiskanen T, Makela PH. 1994. Community-acquired pyelonephritis in adults: characteristics of E. coli isolates in bacteremic and non-bacteremic patients. Scand. J. Infect. Dis. 26:289–296. 10.3109/00365549409011797 [DOI] [PubMed] [Google Scholar]
- 7.Hooton TM. 2001. Recurrent urinary tract infection in women. Int. J. Antimicrob. Agents 17:259–268. 10.1016/S0924-8579(00)00350-2 [DOI] [PubMed] [Google Scholar]
- 8.Russo TA, Stapleton A, Wenderoth S, Hooton TM, Stamm WE. 1995. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J. Infect. Dis. 172:440–445. 10.1093/infdis/172.2.440 [DOI] [PubMed] [Google Scholar]
- 9.Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441. 10.1038/nrurol.2010.101 [DOI] [PubMed] [Google Scholar]
- 10.Reiss D, Engstrom M, Mobley H. 2013. Urinary tract infections, p 323–351 In Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F. (ed), The prokaryotes, 4th ed, vol 5 Human microbiology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 11.Vigil PD, Stapleton AE, Johnson JR, Hooton TM, Hodges AP, He Y, Mobley HL. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2:e00066–11. 10.1128/mBio.00066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spurbeck RR, Dinh PC, Jr, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HL. 2012. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 80:4115–4122. 10.1128/IAI.00752-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busch A, Waksman G. 2012. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1112–1122. 10.1098/rstb.2011.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waksman G, Hultgren SJ. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 7:765–774. 10.1038/nrmicro2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orndorff PE, Falkow S. 1984. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J. Bacteriol. 159:736–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connell I, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93:9827–9832. 10.1073/pnas.93.18.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, Tang CM, Donnenberg MS. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079–1093. 10.1046/j.1365-2958.2002.03078.x [DOI] [PubMed] [Google Scholar]
- 19.Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. 2006. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect. Immun. 74:1387–1393. 10.1128/IAI.74.2.1387-1393.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther NW, IV, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344–3354. 10.1128/IAI.70.7.3344-3354.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kallenius G, Mollby R, Svenson SB, Helin I, Hultberg H, Cedergren B, Winberg J. 1981. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet ii:1369–1372 [DOI] [PubMed] [Google Scholar]
- 22.Hagberg L, Jodal U, Korhonen TK, Lidin-Janson G, Lindberg U, Svanborg Eden C. 1981. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect. Immun. 31:564–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HL. 2011. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect. Immun. 79:4753–4763. 10.1128/IAI.05621-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024. 10.1073/pnas.252529799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010–12. 10.1128/mBio.00010-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191:3469–3481. 10.1128/JB.01717-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigil PD, Wiles TJ, Engstrom MD, Prasov L, Mulvey MA, Mobley HL. 2012. The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect. Immun. 80:493–505. 10.1128/IAI.05713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80:321–332. 10.1128/IAI.05322-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669. 10.1128/IAI.01010-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo JM, Schembri MA. 2012. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. J. Bacteriol. 194:5769–5782. 10.1128/JB.01264-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 73:7657–7668. 10.1128/IAI.73.11.7657-7668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U. S. A. 104:16669–16674. 10.1073/pnas.0607898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644–7656. 10.1128/IAI.73.11.7644-7656.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simms AN, Mobley HL. 2008. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J. Bacteriol. 190:3747–3756. 10.1128/JB.01870-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simms AN, Mobley HL. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 76:4833–4841. 10.1128/IAI.00630-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiss DJ, Mobley HL. 2011. Determination of the target sequence bound by PapX, a repressor of bacterial motility, in the flhD promoter using SELEX and high-throughput sequencing. J. Biol. Chem. 286:44726–44738. 10.1074/jbc.M111.290684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane MC, Simms AN, Mobley HL. 2007. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J. Bacteriol. 189:5523–5533. 10.1128/JB.00434-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper LA, Simmons LA, Mobley HL. 2012. Involvement of mismatch repair in the reciprocal control of motility and adherence of uropathogenic Escherichia coli. Infect. Immun. 80:1969–1979. 10.1128/IAI.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson MM, Mobley HL. 2008. Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol. Microbiol. 69:548–558. 10.1111/j.1365-2958.2008.06307.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch RA, Dellinger EP, Minshew B, Falkow S. 1981. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294:665–667. 10.1038/294665a0 [DOI] [PubMed] [Google Scholar]
- 41.Welch RA. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521–528. 10.1111/j.1365-2958.1991.tb00723.x [DOI] [PubMed] [Google Scholar]
- 42.Gadeberg OV, Orskov I. 1984. In vitro cytotoxic effect of alpha-hemolytic Escherichia coli on human blood granulocytes. Infect. Immun. 45:255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadeberg OV, Orskov I, Rhodes JM. 1983. Cytotoxic effect of an alpha-hemolytic Escherichia coli strain on human blood monocytes and granulocytes in vitro. Infect. Immun. 41:358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith YC, Rasmussen SB, Grande KK, Conran RM, O'Brien AD. 2008. Hemolysin of uropathogenic Escherichia coli evokes extensive shedding of the uroepithelium and hemorrhage in bladder tissue within the first 24 hours after intraurethral inoculation of mice. Infect. Immun. 76:2978–2990. 10.1128/IAI.00075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhakal BK, Mulvey MA. 2012. The UPEC pore-forming toxin alpha-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe 11:58–69. 10.1016/j.chom.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gur C, Coppenhagen-Glazer S, Rosenberg S, Yamin R, Enk J, Glasner A, Bar-On Y, Fleissig O, Naor R, Abed J, Mevorach D, Granot Z, Bachrach G, Mandelboim O. 2013. Natural killer cell-mediated host defense against uropathogenic E. coli is counteracted by bacterial hemolysinA-dependent killing of NK cells. Cell Host Microbe 14:664–674. 10.1016/j.chom.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satchell KJ. 2011. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu. Rev. Microbiol. 65:71–90. 10.1146/annurev-micro-090110-102943 [DOI] [PubMed] [Google Scholar]
- 48.Vigil PD, Alteri CJ, Mobley HL. 2011. Identification of in vivo-induced antigens including an RTX family exoprotein required for uropathogenic Escherichia coli virulence. Infect. Immun. 79:2335–2344. 10.1128/IAI.00110-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battaglioli EJ, Baisa GA, Weeks AE, Schroll RA, Hryckowian AJ, Welch RA. 2011. Isolation of generalized transducing bacteriophages for uropathogenic strains of Escherichia coli. Appl. Environ. Microbiol. 77:6630–6635. 10.1128/AEM.05307-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 52.Charif D, Lobry JR. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis, p 207–232 In Bastolla U, Porto M, Roman HE, Vendruscolo M. (ed), Structural approaches to sequence evolution: molecules, networks, populations. Springer-Verlag, New York, NY. [Google Scholar]
- 53.Pages H, Aboyoun P, Gentleman R, DebRoy S. Biostrings: string objects representing biological sequences, and matching algorithms. R package, version 2.30.1 R Development Core Team, Vienna, Austria [Google Scholar]
- 54.Warnes GR, Bolker B, Gorjanc G, Grothendieck G, Korosec A, Lumley T, MacQueen D, Magnusson A, Rogers J. 2014. gdata: various R programming tools for data manipulation. R package, version 2.13.3 R Development Core Team, Vienna, Austria [Google Scholar]
- 55.Miller J. 1992. A short course in bacterial genetics. Cold Springer Harbor Laboratory Press, Cold Springer Harbor, NY [Google Scholar]
- 56.Holland IB, Schmitt L, Young J. 2005. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review). Mol. Membr. Biol. 22:29–39. 10.1080/09687860500042013 [DOI] [PubMed] [Google Scholar]
- 57.Griessl MH, Schmid B, Kassler K, Braunsmann C, Ritter R, Barlag B, Stierhof YD, Sturm KU, Danzer C, Wagner C, Schaffer TE, Sticht H, Hensel M, Muller YA. 2013. Structural insight into the giant Ca(2)(+)-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells. Structure 21:741–752. 10.1016/j.str.2013.02.020 [DOI] [PubMed] [Google Scholar]
- 58.Zheng Y, Szustakowski JD, Fortnow L, Roberts RJ, Kasif S. 2002. Computational identification of operons in microbial genomes. Genome Res. 12:1221–1230. 10.1101/gr.200601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia Y, Uhlin BE. 1999. Mutational analysis of the PapB transcriptional regulator in Escherichia coli. Regions important for DNA binding and oligomerization. J. Biol. Chem. 274:19723–19730 [DOI] [PubMed] [Google Scholar]
- 60.Hultdin UW, Lindberg S, Grundstrom C, Huang S, Uhlin BE, Sauer-Eriksson AE. 2010. Structure of FocB—a member of a family of transcription factors regulating fimbrial adhesin expression in uropathogenic Escherichia coli. FEBS J. 277:3368–3381. 10.1111/j.1742-4658.2010.07742.x [DOI] [PubMed] [Google Scholar]
- 61.Xia Y, Forsman K, Jass J, Uhlin BE. 1998. Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol. 30:513–523. 10.1046/j.1365-2958.1998.01080.x [DOI] [PubMed] [Google Scholar]
- 62.Lugering A, Benz I, Knochenhauer S, Ruffing M, Schmidt MA. 2003. The Pix pilus adhesin of the uropathogenic Escherichia coli strain X2194 (O2: K(−): H6) is related to Pap pili but exhibits a truncated regulatory region. Microbiology 149:1387–1397. 10.1099/mic.0.26266-0 [DOI] [PubMed] [Google Scholar]
- 63.Xia Y, Gally D, Forsman-Semb K, Uhlin BE. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450–1457. 10.1093/emboj/19.7.1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marklund BI, Tennent JM, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol. Microbiol. 6:2225–2242. 10.1111/j.1365-2958.1992.tb01399.x [DOI] [PubMed] [Google Scholar]
- 65.Wagner C, Barlag B, Gerlach RG, Deiwick J, Hensel M. 2014. The Salmonella enterica giant adhesin SiiE binds to polarized epithelial cells in a lectin-like manner. Cell. Microbiol. 16:962–975. 10.1111/cmi.12253 [DOI] [PubMed] [Google Scholar]
- 66.Stella S, Cascio D, Johnson RC. 2010. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 24:814–826. 10.1101/gad.1900610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuber F, Kotlarz D, Rimsky S, Buc H. 1994. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol. Microbiol. 12:231–240. 10.1111/j.1365-2958.1994.tb01012.x [DOI] [PubMed] [Google Scholar]
- 68.Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CS, Hinton JC, Higgins CF. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255–265. 10.1016/0092-8674(92)90354-F [DOI] [PubMed] [Google Scholar]
- 69.Rice PA, Yang S, Mizuuchi K, Nash HA. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295–1306. 10.1016/S0092-8674(00)81824-3 [DOI] [PubMed] [Google Scholar]
- 70.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 193:2587–2597. 10.1128/JB.01468-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clegg S, Hughes KT. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209–1213. 10.1128/jb.184.4.1209-1213.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823–832. 10.1046/j.1365-2958.2003.03601.x [DOI] [PubMed] [Google Scholar]
- 73.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080–2083. 10.1126/science.1058758 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.