Abstract
In El Tor biotype strains of toxigenic Vibrio cholerae, the CTXϕ prophage often resides adjacent to a chromosomally integrated satellite phage genome, RS1, which produces RS1ϕ particles by using CTX prophage-encoded morphogenesis proteins. RS1 encodes RstC, an antirepressor against the CTXϕ repressor RstR, which cooperates with the host-encoded LexA protein to maintain CTXϕ lysogeny. We found that superinfection of toxigenic El Tor strains with RS1ϕ, followed by inoculation of the transductants into the adult rabbit intestine, caused elimination of the resident CTX prophage-producing nontoxigenic derivatives at a high frequency. Further studies using recA deletion mutants and a cloned rstC gene showed that the excision event was recA dependent and that introduction of additional copies of the cloned rstC gene instead of infection with RS1ϕ was sufficient to enhance CTXϕ elimination. Our data suggest that once it is excised from the chromosome, the elimination of CTX prophage from host cells is driven by the inability to reestablish CTXϕ lysogeny while RstC is overexpressed. However, with eventual loss of the additional copies of rstC, the nontoxigenic derivatives can act as precursors of new toxigenic strains by acquiring the CTX prophage either through reinfection with CTXϕ or by chitin-induced transformation. These results provide new insights into the role of RS1ϕ in V. cholerae evolution and the emergence of highly pathogenic clones, such as the variant strains associated with recent devastating epidemics of cholera in Asia, sub-Saharan Africa, and Haiti.
INTRODUCTION
Vibrio cholerae is the causative agent of cholera, which affects millions of people in the developing countries of Asia, Africa, Latin America, and the Caribbean, with more than 120,000 deaths each year (1, 2). The acute watery diarrhea which is the hallmark of cholera is caused primarily by cholera toxin (CT), produced by V. cholerae after colonization of the host intestine. The genes encoding CT are carried in the genome of CTXϕ, a filamentous bacteriophage (3), which exists as a prophage integrated into the chromosome of toxigenic V. cholerae strains. In El Tor biotype strains, the CTX prophage is inserted at the dif recombination site, near the terminus of DNA replication on the bacterium's large chromosome (4). dif sequences are known to be required for XerC/XerD-mediated resolution of chromosome dimers in Escherichia coli and V. cholerae but are also exploited by various filamentous phages for integration of their genomes into the host chromosome by using XerC/XerD-mediated recombination (5, 6).
The CTXϕ genome consists of a core region that encodes CT as well as functions that are required for virion morphogenesis and an RS2 region that encodes the regulation, replication, and integration functions of CTXϕ (7). The RS2 region consists of three open reading frames (ORFs), namely, rstR, rstA, and rstB, and two intergenic regions, namely, ig1 and ig2. The CTX prophage is often located adjacent to two chromosomally integrated satellite phage genomes. These include the genomes of the RS1 satellite phage (8, 9), originally referred to as the RS1 element (10), and the TLC satellite phage (11, 12). The TLC satellite phage is involved in altering the structure of the V. cholerae dif site to enhance integration of the CTX and RS1 phages (12). The RS1 satellite phage exploits CTXϕ-encoded morphogenesis proteins for its own packaging and secretion as RS1ϕ particles (8, 9). The sequence of RS1 is very similar to that of the RS2 region of CTXϕ, except that RS1 carries an additional ORF, termed rstC, which encodes an antirepressor that counteracts the activity of the phage repressor RstR by binding directly to RstR (9).
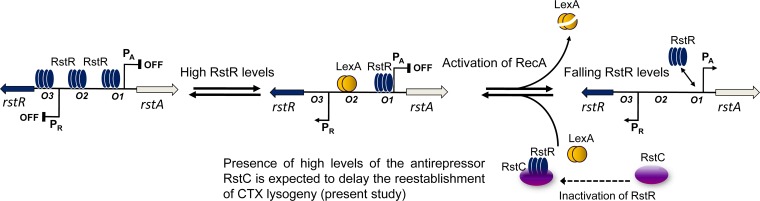
In the CTX prophage, the phage promoter PA, which controls transcription of the genes required for CTXϕ replication and morphogenesis, remains repressed under the control of two DNA-binding proteins: the phage-encoded RstR protein and the host-encoded SOS response regulator LexA (Fig. 1). Although PA is repressed by RstR alone, the repression is enhanced when LexA is also present. Prophage induction in response to DNA damage occurs via the RecA-stimulated autocleavage of LexA, which leads to partial derepression of PA (13, 14). The level of RstR is tightly regulated, since RstR controls transcription of its own gene from the PR promoter (13), and the activation of PR depends exclusively upon LexA being bound to the SOS box (Fig. 1). Thus, the intracellular levels of RstR are maintained via a LexA-dependent auto-feedback loop. The genetic switch of the CTXϕ lysogen ensures that the intracellular levels of RstR are such that PA is repressed but low enough to allow the circuit to respond to changes in the level of LexA. Prophage induction, triggered by agents that cause DNA damage, involves RecA-dependent autocleavage of LexA and subsequent decreased levels of RstR, resulting in derepression of the PA promoter (15–17). Since induction of the CTX prophage does not result in cell lysis and the CTX prophage appears to be stably maintained, presumably the lysogenic program is soon reestablished. However, it was not clear whether the phage antirepressor RstC, which is encoded by the RS1 prophage, has a role in delaying the reestablishment of CTXϕ lysogeny and providing an opportunity for elimination or reassortment of the prophage array. Given that RS1 is carried by most El Tor strains, we further examined the role of RS1 by superinfecting toxigenic V. cholerae strains with a marked RS1 phage (RS1-Kmϕ) and extensively analyzing the transductants. Our results provide new insights into the role of RS1ϕ in the evolution of toxigenic V. cholerae and the generation of new genetic variants.
FIG 1.
Genetic switch of the CTX phage lysogen, based on previous work (13, 16, 35), and role of the antirepressor RstC (this study). Under normal growth conditions, the phage repressor RstR inhibits transcription from the PA promoter by binding to the high-affinity operator, O1, and two weaker operators, O2 and O3, whereas LexA inhibits transcription from PA by binding a single site (SOS box) that overlaps with the O2 operator. RstR (bound to O1) together with LexA activates transcription from PR, which controls the expression of rstR. When RstR levels become too high, RstR displaces LexA from DNA (occupying O2 and O3), and PR is repressed. DNA damage activates the SOS response, which causes RecA-mediated autocleavage of LexA, leading to repression of PR. Eventually, due to falling levels of RstR, derepression of both PA and PR occurs. When the levels of LexA return to normal, the switch is reestablished (16, 35). The presence of high levels of the antirepressor RstC is expected to delay the reestablishment of CTX lysogeny (this study).
MATERIALS AND METHODS
Strains and plasmids.
Toxigenic V. cholerae strains included in the study were from the culture collection of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B), and were originally collected from the stools of cholera patients admitted to the Dhaka or Matlab hospitals of the ICDDR,B. Details of these and other strains used as controls are presented in Table S1 in the supplemental material. The genetically marked phage DNA pRS1-Km was a derivative of the replicative-form (RF) DNA of RS1ϕ in which a kanamycin resistance (Kmr) determinant was introduced into an intergenic NotI site (8). V. cholerae O1 classical biotype strain O395 was transformed with pRS1-Km which carried an rstR gene of the El Tor type. The RS1-Kmϕ phage used in this study was prepared from the culture supernatant of strain O395(pRS1-Km) as described previously (8). The rstC gene, contained in a 454-bp BanII fragment of pRS1-Km, was cloned into pUC18 to construct pRstC (see Table S1).
Probes and hybridization.
PCR-generated amplicons or cloned polynucleotide probes for various genes of the CTX prophage, as described previously (8, 18, 19), were used in hybridization assays. Colony blots or Southern blots were prepared using nylon membranes (Hybond; Amersham Biosciences, Uppsala, Sweden). The blots were hybridized with radioactively labeled probes ([α-32P]dCTP at 3,000 Ci/mmol; PerkinElmer, Boston, MA) and autoradiographed by standard methods (20).
RS1-Kmϕ transduction and pRstC transformation.
Toxigenic V. cholerae strains were exposed to the genetically marked phage RS1-Kmϕ after growing under AKI conditions (21). A genetically marked CTX-Kmϕ phage was also included in the assay to compare the susceptibilities of the strains to both phages. Briefly, bacterial strains were grown statically in AKI medium (21) at 37°C for 4 h and then shifted to overnight shaking at 37°C; the cells were precipitated by centrifugation and were washed in fresh AKI medium. The recipient cells and phage particles were mixed to make approximate final concentrations of 106 bacterial cells and 106 phage particles per ml. The mixture was incubated for 2 h at 37°C, and aliquots of the culture were diluted and plated on Luria agar (LA) plates containing kanamycin (50 μg/ml) to select for Kmr colonies and on plates devoid of kanamycin to determine the total number of colonies. For transformation with pRstC or the empty cloning vector pUC18, recipient V. cholerae strains were electroporated with the appropriate plasmid and selected on LA containing ampicillin (50 μg/ml).
A single colony of RS1 transductants or pRstC transformants derived from each V. cholerae strain was grown in LB for 16 h, and dilutions were plated to prepare replicate colony blots by standard methods (20). In parallel, a derivative of each wild-type strain transformed with the empty cloning vector pUC18 was also processed in an identical way. To detect cells that had lost the CTX prophage and adjacent sequences under in vitro conditions, the blots were hybridized with appropriate probes to identify CTX prophage-negative colonies. The negative colonies were picked and analyzed further by Southern blotting. The chromosomal junctions of the CTX prophage deletion were determined by sequencing the relevant regions in representative CTX-negative derivatives and the corresponding parent strains. To detect the loss of CTX prophage under in vivo conditions, the selected RS1-Kmϕ transductants or pRstC transformants were subjected to passage in the ileal loops of adult rabbits and screened for bacteria that had lost the CTX prophage and related sequences, as described later.
Animal experiments.
The rabbit ileal loop model (22) was used as described previously (23) to examine whether CTX prophage was lost from the pRstC transformants, pUC18 transformants, and RS1 transductants of toxigenic V. cholerae under in vivo conditions. Adult New Zealand White rabbits obtained from the breeding facilities of the Animal Resources Branch of the ICDDR,B were used. A maximum of six ileal loops of approximately 10 cm in length were made in each rabbit, which had previously been fasted for 48 h. Cells grown in LB were precipitated by centrifugation, washed, and resuspended in 10 mM phosphate-buffered saline (PBS), pH 7.4, at a concentration of approximately 1 × 106 cells per ml, and 1-ml aliquots of the cell suspension were inoculated into different sets of rabbit ileal loops. After 16 h, the rabbits were sacrificed and the contents of the ileal loops were collected. The contents of the loops were washed out with 1 ml of 10 mM PBS (pH 7.4) and collected in the same tube. Dilutions of the ileal loop fluids were plated on LA plates. Replicate colony blots were prepared as described above and were hybridized with appropriate probes by standard methods (8, 20) to determine the presence of CTX prophage, RS1, and other relevant sequences flanking the CTX prophage. Total DNAs isolated from representative colonies were also analyzed by Southern blotting. Animal experiments were approved by the Animal Experimentation Ethics Committee of the ICDDR,B.
Chitin-induced transformation.
The recA and lexA mutants of various V. cholerae strains were constructed by using chitin-induced incorporation of the corresponding mutated gene carrying a transposon insertion, obtained by marker exchange from defined mutants in a transposon insertion library (24). Transformation of V. cholerae cells in the presence of chitin was done using the method described by Meibom et al. (25, 26), with modifications. Overnight cultures of the recipient V. cholerae strains were diluted 1:100 in LB medium and grown to an optical density at 600 nm (OD600) of ≈0.3. The bacteria were precipitated by centrifugation, washed, and resuspended in a 0.10 volume of filter-sterilized environmental water or 0.5% sterile sea salt solution (SS). Aliquots of a 2-ml bacterial suspension were dispensed into the wells of a 12-well tissue culture plate containing sterile pieces of shrimp shell. After incubation at 30°C statically for 24 h, the planktonic phase was removed, and fresh water or SS was added. At the same time, 1 to 2 μg of the appropriate DNA was added to the wells. After 24 h, the shrimp shells were removed from the wells, washed in SS, and vortexed in SS to release any attached bacteria. The released bacteria were then plated on LA containing the appropriate antibiotic. Suspected transformants were further analyzed using PCR and hybridization assays.
Illumina sequencing and mapping.
One to 2 μg of genomic DNA was sheared to a length of ∼400 nucleotides by use of a Q500 sonicator (QSonica), and the fragments were used to prepare libraries by using an NEBNext Ultra DNA library preparation kit (New England Biosciences). Libraries were bar coded and sequenced as 50-nucleotide single-end reads on an Illumina HighSeq 2500 flow cell. The total number of reads varied by sample. Reads were aligned to the Haiti H1 strain sequence by using CLC-Genomics Workbench (v. 4.8). In different read data sets, between 26,473 and 1,351,276 reads were found to map within a region that spanned genes that flanked the RS1-CTX-TLC region in the Haiti H1 genome. In the regions of the genome that possessed gaps, sequences were aligned and extended by 20- to 25-nucleotide overlapping windows until the junction was resolved and then reverified from both sequences on either side of the gap. Descriptions of the V. cholerae strains sequenced by Illumina next-generation sequencing to determine the assemblage of phage genetic islands and junctions are presented in Table S2.
Nucleotide sequence accession numbers.
The sequence data have been deposited in the NCBI Sequence Read Archive under the Bioproject accession number PRJNA237589 as sequence Biosamples SAMN02630754 through SAMN02630763.
RESULTS
Toxigenic V. cholerae is susceptible to superinfection by RS1ϕ.
Previous studies have demonstrated heteroimmunity among CTX phages, mediated by diverse CTXϕ repressors encoded by different rstR genes. The existence of at least 3 distinct rstR genes carried by different CTX phages, i.e., CTXϕET, CTXϕClass, and CTXϕCalc, has been recognized (27, 28). Typical El Tor biotype strains carry CTXϕET, whereas classical biotype strains carry CTXϕClass; V. cholerae O139 strains carry CTXϕET, CTXϕCalc, or both. We used genetically marked phages (RS1-Kmϕ and CTX-Kmϕ), both of which carried rstRET, to examine the susceptibility of different strains to these phages. The frequencies of infection of different toxigenic V. cholerae strains by RS1-Kmϕ ranged from 3.2 × 10−4 to 1.2 × 10−2 for El Tor or O139 strains, and this level of susceptibility was ∼50-fold lower than that of classical biotype strains (Table 1). However, compared to the susceptibility of the El Tor or O139 strains to CTX-Kmϕ (2.1 × 10−7 to 1.1 × 10−3), the susceptibility to RS1-Kmϕ was notably higher for most strains. A toxigenic strain carrying CTX prophage is generally resistant to superinfection by a CTXϕ phage carrying the same rstR gene type as that of the resident CTXϕ phage (27). As expected, in this study the classical biotype strains that carried the rstRClass gene were highly susceptible to RS1-Kmϕ (Table 1), which carried the rstRET gene. However, it is significant that El Tor or O139 strains were also superinfected by RS1-Kmϕ at a reasonable frequency, despite the fact that RS1-Kmϕ and the recipient El Tor or O139 strains with resident CTX and RS1 prophages carried the same gene (rstRET).
TABLE 1.
Susceptibilities of different toxigenic Vibrio cholerae strains to superinfection with genetically marked RS1 and CTX phages
| Strain | Description | Presence of prophage or TCP islanda |
Susceptibility to phageb (frequency) |
|||
|---|---|---|---|---|---|---|
| CTX | TCP | RS1 | RS1-Kmϕ | CTX-Kmϕ | ||
| P-27457 | O1, El Tor | + | + | + | 2.3 × 10−3 | 1.1 × 10−6 |
| G-7555 | O1, El Tor | + | + | + | 1.2 × 10−2 | 2.8 × 10−6 |
| G-3985 | O1, El Tor | + | + | + | 6.1 × 10−3 | 1.1 × 10−3 |
| RV508 | O1, classical | + | + | − | 7.5 × 10−1 | 6.9 × 10−1 |
| S-224 | O1, classical | + | + | − | 1.2 × 10−1 | 1.7 × 10−1 |
| Syria-3 | O1, El Tor | + | + | + | 2.7 × 10−3 | 1.2 × 10−6 |
| MG116226 | O1, El Tor | + | + | + | 1.5 × 10−3 | 5.2 × 10−7 |
| MJ1485 | O1, El Tor | + | + | + | 2.1 × 10−3 | 1.2 × 10−6 |
| MG116926 | O1, El Tor | + | + | + | 1.7 × 10−3 | 3.2 × 10−7 |
| AI-1836 | O139 | + | + | + | 2.6 × 10−3 | 1.8 × 10−4 |
| AK-17334 | O139 | + | + | + | 3.6 × 10−3 | 1.1 × 10−4 |
| AR-022-14021 | O139 | + | + | + | 2.4 × 10−3 | 4.5 × 10−7 |
| AL11089 | O139 | + | + | + | 3.2 × 10−4 | 2.1 × 10−7 |
The presence of different representative genes of the prophage or pathogenicity island was detected using DNA probes and PCR assays.
Phage susceptibility was determined using genetically marked phages (see the text for details).
El Tor or O139 strains superinfected with RS1-Kmϕ lose the resident CTX prophage in vivo.
The introduction of additional copies of RS1 into toxigenic V. cholerae cells through infection by RS1-Kmϕ appeared to destabilize the resident CTX prophage and CTX-RS1 arrays in the recipient El Tor or O139 strains. Whereas extensive subculture of wild-type toxigenic strains or passage of the strains in rabbits failed to generate CTX-negative derivatives (data not shown), the presence of pRS1-Km in the RS1-Kmϕ-infected cells led to the production of CTX-negative derivatives at a high frequency when cells were inoculated into the ileal loops of adult rabbits (Table 2). In most cases, these strains also lost the RS1 and CTX prophages together and, infrequently, the TLC prophage as well (Table 3). Similar effects were observed when the replicative form of the RS1-Kmϕ genome, pRS1-Km, was introduced into the toxigenic El Tor or O139 strains by electroporation instead of transduction with RS1-Kmϕ (data not shown).
TABLE 2.
Elimination of CTX prophage from toxigenic V. cholerae strains superinfected with a genetically marked RS1 phage or transformed with a cloned rstC gened
| Parent straina | Description | CTX type | Frequency of loss of CTX prophage upon treatment of parent strain |
|||
|---|---|---|---|---|---|---|
| Infection with RS1-Kmϕ |
Transformation with pRstC |
|||||
| In vitro | In rabbitsb | In vitro | In rabbitsb | |||
| P-27457 | O1, El Tor | CTXET | 6.4 × 10−3 | 8.6 × 10−2 | 1.0 × 10−3 | 2.2 × 10−2 |
| G-7555 | O1, El Tor | CTXET | 9.5 × 10−3 | 2.3 × 10−2 | 1.2 × 10−3 | 6.1 × 10−2 |
| G-3985 | O1, El Tor | CTXET | 5.5 × 10−3 | 9.1 × 10−3 | 1.3 × 10−3 | 4.2 × 10−2 |
| RV508 | O1, classical | CTXClass | <1 × 10−9c | <1 × 10−9c | <1 × 10−9c | <1 × 10−9c |
| S-224 | O1, classical | CTXClass | <1 × 10−9c | <1 × 10−9c | <1 × 10−9c | <1 × 10−9c |
| Syria-3 | O1, El Tor | CTXET | 5.0 × 10−3 | 2.6 × 10−2 | 4.0 × 10−3 | 5.1 × 10−2 |
| MG116226 | O1, El Tor | CTXET | 6.6 × 10−3 | 5.2 × 10−2 | 7.9 × 10−3 | 4.1 × 10−2 |
| AI-1836 | O139 | CTXET | 2.1 × 10−3 | 6.3 × 10−2 | 2.5 × 10−3 | 7.2 × 10−2 |
| AK-17334 | O139 | CTXET | 6.8 × 10−3 | 2.1 × 10−2 | 9.0 × 10−4 | 1.5 × 10−2 |
| AL33457 | O139 | CTXET | 3.2 × 10−3 | 6.8 × 10−3 | 4.3 × 10−3 | 8.3 × 10−3 |
| AR022-14021 | O139 | CTXET, CTXCal | 5.9 × 10−3 | 9.5 × 10−2 | 1.3 × 10−3 | 2.0 × 10−2 |
Extensive subculture of wild-type toxigenic strains in vitro or passage in rabbits without infection with RS1-Kmϕ or transformation with pRstC, or with transformation with the empty cloning vector pUC18 instead of pRstC, did not produce any detectable CTX-negative derivatives.
The CTX-negative strains also lost pRS1-Km and became kanamycin sensitive.
No CTX-negative colonies were found among 109 Kmr recipient cells screened.
For the El Tor and O139 strains, the means of the differences in CTX prophage excision frequencies observed under in vivo conditions compared to in vitro conditions were statistically significant both in the RS1-Kmϕ transductants (P = 0.0102) and in the pRstC transformants (P = 0.0016). Transformation with pRstC caused significantly more CTX prophage excision than infection with RS1-Kmϕ under in vitro conditions (P = 0.0344). However, in the rabbit model, the difference in the frequencies of CTX prophage excision between these two groups was not statistically significant (P = 0.8674).
TABLE 3.
Characteristics of nontoxigenic derivatives of El Tor strains produced by RS1 phage-mediated excision of CTX prophagea
| Parent strain | Presence of genetic region in parent strain |
Presence of genetic region in the nontoxigenic derivative |
Designation of the derivative | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CTX | RS1 | TLC | rtxA | CTX | RS1 | TLC | rtxA | ||
| MG-116926 | + | − | + | + | − | − | − | + | SF2 |
| MG-116226 | + | + | + | + | − | + | + | + | SF3A |
| + | + | + | + | − | − | + | + | SF3B | |
| AR-02214021 | + | + | + | + | − | − | − | + | SF2002 |
| AI-1836 | + | + | + | + | − | − | − | + | SF93 |
| AK-17334 | + | + | + | + | − | − | − | + | SF95 |
| G-7555 | + | + | + | + | − | + | + | + | SF20011 |
| + | + | + | + | − | − | - | + | SF20012 | |
| G-3985 | + | + | + | + | − | − | + | + | SF69-2 |
| P-27457 | + | + | + | + | − | + | + | + | SF20016 |
| + | + | + | + | − | − | + | + | SF20019 | |
The presence of different genes was detected using DNA probes and PCR assays. Selected strains were sequenced to confirm deletion events and identify the junctions.
RstC expressed from a cloned rstC gene is sufficient for CTXϕ excision.
RS1 differs from the RS2 region of CTX prophage in that it carries the antirepressor gene rstC. Hence, we examined whether introducing additional copies of the rstC gene cloned into a plasmid (pRstC) could mimic the effect of superinfection by RS1-Kmϕ. Interestingly, a similar outcome was observed when the V. cholerae cells were transformed with pRstC and introduced into the rabbit ileal loops. Therefore, it appears that excision of the CTX prophage was enhanced due to the activity of the antirepressor RstC, whose gene is also present in pRS1-Km. The elimination of CTX prophage occurred at a higher frequency when cells were introduced into the ileal loops of rabbits than under in vitro laboratory conditions (Table 2). Analysis of the nontoxigenic derivatives for the presence of various genes of the CTX prophage and its adjacent regions by PCR and Southern blot analysis showed that while all nontoxigenic derivatives had lost the CTX prophage, some of the derivatives had also lost the flanking TLC or RS1 phage, or both these prophages, together with the CTX prophage (Table 3; Fig. 2). Unintegrated pRS1-Km or pRstC was also lost at a high frequency (∼90%) when the strains were inoculated into rabbit ileal loops. Sequencing of the relevant genomic region in a representative CTX-negative derivative showed that the strain retained the dif site and the bacterial or phage attachment sequence (see Fig. S1 in the supplemental material).
FIG 2.
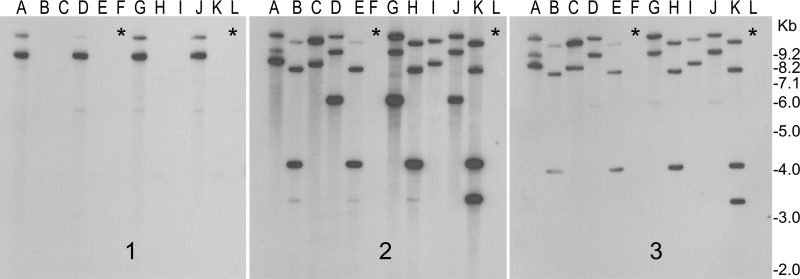
RS1ϕ-induced elimination of CTX prophage from toxigenic V. cholerae. Toxigenic strains were exposed to RS1-Kmϕ and screened for the production of CTX-negative derivatives (see the text for details). DNAs isolated from representative colonies were digested with BglI and analyzed by Southern blotting with probes for the ctxAB (1), rstR (2), and rstC (3) genes. Lanes A, P27457 wild type; lanes B and C, P27457 infected with RS1-Kmϕ; lanes D, G7555 wild type; lanes E and F, G7555 infected with RS1-Kmϕ; lanes G, strain G3985 wild type; lanes H and I, G3985 infected with RS1-KmΦ; lanes J, Syria-3 wild type; lanes K and L, Syria-3 infected with RS1-KmΦ. Lanes marked with asterisks show CTX-negative derivatives which simultaneously lost RS1 and, hence, all rstR- and rstC-hybridizing sequences.
RS1-mediated excision of CTX prophage is recA dependent.
The induction of CTX prophage is known to involve recA-mediated cleavage of the SOS response regulator LexA (14). We used a set of recA or lexA mutants to determine whether the CTX excision event in the presence of additional copies of RS1 or cloned rstC also occurred through the same mechanism. Although in this process the parent strains were found to lose the CTX prophage at a high frequency, the corresponding recA mutants mostly retained the CTX prophage (Table 4). The lexA mutants, however, lost the CTX prophage, and the frequency of loss was comparable to that of the parent strains. Hence, the RecA activity was essential to the induction of chromosomally integrated CTX prophage.
TABLE 4.
Effects of different recA or lexA mutations on RS1-mediated excision of CTX prophage from toxigenic V. cholerae strains
| Straina | Frequency of loss of CTX prophage upon treatment of parent strain |
|
|---|---|---|
| Infection with RS1-Cmϕ | Transformation with pRstC | |
| G-7555 | 2.3 × 10−2 | 6.1 × 10−2 |
| G-7555 recA::TnFGL3 | <1 × 10−7b | <1 × 10−7b |
| G-7555 lexA::TnFGL3 | 1.7 × 10−2 | 2.3 × 10−2 |
| G-3985 | 9.1 × 10−3 | 4.2 × 10−2 |
| G-3985 recA::TnFGL3 | <1 × 10−7b | <1 × 10−7b |
| G-3985 lexA::TnFGL3 | 2.4 × 10−2 | 1.3 × 10−2 |
| AL33457 | 6.8 × 10−3 | 8.3 × 10−3 |
| AL33457 recA::TnFGL3 | <1 × 10−7b | <1 × 10−7b |
| AL33457 lexA::TnFGL3 | 5.2 × 10−3 | 2.2 × 10−2 |
Extensive subculture of all wild-type toxigenic strains in vitro or passage in rabbits without infection with RS1-Kmϕ or transformation with pRstC did not produce any detectable CTX-negative derivatives.
No CTX-negative colonies were found among 107 Kmr recipient cells screened.
RS1-mediated nontoxigenic derivatives are susceptible to CTXϕ reinfection.
We tested whether the nontoxigenic derivatives recovered from the assays in rabbits following transduction with RS1-Kmϕ could act as progenitors of fresh toxigenic strains by reinfection with CTXϕ. Most of the nontoxigenic derivatives were susceptible to a genetically marked phage (CTX-Kmϕ) (see Table S3 in the supplemental material). Further analysis confirmed that in most of the strains, the CTX phage genome was integrated into the chromosome, forming stable lysogens.
Nontoxigenic El Tor strains acquire classical CTX prophage.
Typically, classical biotype strains that carry CTXClass prophage do not produce infectious CTXClassϕ particles (29), and hence, the transfer of the CTXClass genome to recipient strains is unlikely to occur through infectious phages. However, CTXClass prophage has been detected in strains other than the classical biotype of V. cholerae O1 (18, 30). We tested whether the CTX-negative El Tor strains could acquire CTXClass prophage by using an alternative mechanism, e.g., chitin-induced transformation (25) with genomic fragments of a classical biotype strain (O395-NT) (31) which carries a kanamycin resistance marker in the CTX prophage. The induction with chitin caused uptake and recombination of the Kmr-marked CTXClass DNA, resulting in variants of El Tor strains that carry the CTXClass prophage. The frequencies of transformation of various recipient strains are presented in Table S4 in the supplemental material. Both the infection of Kmr-marked CTXϕ and chitin-induced uptake of classical O395 DNA introduced the CTX phage genome between maintained attP and dif sites in strains deleted for CTX and the CTX-RS1-TLC region (see Fig. S1).
DISCUSSION
The results described here provide new insights into the role of the RS1 satellite phage in V. cholerae evolution, and presumably that of other bacteria which carry phages with similar antirepressors involved in lysogeny (32, 33). Understanding the CTX genetic switch may lead to comprehension of possible thematic insights about lysogenic phages which show a reversible pattern of induction and reestablishment of lysogeny. The typical “genetic switch” of phage λ is controlled by λCI, a phage-encoded DNA-binding repressor protein (34). In response to agents that cause DNA damage, RecA stimulates the autocleavage of λCI, which leads to derepression of the lysis genes (17). Unlike the genetic switch of λ phage, which, once flipped, irreversibly commits the phage to the lysis pathway, the CTXϕ switch should have distinct features that enable it to display reversible behavior upon induction and eventually to reestablish lysogeny. Since the heteroimmunity of V. cholerae to CTX and related phages is dependent on the diversity of rstR, the role of the antirepressor RstC is particularly important. In this study, the higher susceptibility of El Tor strains to RS1-Kmϕ than to CTX-Kmϕ (Table 1) occurred presumably because RS1 encodes the RstC antirepressor that can counteract the phage repressor RstR (9). Thus, the rstC gene carried by RS1ϕ may also contribute to this element's dissemination by overriding established CTXϕ and RS1ϕ prophage immunity and may allow RS1ϕ to successfully superinfect CTX- and RS2-positive toxigenic strains. As shown here, this superinfection can drive the eventual elimination or reassortment of the resident prophages related to CTXϕ and RS1ϕ.
McLeod and colleagues (16) proposed a model for maintaining CTX lysogen by toxigenic V. cholerae (35). In this context, RstR inhibits transcription from PA by binding to the high-affinity operator, O1, and two weaker operators, O2 and O3, whereas LexA inhibits transcription from PA by binding a single site (SOS box) that overlaps with the O2 operator. As expected based on this overlap, LexA binding to the SOS box and RstR binding to the O2 operator are mutually exclusive. In this model, the intracellular levels of LexA and RstR are proposed to be such that LexA occupies the SOS box DNA. Furthermore, because the accumulation of high levels of RstR would cause RstR to occupy the O2 site and prevent LexA from binding to the SOS box, RstR regulates the expression of its own gene, which involves a LexA-dependent auto-feedback loop (13). In particular, when the concentration of RstR is such that RstR occupies only O1, RstR activates transcription of PR. Thus, when the concentration of RstR is sufficiently high that O1, O2, and O3 are occupied, RstR represses PR transcription. However, the ability of RstR to activate transcription from PR depends strictly upon LexA being bound to the SOS box (Fig. 1).
This feature of the CTXϕ switch ensures that the intracellular levels of RstR are such that PA is repressed (i.e., O1 is occupied by RstR) but are low enough to allow the circuit to respond to changes in the levels of LexA (i.e., O2 is not occupied by RstR). Once the CTX prophage is induced, reestablishment of CTXϕ lysogeny would presumably require a basal amount of RstR to remain present in the cell when LexA levels are restored. The role of the antirepressor RstC was not fully addressed in previous studies of this complex regulatory circuit. It appears from our data that the presence of high levels of RstC as a result of RS1 superinfection delays the reestablishment of lysogeny and that this eventually leads to the loss of CTX prophage in a proportion of V. cholerae cells, particularly under in vivo conditions. Since the structures of RstR proteins in classical and El Tor strains are significantly different (27, 28), the RstC protein is expected to inactivate the RstR protein of the El Tor type (RstRET), not the classical type (RstRClass). Consequently, RstC does not inflict the same effect on the lysogeny of the CTXClass prophage as on the CTXET prophage carried by El Tor or O139 strains. As expected, the classical biotype strains included in this study did not lose the CTX prophage under the same conditions that caused El Tor strains to become nontoxigenic (Table 1).
In addition, our results address several other issues related to lysogeny and induction of CTX phage. Early studies proposed that the induction of CTX prophage occurs inside the host intestine, based on observed transduction of recipient strains in an infant mouse model (3). It was also suggested that CTX prophage is maintained in toxigenic V. cholerae by continuous reinfection (36, 37), since toxigenic strains very rarely become nontoxigenic spontaneously. On the other hand, we previously showed that the CTXϕ genome was unstable in V. cholerae strains which lack the attachment sequence attRS, which is required for chromosomal integration of CTXϕ (23). Such strains carry the RF of the CTX phage genome, i.e., pCTX, which is rapidly lost when the host strain is subjected to passage in the ileal loops of adult rabbits (23). Similarly, in the present study, the toxigenic strains lost the CTX prophage when inoculated into rabbit ileal loops after superinfection with RS1ϕ or transformation with pRstC (Table 2). These findings provide further support for our model in which CTX phage is generally excised from the chromosome in a proportion of cells but the phage genome fails to efficiently reestablish lysogeny in the presence of excessive RstC. The extrachromosomal pCTX is then eliminated from the bacterial cells in the intestinal environment, as demonstrated previously for attRS-negative strains carrying pCTX (23). These dynamic behaviors of the CTXϕ and RS1ϕ prophages may explain, in part, the apparent amplification of these genomic elements under in vivo conditions (38).
Davis and Waldor proposed that production of RF of the CTX phage genome, i.e., pCTX, can occur from an array of prophages, without excision of the prophage from the chromosome (39). As a result, toxigenic strains with integrated CTX prophage appear to stably maintain the prophage. However, our results clearly indicate that the CTX prophage is physically excised from the chromosome in the presence of additional copies of RS1 or simply the presence of a plasmid expressing the anti-RstR repressor encoded by the rstC gene. We conclude that there is more than one mechanism for the induction and production of infectious phage particles from CTXϕ lysogens and that some of these may lead to excision of the CTXϕ prophage.
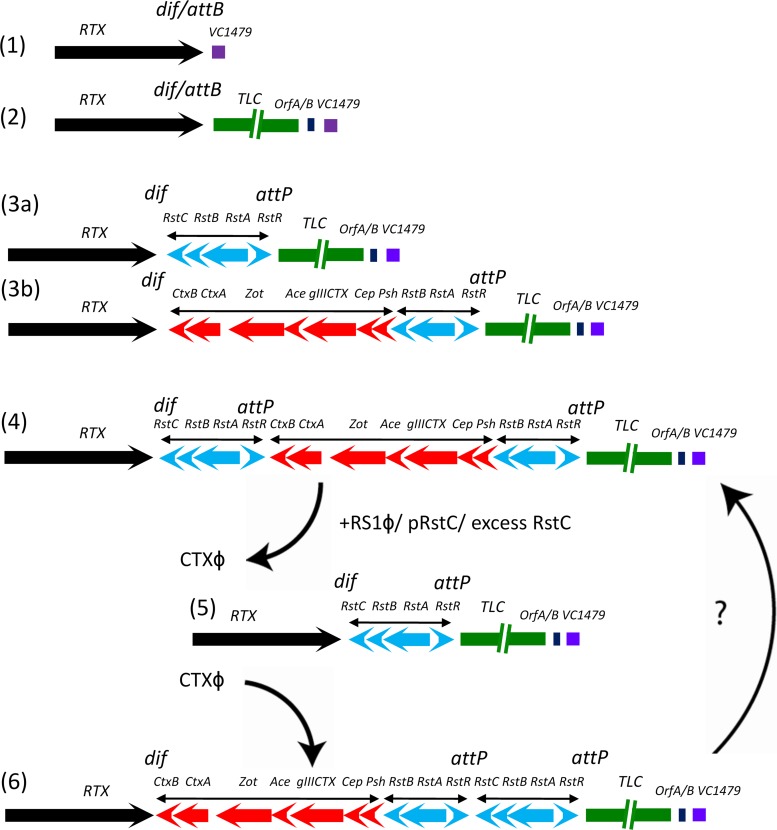
Recent epidemiological studies of cholera have described emerging variants of El Tor biotype strains which carry the CTXClass prophage, characterized by the presence of the rstRClass gene in the prophage, and these strains are now popularly known as “variant” 7th pandemic El Tor strains (18, 30). The elimination of CTX phage in the current studies resulted in the generation of a variety of nontoxigenic derivatives (Table 3). Because most of these were also susceptible to reinfection by CTX phage, we propose that these nontoxigenic derivatives can act as precursors of new variants of toxigenic strains by acquiring a CTX phage type that is the same or different from the one they originally carried. Although induction of infectious CTXClass phage has not been demonstrated, some of the nontoxigenic derivatives of El Tor biotype strains were able to acquire CTXClass prophage through chitin-induced transformation (see Table S4 in the supplemental material). We previously proposed the entire pathway involved in the toxigenic conversion of nontoxigenic progenitor strains by CTXϕ and the roles of various satellite phages (12). In the present study, we show that superinfection by RS1ϕ or overexpression of the antirepressor RstC in a cell which carries chromosomally integrated CTXϕ and RS1 islands can result in excision of CTXϕ; reintegration of CTXϕ may result in rearrangement of the CTX prophage and RS1 array (Fig. 3).
FIG 3.
Model of the integration of TLC (2) and RS1 (3a) or CTX (3b) into the attB site of the replication terminus of chromosome I and orientation of the genomic region commonly found in 7th pandemic El Tor strains (4). (5) Superinfection of RS1 or excess expression of the RstC antirepressor in a cell possessing genomic CTXϕ and RS1 islands can result in excision of CTXϕ. (6) Reintegration of CTXϕ may result in rearrangement, though reorganization between the genomes in panels 4 and 6 is unclear.
Taken together, the results of the present study suggest additional and important roles of the RS1 satellite phage genome in the evolution of V. cholerae. In this regard, it is interesting that Camilli and colleagues have established that at least one lytic vibriophage can inactivate phage-antagonistic chromosomal islands by encoding CRISPR elements that target the DNA in these elements (40). It seems that the battle for fitness between prophages extends well beyond superinfection immunity, which suggests that the competition between virulence-associated phages and chromosomal elements may be much more complex than previously appreciated.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded in part by National Institutes of Health grant 2RO1-GM068851-9, under a subagreement between the Harvard Medical School and the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B). The ICDDR,B gratefully acknowledges the following donors who provided unrestricted support: Australian Agency for International Development (AusAID), Government of the People's Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), and the Department for International Development, United Kingdom (DFID).
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01699-14.
REFERENCES
- 1.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science 272:1910–1914. 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- 4.Huber KE, Waldor MK. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656–659. 10.1038/nature00782 [DOI] [PubMed] [Google Scholar]
- 5.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. 2005. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19:559–566. 10.1016/j.molcel.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 6.Das B, Bischerour J, Val ME, Barre FX. 2010. Molecular keys of the tropism of integration of the cholera toxin phage. Proc. Natl. Acad. Sci. U. S. A. 107:4377–4382. 10.1073/pnas.0910212107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldor MK, Rubin EJ, Pearson GD, Kimsey H, Mekalanos JJ. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXΦ are encoded by region RS2. Mol. Microbiol. 24:917–926. 10.1046/j.1365-2958.1997.3911758.x [DOI] [PubMed] [Google Scholar]
- 8.Faruque SM, Asadulghani Kamruzzaman M, Nandi RK, Ghosh AN, Nair GB, Mekalanos JJ, Sack DA. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXphi. Infect. Immun. 70:163–170. 10.1128/IAI.70.1.163-170.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis BM, Kimsey HH, Kane AV, Waldor MK. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240–4249. 10.1093/emboj/cdf427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson GD, Woods A, Chiang SL, Mekalanos JJ. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. U. S. A. 90:3750–3754. 10.1073/pnas.90.8.3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin EJ, Lin W, Mekalanos JJ, Waldor MK. 1998. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28:1247–1254. 10.1046/j.1365-2958.1998.00889.x [DOI] [PubMed] [Google Scholar]
- 12.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. 2010. Satellite phage TLCϕ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467:982–985. 10.1038/nature09469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimsey HH, Waldor MK. 2009. Vibrio cholerae LexA coordinates CTX prophage gene expression. J. Bacteriol. 191:6788–6795. 10.1128/JB.00682-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinones M, Kimsey HH, Waldor MK. 2005. LexA cleavage is required for CTX prophage induction. Mol. Cell 17:291–300. 10.1016/j.molcel.2004.11.046 [DOI] [PubMed] [Google Scholar]
- 15.Davis BM, Lawson EH, Sandkvist M, Ali A, Sozhamannan S, Waldor MK. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science 288:333–335. 10.1126/science.288.5464.333 [DOI] [PubMed] [Google Scholar]
- 16.McLeod SM, Kimsey HH, Davis BM, Waldor MK. 2005. CTXphi and Vibrio cholerae: exploring a newly recognized type of phage-host cell relationship. Mol. Microbiol. 57:347–356. 10.1111/j.1365-2958.2005.04676.x [DOI] [PubMed] [Google Scholar]
- 17.Waldor MK, Friedman DI. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459–465. 10.1016/j.mib.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 18.Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296–3299. 10.1128/JCM.40.9.3296-3299.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque SM, Chowdhury N, Kamruzzaman M, Ahmad QS, Faruque AS, Salam MA, Ramamurthy T, Nair GB, Weintraub A, Sack DA. 2003. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 9:1116–1122. 10.3201/eid0909.020443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 21.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. 1986. Culture condition for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075–1083. 10.1111/j.1348-0421.1986.tb03037.x [DOI] [PubMed] [Google Scholar]
- 22.De SN, Chatterje DN. 1953. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J. Pathol. Bacteriol. 46:559–562 [DOI] [PubMed] [Google Scholar]
- 23.Faruque SM, Rahman MM, Hasan AK, Nair GB, Mekalanos JJ, Sack DA. 2001. Diminished diarrheal response to Vibrio cholerae strains carrying the replicative form of the CTXΦ genome instead of CTXΦ lysogens in adult rabbits. Infect. Immun. 69:6084–6090. 10.1128/IAI.69.10.6084-6090.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 105:8736–8741. 10.1073/pnas.0803281105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meibom KL, Blokesch M, Dolganov NA, Wu C, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. 10.1126/science.1120096 [DOI] [PubMed] [Google Scholar]
- 26.Udden SMN, Zahid MSH, Biswas K, Ahmad QS, Cravioto A, Nair GB, Mekalanos JJ, Faruque SM. 2008. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc. Natl. Acad. Sci. U. S. A. 23:11951–11958. 10.1073/pnas.0805560105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimsey HH, Waldor MK. 1998. CTXΦ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. U. S. A. 95:7035–7039. 10.1073/pnas.95.12.7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis BM, Kimsey HH, Chang W, Waldor MK. 1999. The Vibrio cholerae O139 Calcutta bacteriophage CTXΦ is infectious and encodes a novel repressor. J. Bacteriol. 181:6779–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis BM, Moyer KE, Boyd EF, Waldor MK. 2000. CTX prophages in classical biotype of Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992–6998. 10.1128/JB.182.24.6992-6998.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansaruzzaman M, Bhuiyan NA, Nair GB, Sack DA, Lucas M, Deen JL, Ampuero JC, Chaignat CL. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10:2057–2059. 10.3201/eid1011.040682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, de Wilde M. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557. 10.1038/306551a0 [DOI] [PubMed] [Google Scholar]
- 32.Nguyen DT, Nguyen BM, Tran HH, Ngo TC, Le TH, Nguyen HT, Albert MJ, Iwami M, Ehara M. 2008. Filamentous vibriophage fs2 encoding the rstC gene integrates into the same chromosomal region as the CTX phage. FEMS Microbiol. Lett. 284:225–230. 10.1111/j.1574-6968.2008.01200.x [DOI] [PubMed] [Google Scholar]
- 33.Lemire S, Figueroa-Bossi N, Bossi L. 2011. Bacteriophage crosstalk: coordination of prophage induction by trans-acting antirepressors. PLoS Genet. 7:e1002149. 10.1371/journal.pgen.1002149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochschild A, Lewis M. 2009. The bacteriophage lambda CI protein finds an asymmetric solution. Curr. Opin. Struct. Biol. 19:79–86. 10.1016/j.sbi.2008.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickels BE. 2009. A new twist on a classic paradigm: illumination of a genetic switch in Vibrio cholerae phage CTX Phi. J. Bacteriol. 191:6779–6781. 10.1128/JB.01150-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faruque SM, Asadulghani Saha MN, Alim ARMA, Albert MJ, Islam KMN, Mekalanos JJ. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect. Immun. 66:5819–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazar S, Waldor MK. 1998. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect. Immun. 66:394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mekalanos JJ. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253–263. 10.1016/0092-8674(83)90228-3 [DOI] [PubMed] [Google Scholar]
- 39.Davis BM, Waldor MK. 2000. CTXΦ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. U. S. A. 97:8572–8577. 10.1073/pnas.140109997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. 10.1038/nature11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.