Abstract
The polysaccharide capsule surrounding Streptococcus pneumoniae is essential for virulence. Recently, Streptococcus mitis, a human commensal and a close relative of S. pneumoniae, was also shown to have a capsule. In this study, the S. mitis type strain switched capsule by acquisition of the serotype 4 capsule locus of S. pneumoniae TIGR4, following induction of competence for natural transformation. Comparison of the wild type with the capsule-switching mutant and with a capsule deletion mutant showed that the capsule protected S. mitis against phagocytosis by RAW 264.7 macrophages. This effect was enhanced in the S. mitis strain expressing the S. pneumoniae capsule, which showed, in addition, increased resistance against early clearance in a mouse model of lung infection. Expression of both capsules also favored survival in human blood, and the effect was again more pronounced for the capsule-switching mutant. S. mitis survival in horse blood or in a mouse model of bacteremia was not significantly different between the wild type and the mutant strains. In all models, S. pneumoniae TIGR4 showed higher rates of survival than the S. mitis type strain or the capsule-switching mutant, except in the lung model, in which significant differences between S. pneumoniae TIGR4 and the capsule-switching mutant were not observed. Thus, we identified conditions that showed a protective function for the capsule in S. mitis. Under such conditions, S. mitis resistance to clearance could be enhanced by capsule switching to serotype 4, but it was enhanced to levels lower than those for the virulent strain S. pneumoniae TIGR4.
INTRODUCTION
Eighty-five years have passed since Griffith's publication of the “transforming principle” (1). In his studies, Streptococcus pneumoniae underwent in vivo transformation, changing from an avirulent strain devoid of a capsule to a virulent encapsulated strain (1). More than 90 S. pneumoniae capsular serotypes have now been identified, and most of these are encoded by a single genetic locus varying from 10 kb to 30 kb in length (2). Serotype variants of single clones are not uncommon and are thought to arise from natural transformation events. In such variants, the acquisition of DNA segments of more than 39 kb involving the capsule locus region has been reported (3–5).
A close relative of S. pneumoniae is Streptococcus mitis, a predominantly commensal that colonizes various oral niches, including the teeth, tongue, cheeks, subgingival sites, and tonsils, as well as the nasopharynx (6), where it interacts with several other bacteria (7). The common presence of S. mitis in bronchoalveolar lavage fluid (BALF) indicates that S. mitis is, in addition, an important component of the human lung microbiome (8). S. pneumoniae is also a nasopharyngeal commensal but, unlike S. mitis, has a high pathogenic potential and is responsible for an estimated 14.5 million infections and 1 million deaths in children under 5 years of age annually (9). Most of the deaths caused by S. pneumoniae are a result of pneumonia, bacteremia, or meningitis, and in virtually all cases, these diseases are caused by encapsulated strains. In contrast, S. mitis is only occasionally associated with diseases (10–13). Its potential as a probiotic microorganism has been previously reported (14), but concerns about its pathogenic potential have been raised (15). Despite the ubiquitous prevalence of S. mitis as a human colonizer, few studies have addressed the molecular mechanisms involved in S. mitis commensalism or pathogenicity (12).
The S. pneumoniae capsule plays a significant role in colonization and invasive infections by preventing both phagocytosis and entrapment in the respiratory tract mucus (16–18). Recently, capsule loci with sequences indicative of different capsule types were identified in strains of S. mitis (19–21). Such variation in capsule types may result from adaptations to immune responses and other selective environmental pressures, as suggested for S. pneumoniae (22). In S. pneumoniae, variations in pathogenic potential are observed among different serotypes. Some capsule types, independently of their genetic background, are thought to provide higher resistance to phagocytosis (23) or may be expressed at levels that affect the accessibility of surface proteins, therefore affecting the ability of S. pneumoniae to colonize and cause disease (24). In S. mitis, the role of the capsule remains mostly unknown, except for its negative effect on biofilm formation, demonstrated in vitro (20).
In S. pneumoniae and S. mitis, the capsule locus is flanked by the dexB and aliA genes. In addition, the cpsABCDE genes in the 5′ end of the capsule locus are conserved in both species (20). This raises the question of whether S. mitis and S. pneumoniae may exchange capsules. For S. mitis, acquisition of the S. pneumoniae capsule would have the potential to alter its virulence. In S. pneumoniae, intraspecies capsule switch has occasionally been observed both before and after the introduction of the 7-valent pneumococcal vaccine (3–5). This most often involves the exchange of long stretches of DNA, an expected finding, given the large sets of specific enzymes usually required for expression of different capsule structures. The role of S. mitis as a reservoir of genetic diversity for S. pneumoniae, including antibiotic resistance determinants, is well documented (25–27). It remains to be determined whether the capsule locus may also provide a pool of sequences that may be exchanged between S. mitis and S. pneumoniae. In clinical isolates, proper assessment is still limited by the high variability and relatively few sequenced genomes of S. mitis strains.
The first aim of the present study was to examine whether exposure to whole DNA from the virulent strain S. pneumoniae TIGR4 may lead to capsule switching in competent S. mitis. The second aim was to investigate the role of the native capsule or the capsule from S. pneumoniae in (i) S. mitis NCTC 12261T resistance to phagocytosis in vitro, (ii) S. mitis NCTC 12261T resistance to clearance in mouse models of early lung infection and bacteremia, and (iii) the ability of S. mitis NCTC 12261T to grow ex vivo in blood. We found that the TIGR4 capsule locus could be taken up and could be expressed by the S. mitis type strain. Both the native capsule and the TIGR4 capsule in S. mitis provided resistance to phagocytosis in vitro and resistance to clearance in a mouse model of lung infection, with higher resistance levels being conferred by the TIGR4 capsule. In the bacteremia model, no significant differences were observed between the wild type and mutants. Survival in human whole blood was, however, favored by the TIGR4 capsule and was significantly reduced or abolished in a capsule deletion mutant.
MATERIALS AND METHODS
Bacteria and media.
All bacteria used and their characteristics are listed in Table 1. The bacteria were stored at −80°C in Todd-Hewitt broth (THB; Becton, Dickinson and Company, Le Pont de Claix, France) supplemented with 15% glycerol. Precultures used in the transformation experiments described below were made from overnight (ON) cultures incubated at 37°C in a 5% CO2-supplemented atmosphere, diluted 1:25 in THB, and grown until the cultures reached an absorbance at 600 nm (optical density at 600 nm [OD600]; Biophotometer; Eppendorf) of 0.3, before storage at −80°C in 15% glycerol. Frozen stocks used for bacterial growth in human serum, human and horse whole blood, and the mouse challenge experiments were made by growing the strains in THB (Becton, Dickinson and Company, Le Pont de Claix, France) supplemented with 5% yeast extract (Becton, Dickinson and Company, Le Pont de Claix, France) to an OD600 of approximately 0.5. The medium used in the transformation assays of S. mitis and S. pneumoniae was THB without and with 0.2% yeast extract (Becton, Dickinson and Company), respectively. For growth on agar plates, blood agar base no. 2 (Oxoid, Hampshire, England) supplemented with 5% defibrinated sheep blood (TCS Biosciences Ltd., Buckingham, United Kingdom) was used. Incubation was performed at 37°C under 5% CO2-supplemented atmospheric conditions ON. Selective levels of antibiotics were 500 μg ml−1 kanamycin and 20 μg ml−1 erythromycin.
TABLE 1.
Strains used in this study
| Strain | Description | Sourcea |
|---|---|---|
| S. mitis | ||
| CCUG31611T | Wild-type S. mitis biovar 1 type strain, which corresponds to NCTC 12261; encapsulated, transformable strain; sequenced genome; Kms Erms | CCUG |
| MI016 Δcps | CCUG31611T cps::km Kmr | 20 |
| TIGR4cps (MI030) | CCUG31611T Ω TIGR4 cps locus; Kmr Ermr | This study |
| MI067 | CCUG31611T Ω TIGR4 cps locus; Kmr Ermr; similar to MI030, but from independent expts | |
| MI051 ΔpabB | CCUG31611T pabB::spec Specr | This study |
| SK575 | CCUG62644; cps locus positive | |
| SK579 | CCUG62641; cps locus positive | CCUG |
| SK597 | CCUG55094; cps locus positive, lytA positive, ply positive | |
| S. pneumoniae | ||
| TIGR4 | Wild-type serotype 4, transformable strain; sequenced genome | 45 |
| TIGR4 Δcps (SP011) | TIGR4 cps::erm Ermr | This study |
| TIGR4 Δcps (FP23) | TIGR4 cps::km Kmr | 46 |
| SP026.1 | Insertion of kanamycin resistance cassette downstream of dexB; Kmr | This study |
| SP029.1 | Insertion of erythromycin resistance cassette upstream of aliA; Ermr | This study |
| SP033 | SP029.1 transformed with DNA from SP026.1; Kmr Ermr | This study |
| ΔpabB | TIGR4 pabB::km Kmr | 16 |
| D39 | Wild-type serotype 2, transformable strain; sequenced genome; NCTC 7466 | NCTC |
| R36A | D39 derivative, highly passaged spontaneous unencapsulated strain of NCTC 10319 | NCTC |
| Serotype 1 | Corresponds to sequence type 306 | Clinical isolate, Norwegian Institute of Public Health |
CCUG, culture collection, University of Göteborg; NCTC, National Collection of Type Cultures.
Construction of mutants.
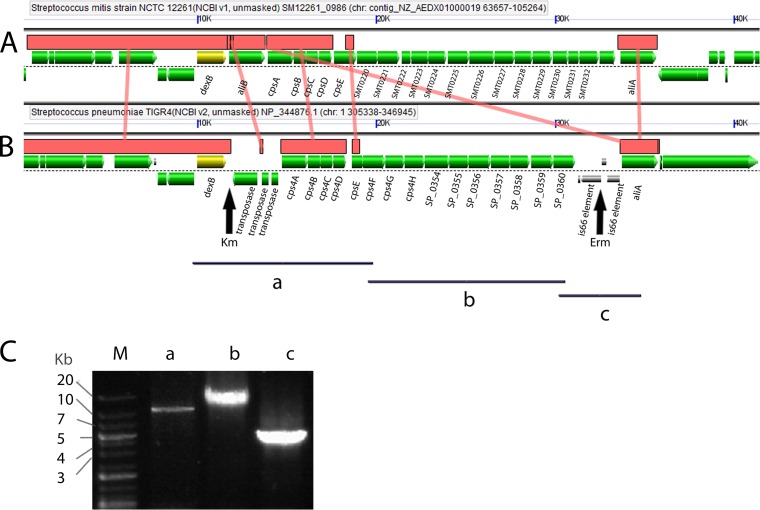
The capsule locus deletion strategy for S. pneumoniae was performed as described previously for S. mitis (20) following a procedure adapted from Pearce et al. (28), using the PCR ligation mutagenesis strategy (29). Primer pairs are listed in Table 2 and were constructed on the basis of the genome sequences available for S. pneumoniae TIGR4 (GenBank accession number NC_003028.3) and S. mitis NCTC 12261T (GenBank accession number NZ_AEDX00000000.1). The PCR ligation mutagenesis strategy was also used to investigate the possibility of capsule switching in competent S. mitis (Fig. 1). Primer pairs designed to insert the markers within the capsule flanking intergenic regions of S. pneumoniae were used. For the introduction of the kanamycin resistance marker upstream of the capsule locus, we used primer pairs FP585-FP586 and FP587-FP588, and for the introduction of the erythromycin resistance cassette downstream of the capsule locus, we used primer pairs FP593-FP594 and FP595-FP596. Ligation and purification of the PCR products were performed using T4 DNA ligase (Fermentas) and a PCR purification kit (Qiagen), respectively. Insertion of the antibiotic resistance markers in the capsule locus flanking regions followed two steps. The kanamycin or erythromycin resistance marker was first inserted into the wild type, creating mutants SP26.1 and SP29.1, respectively. This was followed by the second step, in which DNA from SP29.1 was used to transform the SP26.1 mutant. The resultant kanamycin and erythromycin doubly resistant mutant, SP033, was used as the DNA donor in the capsule switch experiments.
TABLE 2.
Primers used in the study
| Primer | Sequencea | Purpose |
|---|---|---|
| FP415 | CGTGAGGCTCTTGAAAAAGG | S. pneumoniae SP011 Δcps construction |
| FP416 | AGGCGCGCCGTCAGGCCTGCACTAAGGAC | |
| FP417 | AGGCCGGCCAGGAGGAGCTCCATGAATGA | |
| FP418 | GCTTTGGCAAAATCATCTCC | |
| FP585 | GACCAAGAATACCGCGAAAA | SP026.1 construction |
| FP586 | AGGCGCGCCTCTGTCTTTTATACAGTCCT | |
| FP587 | AGGCCGGCCCTTTCATTCTTGAAATTCAATTAAC | |
| FP588 | TTTATCGTGGACGTGGTCAA | |
| FP593 | CATGAACGTTTGGGATTGAA | SP029.1 construction |
| FP594 | AGGCGCGCCCGCAAAAACAAGCCAAAAAT | |
| FP595 | AGGCCGGCCGACGCGCTTACGATATTCAT | |
| FP596 | GCTTTGGCAAAATCATCTCC | |
| FP617 | AGCCAGAACCATTCAGGTTG | Verification of capsule switch event in S. mitis TIGR4cps |
| FP618 | CCTGCCGCCATATAAGATTG | |
| FP623 | GTGGGAACACGTCCAGAGAT | |
| FP624 | TTGTGCCAAGCTAACAGCAG | |
| FP643 | GAAAAATGGTGGCACAATGC | |
| FP644 | GACTGCCTTGTCTTGAAGTTCC | |
| FP789 | CGCCAATTCTTTGAGAATCA | Screening of region taken up by S. mitis MI030 and MI067 |
| FP790 | CTAGCACTGCAAGCGAATCA | |
| FP795 | TAGTCTAAATTTTTTAAAACAATG | |
| FP796 | TGCCTGTTGTGGCATTTGC | |
| FP805 | AGGACCAAAAGGAGACA | |
| FP806 | TGAGCGAATCCTTGATAACA | |
| FP803 | TGATTGCCATTAATGCTAG | |
| FP804 | TTGCCCATGCCTTCTGGG | |
| FP791 | GGTTAATCCAAATTACGAGATG | |
| FP792 | CCAAGACCTGTTTTTCAAGT | |
| FP793 | CTCCAGAGATTGCCAAGTTC | |
| FP794 | CCAAGACCTGTTTTTCAAGT | |
| FP787 | AATGATAACGCAGGCAAACC | |
| FP788 | TGTTTCAGGAGCAGCAACAC | |
| FP797 | ATATCCGTTTCACGCACCTC | |
| FP798 | CAATGCTAACTGCACCGAAA | |
| FP799 | GAGCTGTTGGGAATGGTGTT | |
| FP800 | TGAATCCCTCCGAAGAAATG | |
| FP725 | AAGGCAACTGGAATCACAGC | S. mitis MI051 ΔpabB construction |
| FP726 | AGGCGCGCCTGTATCTCTCGCCCAAAGC | |
| FP727 | AGGCCGGCCCTACACACCGCCTATCCGACT | |
| FP728 | AAGACATGAGGTCCATCTGGT | |
| FP001 | AGGCGCGCCGTTTGATTTTTAATG | Amplification of the Km resistance cassette |
| FP068 | AGGCCGGCCTAGGTACTAAAACAATTCATCCAGTA | |
| FP015 | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT | Amplification of the Erm resistance cassette |
| FP016 | GGCCGGCCAGTCGGCAGCGACTCATAGAAT | |
| FP676 | AGGCGCGCCGTAACGTGACTGGCAAGAGATATT | Amplification of the Sp resistance cassette |
| FP677 | AGGCCGGCCAAATAATAAAACAAAAAAATT |
Primers are listed 5′ to 3′; restriction sites are underlined.
FIG 1.
Schematic representation of the S. mitis and S. pneumoniae capsule locus and flanking regions. The interspecies homologous regions between S. mitis NCTC 12261T (A) and S. pneumoniae TIGR4 (B) are shown as horizontal brown bars connected by vertical brown lines. The alignment and graphical representation were obtained using the GeVo program, available at https://genomevolution.org/CoGe/GEvo.pl. The dexB gene, shown in yellow, was chosen as the reference gene for alignment. The vertical arrows show the sites of insertion of the erythromycin (Erm) and kanamycin (Km) resistance markers in S. pneumoniae TIGR4, used as the donor of the capsule locus. Black bars, the PCR-amplified regions used to confirm the capsule switch event leading to the insertion of the S. pneumoniae capsule locus into the S. mitis genome. The primer pair used for amplification of fragment a was FP643-FP618, that used for amplification of fragment b was FP617-FP624, and that used for amplification of fragment c was FP623-FP644. (C) Agarose gel electrophoresis imaging of amplicons a, b, and c. Lane M, DNA ladder molecular size marker.
For construction of the S. mitis ΔpabB mutant, the ligation mutagenesis strategy described above was used. The pabB flanking regions were amplified using primer pairs FP725-FP726 and FP727-FP728, and the spectinomycin resistance marker (a kind gift from D. Morrison) was amplified using primer pair FP676-FP677.
Transformation.
Transformation of S. pneumoniae was done as described by Bricker and Camilli (30). Briefly we used 9 ml of THB supplemented with yeast extract, 1 ml 5% glycine (Calbiochem), and 50 μl 1.0 M HCl (Sigma), to which 200 μl of preculture was added. The bacteria were grown at 37°C in a 5% CO2-supplemented atmosphere until the OD600 reached 0.03. At this density, the following substrates were added to the cultures (final concentrations): 10 mM NaOH (Sigma), 0.02% bovine serum albumin (Sigma), 1 mM CaCl2 (Merck), and 100 ng ml−1 S. pneumoniae TIGR4 competence-stimulating peptide (CSP; EMRISRIILDFLFLRKK; GenScript). Then, 1 ml of the mixtures was transferred to 15-ml Falcon tubes, and DNA was added. For ligation mutagenesis, 5 μl of the purified final product from the PCR was used.
Transformation of S. mitis was carried out as previously described, with slight modifications (5). Briefly, precultures of S. mitis were diluted 1:10 in TSB. Competence-stimulating peptide for S. mitis CCUG31611T (EIRQTHNIFFNFFKRR; GenScript) was added at a final concentration of 125 nM. For the capsule switch experiments (Table 3), the bacteria were allowed to grow to an OD600 of 0.03 before addition of chromosomal DNA at a concentration of 20 to 30 μg per ml culture and CSP at a final concentration of 250 nM. The cultures were incubated under aerobic conditions at 37°C for 4 h. Mutants were selected on blood agar plates supplemented with the appropriate antibiotics.
TABLE 3.
S. mitis transformation data for independent experimentsb
Colonies selected for capsule switch characterization (S. mitis MI030 and MI067).
The recipient strain was S. mitis NCTC 12261T, and the DNA source was S. pneumoniae SP033. ND, not determined; Erm, erythromycin; Km, kanamycin.
The correct insertion sites of the antibiotic resistance markers in the S. pneumoniae and S. mitis mutants were verified by PCR using primer pairs complementary to sequences within the antibiotic resistance genes combined with primers in regions up- or downstream from the insertion. To verify the capsule switch event in S. mitis, DNA from the S. mitis MI030 mutant was used in PCRs using primers amplifying overlapping sequences of the S. pneumoniae capsule locus. For this purpose, Accutaq LA DNA polymerase (Sigma) was used according to the manufacturer's recommendations along with primer pairs FP643-FP618, FP617-FP624, and FP623-FP644, as illustrated in Fig. 1. To verify the extension of genetic exchange in the vicinity of the genes flanking dexB and aliA, primers were constructed to amplify genes upstream of dexB and genes downstream of aliA in S. pneumoniae. For the region upstream of and within dexB, we used the following primer pairs: FP789-FP790 (SP_0329), FP795-FP796 (SP_0338), FP805-FP806 (SP_0340, luxS), FP803-FP804 (SP_0341), FP791-FP792 (SP_0342, dexB), and FP793-FP794 (SP_0342, dexB). For the region downstream of aliA, we used the following primer pairs: FP787-FP788 (SP_0368), FP797-FP798 (SP_0379), and FP799-FP800 (SP_0394).
Capsular test.
The Neufeld capsular reaction test was performed according to the manufacturer's recommendations. Briefly, bacteria were grown on blood agar at 37°C in a 5% CO2-supplemented atmosphere ON. For the test, bacteria were transferred and mixed with 2.5 μl phosphate-buffered saline (PBS) and 2.5 μl pool A antiserum (Statens Serum Institute, Denmark) using 1-μl disposable loops. For the control, 5 μl PBS was used. Images were acquired using a Zeiss Axiovert Observer Z1 microscope coupled to an Axiocam Mrm camera (Zeiss). We also performed a dot blot assay using serial dilutions of the bacterial strains and reaction of the strains with anti-type 4 serum (Statens Serum Institute) and performed Ponceau staining as a control. The blot was scanned, and capsule data were normalized for Ponceau staining values.
Optochin test.
Bacteria were grown in TSB at 37°C in a 5% CO2-supplemented atmosphere ON. The bacteria were then streaked on blood agar plates using 1-μl disposable loops. Optochin discs (Fluka Analytical) were placed in the center of the streak, and the plates were incubated at 37°C in a 5% CO2-supplemented atmosphere ON.
Phagocytosis assay.
Cells of the murine macrophage cell line RAW 264.7 were grown in RPMI medium (Lonza, Verviers, Belgium) with 10% heat-inactivated fetal calf serum (FCS; Lonza or Sigma) and 100 U ml−1 of streptomycin and penicillin (Lonza or Sigma). They were grown to 90% confluence in 12-well multidishes (Cornerstone), washed 2 times with prewarmed RPMI or PBS, pH 7.4, and inoculated with bacteria at a multiplicity of infection (MOI) of 10:1. The bacteria were incubated with the macrophages for 1 h at 37°C in a 5% CO2-supplemented atmosphere by a modification of the procedure described by Peppoloni et al. (31). To measure the amount of bacteria within the macrophages, the cells were washed with RPMI, followed by incubation with 1 ml RPMI, 1% FCS, 10 μg ml−1 penicillin (Sigma), and 200 μg ml−1 gentamicin (Sigma) for an additional 45 min at 37°C in a 5% CO2-supplemented atmosphere. This procedure killed 100% of the S. mitis organisms cultured in the absence of macrophages (data not shown). After washing the cells twice with PBS, lysis of the macrophages was performed using 1 ml water, followed by serial dilution and plating. To estimate bacterial killing by the macrophages over time, the number of CFU was also determined for the next 2 h at 1-h intervals following the initial measurement of internalized bacteria. At each time point, the wells were washed two times with PBS, followed by lysis and plating. Phagocytosis by RAW 264.7 macrophages was also investigated using flow cytometry, as previously described (32). This method provides information on the association of streptococci with macrophages. Briefly, the bacteria were labeled with 6-carboxyfluorescein succinimidyl ester (FAMSE; Molecular Probes) for 60 min at 37°C and then washed with PBS until no residual FAMSE was visible in the supernatant. The bacteria were frozen at −80°C in 10% glycerol until use. The bacteria were incubated with RAW 264.7 macrophages at an MOI of 10:1 at 37°C in a 5% CO2-supplemented atmosphere for 1 h. The cells were washed with PBS and trypsinized for 7 min, followed by addition of FBS and 100 μl 4% paraformaldehyde (PFA) in PBS. The samples were kept in the dark at 4°C until analysis using a FACSCalibur flow cytometer (BD Biosciences). The proportion of macrophages associated with bacteria was determined in the flow cytometer by analyzing a minimum of 25,000 cells.
For transmission electron microscopy (TEM), the bacteria were grown as described above. RAW 264.7 macrophages were grown in 6-cm petri dishes as described above at an MOI of 10:1. The bacteria were incubated with the macrophages for 1 h at 37°C in a 5% CO2-supplemented atmosphere. The cells were then fixed by adding 2% glutaraldehyde (GA; Electron Microscopy Sciences, PA) in 100 mM phosphate buffer, pH 7.4, to the culture medium at a 1:1 volume ratio. After 5 min, the supernatant was discarded and replaced with fresh 1% GA in phosphate buffer. After overnight incubation at 4°C, the cells were washed with 100 mM cacodylate buffer, postfixed with 2% OsO4 (EMS, PA) solution containing 1.5% potassium ferricyanide for 1 h, and stained en block with 1.5% aqueous uranyl acetate for 30 min. The cells were then dehydrated using a graded ethanol series and propylene oxide and embedded in epoxy resin (Sigma-Aldrich, St. Louis, MO). Ultrathin sections of 60 to 70 nm were cut with an ultramicrotome (Ultracut UCT; Leica, Wetzlar, Germany), stained with 0.2% lead citrate (Taab, Berks, England) in 0.1 M NaOH for 20 s, and examined in a Philips CM100 transmission electron microscope.
Mouse model of early lung infection.
The mouse model of early lung infection was performed as previously described (24). Briefly, outbred female CD1 mice (age, 6 weeks; Charles River Breeders) were infected intranasally (i.n.) with a bacterial inoculum corresponding to 5 × 106 CFU in 50 μl PBS while they were under general halothane anesthesia (Zeneca). Bronchoalveolar lavage fluid (BALF) was collected and used to determine the number of bacterial CFU after dilution and plating on blood agar.
Mouse model of bacteremia.
Female BALB/c mice (age, 8 weeks; Charles River Italia, Lecco, Italy) were infected intravenously with the S. mitis wild type and capsule mutants and S. pneumoniae TIGR4 and a capsule mutant (FP23) at amounts corresponding to 5 × 106 CFU per mouse. The experiment was repeated with a 10-fold higher dose (5 × 107 CFU per mouse) for the S. mitis wild type and capsule mutants (33). At 30 min, blood samples were collected from the submandibular vein of the mice. To prevent blood coagulation, 100 U ml−1 of heparin (MS Pharma, Milan, Italy) was added.
Ex vivo growth in blood.
Human blood and serum were obtained with written consent from healthy human volunteers under ethical approval granted by the local University College London ethics committee (application 3076/001). The blood was prevented from clotting using a final concentration of 0.1 M sodium citrate. Growth in blood and serum was investigated using an inoculum of bacteria corresponding to approximately 5 × 106 CFU per ml. The samples were incubated at 37°C for up to 6 h in a 5% CO2-supplemented atmosphere before serial dilution and plating on blood agar plates.
Ethical statements.
Experiments with the mouse model of bacteremia were performed according to institutional guidelines, authorized by the Comitato Etico of the Azienda Ospedaliera Universitaria Senese, and approved by the Ministry of Health of Italy. Experiments with the mouse model of lung infection were approved by the UCL Biological Services Ethical Committee and the United Kingdom Home Office (project license PPL70/6510) and were performed according to United Kingdom national guidelines for animal use and care under a United Kingdom Home Office license.
Statistical analysis.
For the in vitro assays, comparisons were conducted using one-way analysis of variance (ANOVA), followed by the Holm-Sidak test. For the bacteremia model, one-way analysis of variance on ranks followed by the Kruskal-Wallis test was used. For the lung infection assays and growth in human whole blood, ANOVA followed by Dunnett's test with comparison against the results for the S. mitis wild type or S. pneumoniae TIGR4 was used. Significance was set at a P value of <0.05.
RESULTS
S. mitis capsule switching to serotype 4.
S. mitis acquisition of genetic material from related streptococci has recently been suggested, but the extent and the location of the recombination events have so far been analyzed in only a few strains whose genomes have been sequenced (34). We wanted to experimentally test whether the S. mitis native capsule could be replaced by the S. pneumoniae TIGR4 capsule during S. mitis competence for natural transformation. S. pneumoniae TIGR4 was chosen because it is a well-characterized strain and highly invasive in mice and humans. To track this event, we introduced kanamycin and erythromycin resistance cassettes in the S. pneumoniae TIGR4 capsule flanking regions as markers of recombination (Fig. 1; Table 1). The kanamycin resistance cassette was inserted immediately downstream of the dexB gene, while the erythromycin resistance cassette was inserted upstream of aliA. Transformants were selected on agar plates containing the appropriate antibiotics. S. pneumoniae SP033 DNA with the antibiotic resistance markers in the flanking regions was used to select candidate mutants for capsule switch in S. mitis (Table 1). Transformation resulting in the incorporation of both antibiotic resistance markers yielded positive results in four of five experiments (Table 3). A control experiment using DNA from an S. mitis strain with antibiotic resistance markers in the flanking regions showed that transformation levels were at least 56-fold higher (data not shown) than those with DNA from S. pneumoniae (Table 3). Two S. mitis mutants (Table 3) resistant to the combination of antibiotics (MI030 and MI067) were randomly selected from experiments 1 and 5 for (i) characterization of the capsule flanking regions that were incorporated by S. mitis into its genome, (ii) assessment of the growth rate, and (iii) assessment of TIGR4 capsule expression. PCR amplification using primers that were designed to amplify S. pneumoniae but not S. mitis sequences in the genes upstream and downstream of the dexB-cps-aliA region (Table 2) was conducted. The genes upstream of dexB screened were SP_0329, SP_0338, and SP_0340 (luxS), and the genes downstream of aliA screened were SP_0368, SP_0379, and SP_0394. The results showed that these genes were present in the S. pneumoniae TIGR4 positive control but not in S. mitis capsule-switching mutant MI030 or MI067. To verify the incorporation of the S. pneumoniae capsule locus, primers designed to amplify overlapping regions were used. The results showed that the whole locus of S. pneumoniae TIGR4 had been transferred to the S. mitis type strain (Fig. 1).
To verify whether the recombination event leading to the transfer of the TIGR4 capsule to S. mitis was accompanied by the production of a type 4 capsule, the Neufeld capsular reaction test was used. Similar to the findings for S. pneumoniae TIGR4, the MI030 and MI067 capsule mutants showed strong agglutination with pool A antiserum, whereas no agglutination was observed with the S. mitis wild type. Images of the capsular reaction tests using the S. mitis wild type, S. mitis MI030, and S. pneumoniae TIGR4 are shown in Fig. 2A to C, respectively. To confirm the species identity of the recombinant S. mitis strain, the optochin test was used. As expected, the test showed a clear inhibition zone around the optochin discs for S. pneumoniae but not for S. mitis expressing the native or the TIGR4 capsules (Fig. 2D). The dot blot assay using anti-type 4 serum confirmed the expression of the TIGR4 capsule by MI030 (data not shown). S. mitis MI030 showed values that were approximately 13% lower than those for S. pneumoniae TIGR4. This indicates that S. mitis MI030 may produce slightly less of the serotype 4 capsule or that anti-type 4 serum reacts more with S. pneumoniae TIGR4, because whole S. pneumoniae is used for antiserum production (Statens Serum Institute, Denmark).
FIG 2.
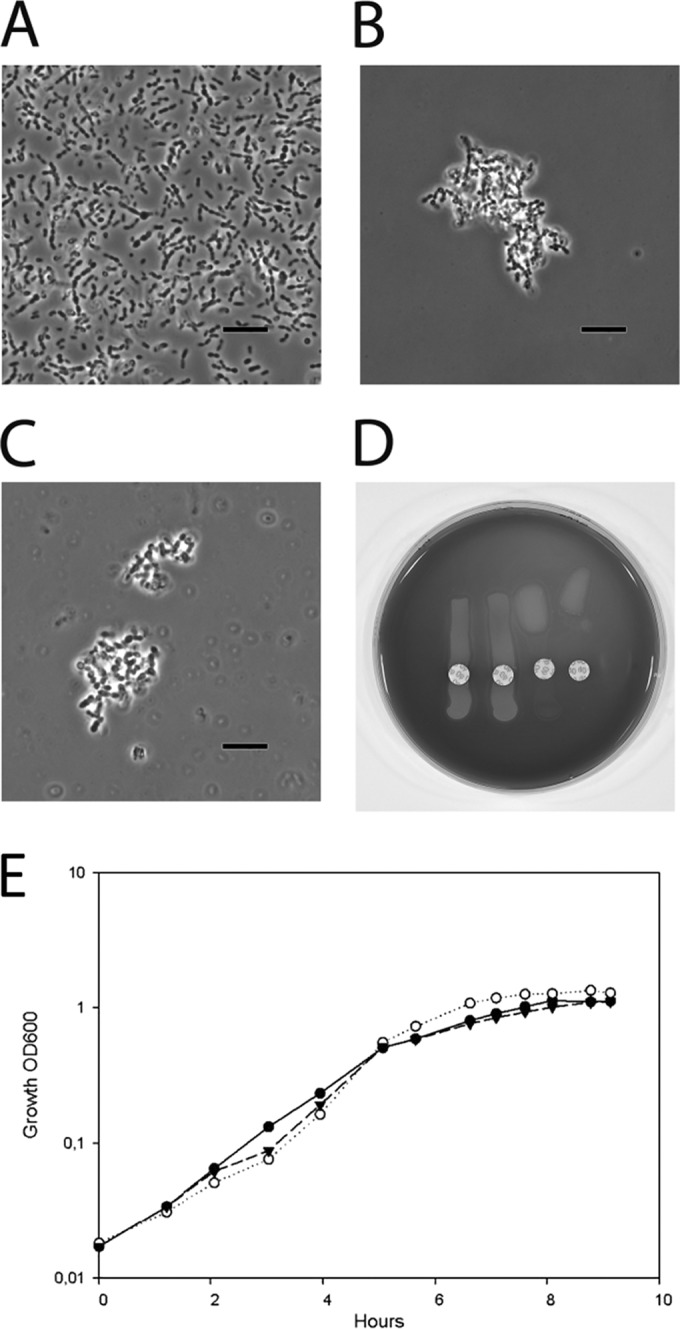
TIGR4 capsule expression by S. mitis. The capsule reaction test (Neufeld test) with pool A antiserum (Statens Serum Institute, Denmark) and the optochin sensitivity test were performed. (A to C) Lack of agglutination of the S. mitis wild type (A), agglutination of the S. mitis TIGR4cps isogenic mutant that acquired the type 4 capsule (B), and agglutination of S. pneumoniae TIGR4 serotype 4 (C). The images in panels A to C were taken with phase-contrast and oil immersion. Bars, 5 μm. (D) Optochin sensitivity test confirming the species identification of (from the left) the S. mitis wild type, S. mitis TIGR4cps, the S. pneumoniae TIGR4 wild type, and S. pneumoniae SP033 (TIGR4 capsule donor). (E) Growth curve of the S. mitis wild type (black circles), S. mitis Δcps (MI016; white circles), and S. mitis TIGR4cps (MI030; black inverted triangles).
To investigate whether capsule deletion or capsule switch could have a deleterious effect on growth, we compared the growth rate of MI030 and MI067 to that of the wild-type strain and S. mitis Δcps during their exponential phases in TSB medium. The average doubling time was 60 min (standard deviation [SD], 3.8 min) for S. mitis, 49 min (SD, 4.1 min) for S. mitis Δcps, 62 min (SD, 12.2 min) for MI030, and 70 min (SD, 10.8 min) for MI067. The results were from 3 to 4 independent experiments. Figure 2E shows the growth curves of the wild type and MI030 in a representative experiment. Comparison using one-way ANOVA did not reveal significant differences in growth rates between the S. mitis wild type and the mutants.
Taken together, the results revealed that the entire S. pneumoniae TIGR4 capsule locus was taken up and expressed by the S. mitis type strain, without compromising growth in TSB. In the experiments performed to characterize the protective role of the S. mitis capsule, described below, MI030 was included to initially address the possibility that capsule type may impact S. mitis resistance to phagocytosis or clearance from lungs and blood.
The native and the TIGR4 capsules in S. mitis provide different levels of resistance against phagocytosis.
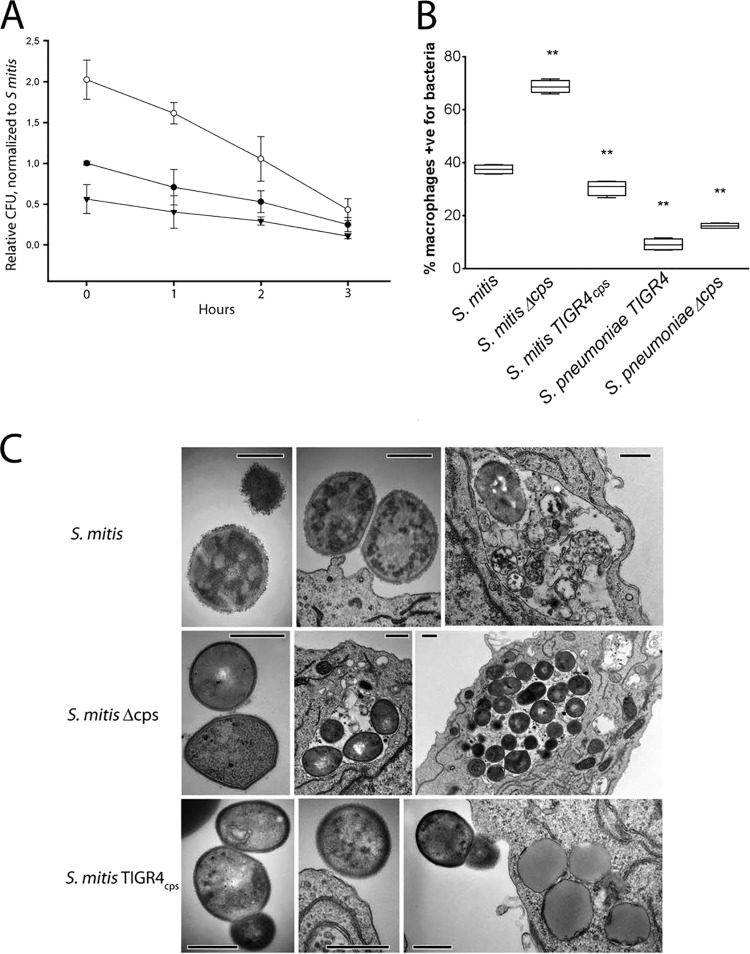
It is not known whether capsule expression by S. mitis may offer resistance against phagocytosis or intracellular killing by macrophages. To address this question, RAW 264.7 macrophages were used in phagocytic assays that compared the uptake and killing of the wild-type strain with those of the capsule locus (MI016) and the capsule-switching mutant S. mitis TIGR4cps. The capsule deletion mutant was present within the macrophages in larger amounts than the S. mitis encapsulated strains, measured by determination of the number of CFU, and within 3 h of incubation it showed a steeper decline than the S. mitis encapsulated strains (Fig. 3A; P < 0.05). The different resistance levels against S. mitis phagocytosis offered by the native and the TIGR4 capsules were confirmed by flow cytometry (Fig. 3B). Both the encapsulated and unencapsulated S. pneumoniae strains were more resistant to phagocytosis than the S. mitis wild-type strain and the S. mitis TIGR4cps strain. To verify the intracellular localization of S. mitis, TEM images were obtained (Fig. 3C). The results confirmed the streptococcal intracellular localization. Notably, S. mitis TIGR4cps (MI030) was seldom observed within macrophages, whereas the capsule deletion mutant was often found in high numbers within single macrophages (Fig. 3C). Taken together, the intracellular viability, the flow cytometry, and the image analysis results indicate that the capsule increases S. mitis resistance to phagocytosis and that both the streptococcal capsule type and the genetic background play important roles in resistance against phagocytosis.
FIG 3.
Effect of capsule locus deletion and interspecies capsule switch on S. mitis phagocytosis and intracellular killing by RAW 264.7 macrophages. (A) The S. mitis wild type, S. mitis Δcps, and S. mitis TIGR4cps were incubated with RAW 264.7 macrophages for 1 h to allow phagocytosis. The number of CFU was used to determine intracellular killing over 3 h. The numbers of CFU were normalized to the baseline values determined for the S. mitis wild type. The means and standard deviations from three independent experiments are presented. (B) The wild type and mutants were labeled with FAMSE and then incubated with RAW 264.7 macrophages for 1 h to allow phagocytosis, before the samples were analyzed by flow cytometry. The median proportion of macrophages positive (+ve) for S. mitis is shown (n = 4 or 5). Lines in center of boxes, median values; central boxes, values from the lower to the upper quartiles (25th to 75th percentiles); vertical lines extend from the minimum to the maximum value. **, significantly different from the S. mitis wild type (P < 0.01). (C) TEM images of internalized and extracellular S. mitis and capsule mutants. Bars, 500 nm.
The capsule protects S. mitis from clearance in a mouse model of early lung infection.
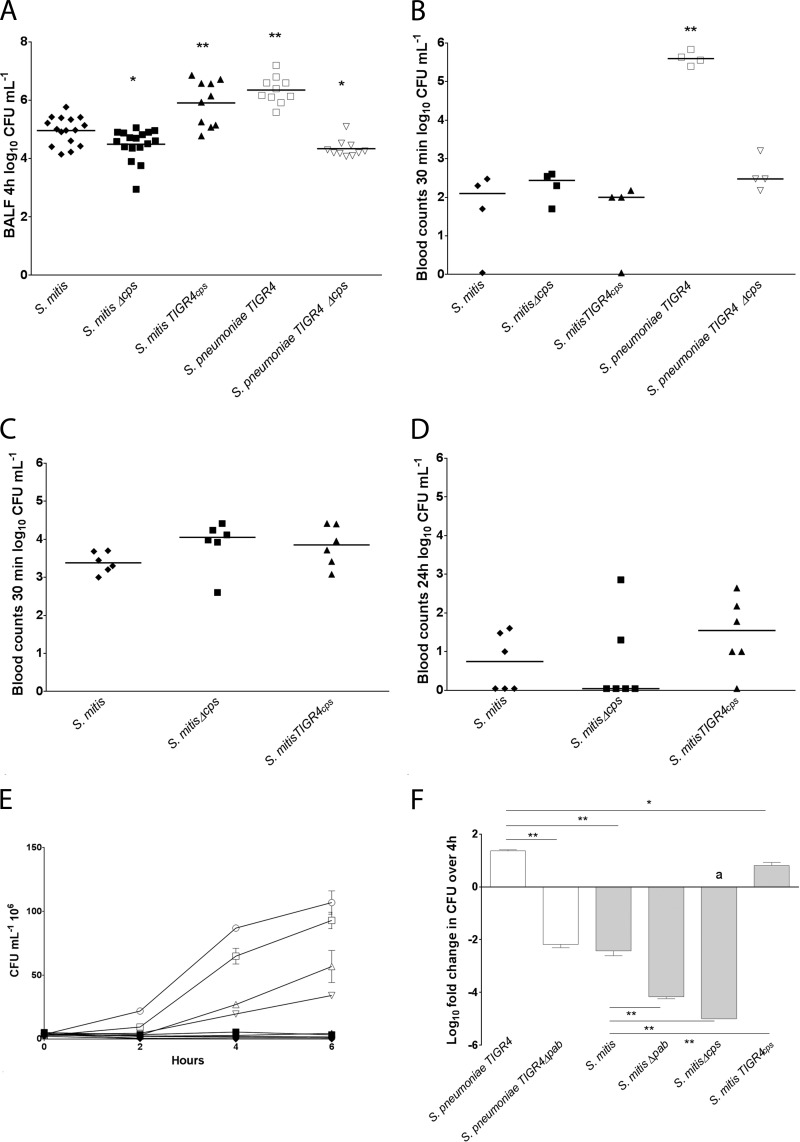
S. mitis is commonly present in BALF as part of the lung microbiome (8). Mechanisms that may affect the ability of S. mitis to resist clearance from the lungs have, however, never been explored. We therefore sought to investigate whether the capsule may have a protective effect against clearance using a mouse model of early lung infection (Fig. 4A). The encapsulated strains of S. mitis were recovered from BALF at significantly higher levels than the nonencapsulated mutant, with the TIGR4 capsule mutant exhibiting the highest values. Resistance was higher for S. pneumoniae TIGR4 than for the S. mitis type strain. No significant differences were observed for S. mitis TIGR4cps and S. pneumoniae TIGR4. The results indicate that the capsule protects S. mitis from lung clearance and that the capsule type may play an important role.
FIG 4.
Effect of capsule locus deletion and interspecies capsule switch on S. mitis clearance in mouse models of lung infection and bacteremia and in ex vivo models of blood growth. (A) BALF samples were collected after 4 h of infection. The numbers of CFU ml−1 recovered in BALF are presented. Data were analyzed using one-way ANOVA followed by Dunnett's test using S. mitis as a control. **, P < 0.01; *, P < 0.50. (B and C) Bacterial counts (number of CFU ml−1) in blood of mice challenged intravenously with S. mitis or S. pneumoniae. Inoculum sizes of 5 × 106 CFU ml−1 (B) and 5 × 107 CFU ml−1 (C and D) were used. Blood samples were collected 30 min (C) and 24 h (D) following infection and spread on blood agar plates for determination of viable counts. (A to D) The dot plots show the numbers of CFU obtained for each mouse, with horizontal lines corresponding to median values. (E and F) S. mitis and S. pneumoniae growth and survival in blood. (E) Growth of S. mitis and S. pneumoniae in horse blood. Black symbols, S. mitis strains CCUG12261 (crossed circles), MI016 (circles), MI30.1 (inverted triangles) SK575 (triangles), SK579 (diamonds), and SK597 (squares); white symbols, S. pneumoniae strains TIGR4 (squares), SP035 (inverted triangles), D39 (triangles), and serotype 1 clinical isolate ST306 (circles). (F) S. mitis and S. pneumoniae TIGR4 survival in human blood. The bars are averages from two to three experiments with two to three parallel samples each and represent the fold change in the number of CFU ml−1 after 4 h of incubation in relation to the number of CFU ml−1 in the inoculum. Error bars represent standard errors of the means. Data were analyzed by one-way ANOVA followed by Dunnett's test using S. pneumoniae TIGR4 as a control. *, P < 0.05; **, P < 0.01; a, no colonies were recovered.
Capsule expression by S. mitis has no significant role in a mouse model of bacteremia.
S. mitis is occasionally associated with septicemia, particularly in immunocompromised individuals (35). We therefore sought to investigate whether the capsule protected S. mitis from clearance in a mouse model of bacteremia. Inoculation of mice with 5 × 106 bacteria resulted in approximately 99% clearance from the blood after 30 min, whereas the level of clearance from the blood was 82% for S. pneumoniae (Fig. 4B). We therefore increased the S. mitis inoculum by a factor of 10 (Fig. 4C and D). Blood count comparisons between the wild type, the S. mitis Δcps strain, and S. mitis TIGR4cps revealed no significant differences after 30 min (Fig. 4C) or 24 h (Fig. 4D) (P = 0.18). The results suggest that the genetic background of the S. mitis type strain may play a more important role than the capsule in clearance from blood in vivo.
S. mitis shows reduced growth in blood and high susceptibility to blood bactericidal action.
The effective clearance of the S. mitis type strain in the mouse model of bacteremia compared to the clearance of S. pneumoniae TIGR4 led us to hypothesize that S. mitis may have a reduced ability to grow in blood compared to S. pneumoniae. To start addressing this hypothesis, we extended the comparisons between S. mitis and S. pneumoniae to include other strains with different or potentially different capsular types in an ex vivo model of blood growth. Since blood samples from individual mice are limited to small volumes, we chose to use horse blood for this screening assay. The growth of the four wild-type strains of S. mitis was suppressed, whereas significant growth was observed for the three wild-type strains of S. pneumoniae representing serotypes 1, 2, and 4 (Fig. 4E). The S. pneumoniae Δcps strain showed reduced growth compared with the wild type, supporting a protective role of the serotype 4 capsule for S. pneumoniae. Notably, suppression of S. mitis growth was not reversed by expression of the S. pneumoniae serotype 4 capsule. The results support the hypothesis that genetic differences other than capsule type may have a major role in the different abilities of S. mitis and S. pneumoniae to grow in blood.
In human blood, the capsule provides a survival advantage to S. mitis that is enhanced in the capsule-switching mutant.
To account for possible effects on bacterial growth that may depend on the species that is used as the source of blood, we investigated the role of the capsule on the growth of S. mitis in human blood as well (Fig. 4F). After 4 h incubation in human blood, no S. mitis Δcps colonies were recovered, despite the finding that the growth of the S. mitis Δcps strain in serum was superior to that of the encapsulated strains (data not shown). For the S. mitis wild type, there was a greater than 100-fold reduction in the number of CFU in blood compared to that in the inoculum. In contrast, the S. mitis TIGR4cps strain demonstrated some increase in the number of CFU, but the increase was not as pronounced as that for S. pneumoniae TIGR4. The results therefore show that the capsule provided a survival advantage to S. mitis in human whole blood that was not found in horse blood. This indicates that the advantage provided by the capsule in the S. mitis type strain may depend on the blood source.
Since the ability to grow in blood may be an important factor behind the differences in the pathogenic potential of S. mitis and S. pneumoniae, we decided to further investigate the influence of blood bactericidal actions on growth, without the effect of bacterial replication. For this purpose, pabB deletion mutants of S. mitis and S. pneumoniae were included in the blood growth assay using human blood. Since for replication pab deletion mutants depend on the presence of para-aminobenzoic acid (PABA), which is absent in whole blood, growth is abolished (36). In serum, the numbers of CFU for the Δpab mutants after 4 h of incubation were similar to those in the inoculum at time zero, indicating that human serum had no detectable bacterial killing activity. In whole blood, the reduction in the number of CFU in relation to that in the wild-type strains was stronger for S. mitis than for the S. pneumoniae Δpab mutants, indicating that S. mitis may be more susceptible to the bactericidal actions in human whole blood than S. pneumoniae.
DISCUSSION
The recent focus on human microbiomes and the realization that ecological disturbances within microbial communities may be the culprit behind several diseases have increased the interest in commensals. In the oral cavity, S. mitis is a commensal that appears to be unique in its ability to colonize virtually all oral sites, in addition to the oro- and nasopharynxes, as well as the lungs. The molecular mechanisms involved in S. mitis colonization and host interactions are, however, still poorly characterized. Although several of the methodologies developed for the study of S. pneumoniae may be adapted to S. mitis, the challenges of working with a species that has not been extensively characterized should not be underestimated. The genetic variability between strains of S. mitis is well documented, and a full understanding of its molecular mechanisms involved in the commensal and occasional pathogenic interactions with the host will require the characterization of several strains. However, the natural transformation of S. mitis has been reported for only a couple of strains (20, 37), and in our hands, the transformation of isolates other than the type strain, including the SK575, SK579, and SK597 strains used in this study, has so far been unsuccessful with or without addition of their respective CSPs (data not shown). In contrast to S. pneumoniae strains, which are mostly grouped into two CSP pherotypes, each S. mitis strain appears to use its own specific CSP (19), a methodological issue that should be considered when using strains that have not yet been sequenced. Animal models have been extensively used for modeling S. pneumoniae infection and colonization, but the models may need to be adapted to allow the study of S. mitis strains. In the bacteremia model used in our study, for instance, the S. mitis inoculum size had to be increased in relation to that for S. pneumoniae TIGR4 to better investigate the role of the capsule. The reason was that S. mitis was rapidly cleared from the blood at inoculum sizes that in S. pneumoniae TIGR4 resulted in recoveries approximately 3 log units higher (Fig. 4B). The results presented here begin to address the gap in knowledge on the molecular biology of S. mitis by revealing the contribution of the capsule and the capsule type to S. mitis NCTC 12261T resistance to phagocytosis by RAW 264.7 macrophages, resistance to clearance from the lungs in a mouse model, and survival in human blood.
S. mitis resistance to lung clearance was enhanced in the encapsulated strains, with S. mitis TIGR4 showing the highest values. These values were comparable to those obtained for S. pneumoniae TIGR4. This indicates that the expression of the TIGR4 capsule by S. mitis may compensate for the reduced ability of the S. mitis wild type to resist lung clearance compared with that of S. pneumoniae TIGR4. For S. pneumoniae TIGR4, the unencapsulated isogenic mutant was also cleared more efficiently than the wild type. This is in accordance with previous findings showing that the capsule protects S. pneumoniae TIGR4 and D39 from early clearance from the lungs (38).
In a bacteremia model of infection, the capsule protects S. pneumoniae from clearance from blood (39). This was confirmed by our results showing that for the S. pneumoniae wild type the number of CFU recovered 30 min after inoculation compared to the number of CFU in the initial inoculum was 5-fold reduced, whereas for the S. pneumoniae Δcps strain, a reduction of almost 10,000 was observed. We were not able to demonstrate an effect of the capsule on the ability of S. mitis to resist clearance, even after increasing the inoculum size 10-fold. We also conducted experiments in which we measured clearance 24 h after intravenous inoculation, but again, no significant differences were observed. It remains to be determined whether this is a phenomenon restricted to the type strain or a more general characteristic within the S. mitis species.
In human blood, S. mitis Δcps had a remarkable disadvantage over the wild type, as no colonies of this strain were recovered after 4 h of incubation, whereas S. mitis TIGR4 was recovered in higher numbers than the S. mitis wild-type strain. In contrast, growth of the S. mitis wild type and mutants was severely affected in the ex vivo experiments using horse blood. Since S. mitis is a predominant human colonizer, the results in human blood are of particular relevance. Results using blood from other animals are, however, also important, since they may reveal differences less likely to be associated with a life history of high levels of exposure to the microorganisms examined. Although alpha streptococci are described in mice and horses, they appear to constitute a significantly smaller proportion of their microbiota (40, 41). It was interesting, therefore, to observe that the poor growth of S. mitis in blood extended to both human and horse blood and was in contrast to the higher growth values found with S. pneumoniae. When bacterial replication was prevented by the pabB deletion, viability after 4 h in human blood was close to zero for S. mitis; in contrast, for S. pneumoniae Δpab, viability was only slightly affected. Further investigations on the mechanisms behind such differences are warranted, not only in view of its occasional association with septicemia in neutropenic patients and endocarditis but also in relation to its potential use in future probiotic applications (12).
S. pneumoniae and S. mitis are thought to have evolved from a common ancestor that more closely resembles S. pneumoniae, and the loss of virulence factors that allowed S. mitis to better adapt to a commensal lifestyle is thought to have occurred (19). It will be interesting to find out the factor that S. mitis may have lost or perhaps gained that contributed to the establishment of a more successful relationship with the host. Although our study focused on the role of the capsule in S. mitis, comparisons with S. pneumoniae showed for the first time important differences in blood survival between the two species. S. pneumoniae showed higher survival rates than S. mitis, and this finding was not restricted to the S. mitis type strain and the S. pneumoniae TIGR4 wild type or capsule isogenic mutants but was also a characteristic of 3 other wild-type isolates of S. mitis and 2 other isolates of S. pneumoniae representing serotypes 1 and 2. The superior ability of S. pneumoniae TIGR4 to survive in blood was also confirmed in experiments using human blood and in the bacteremia model. This raises the hypothesis that S. mitis and S. pneumoniae have species-specific differences in their ability to grow and/or survive in blood that may contribute to their differences in pathogenic potential. Understanding the mechanisms behind the commensal nature of oral streptococci is particularly relevant in view of their potential to modulate ecological relationships within microbial communities that may be beneficial to the human host (7).
Capsule expression by S. mitis is supported both by microscopic approaches comparing the wild type of the type strain to a capsule locus deletion mutant (20) and by biochemical means in SK137 (42). In addition, the capsule locus has been identified in a range of S. mitis strains (19), including 11 of the 15 available S. mitis genomes. Comparative analysis of the capsule locus indicates that variation in capsule types may not be restricted to S. pneumoniae but may also be a common feature in S. mitis (20). In these sequenced strains, the wzy polymerase genes differed among the strains and had no significant homologues in S. pneumoniae. As discussed above, strains belonging to the S. mitis species show a remarkable diversity, and in view of the few genomes sequenced, there is still limited information on the range of capsule types that S. mitis may produce. To date, 25 complete and 220 unfinished genomes of S. pneumoniae are publically available (NCBI), whereas for S. mitis, of the 15 genomes available at present, the complete genome sequence is available for only 1 (43). Future studies should determine whether regions within the capsule locus or even the entire locus may be exchanged between S. mitis and S. pneumoniae. Our study indicates that exchange of the entire locus is possible. A recent report indeed suggests a similar or identical S. pneumoniae serotype 19C capsule in an S. mitis strain (44). PCR serotyping has also revealed amplicons in S. mitis strains isolated from the pharynx whose sequences are similar to those in the capsule variable region of S. pneumoniae (21). In the previous study, the investigators recovered two different strains of S. mitis that were PCR positive for S. pneumoniae serotype-specific loci and that had sequences nearly identical to those from S. pneumoniae serotype 5 and serogroup 18, respectively. Such findings are consistent with the presence of the corresponding serotypes in S. mitis. Although the TIGR4 capsule provided a survival advantage to the S. mitis type strain in in vitro phagocytosis, as well as in the human blood growth assay and in the lung model of infection, significant advantages over the native capsule were not observed using the mouse model of bacteremia. The natural setting for recombination between S. pneumoniae and S. mitis is most probably during colonization of the respiratory tract and oral cavity, where favorable traits may be acquired. Thus, it will be interesting in future studies to investigate the interplay between the two species and the role of different S. mitis capsule types under colonization conditions.
ACKNOWLEDGMENTS
We are grateful for the excellent technical assistance from Heidi Aarø Åmdal. The antibiotic resistance cassettes were a kind gift from Donald Morrison. We thank Ingeborg Aaberge for the S. pneumoniae TIGR4 strain and Didrik F. Vestrheim for the serotype 1 strain. We thank also the Electron Microscopy Unit for Biological Sciences, Department of Biosciences, University of Oslo.
The work was partially funded by the Faculty of Dentistry, University of Oslo (to H.V.R.); the Department of Health's NIHR Biomedical Research Centre's funding scheme, BBSRC (http://www.bbsrc.ac.uk/home/home.aspx; to H.M.); MRC (http://www.mrc.ac.uk/index.htm MR/K00168X/1; to J.P.); the Wellcome Trust (http://www.wellcome.ac.uk/WT097216MA; to R.J.J.); and NFR grant 213830/F20 (http://www.forskningsradet.no/; to R.S.K. and U.R.). H.V.R. was the recipient of a research travel grant from MLS-UIO.
Footnotes
Published ahead of print 23 June 2014
REFERENCES
- 1.Griffith F. 1928. The significance of pneumococcal types. J. Hyg. (Lond.) 27:113–159. 10.1017/S0022172400031879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:262–269. 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. 10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golubchik T, Brueggemann AB, Street T, Gertz RE, Jr, Spencer CC, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, Peto TE, Moore MR, Donnelly P, Crook DW, Bowden R. 2012. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat. Genet. 44:352–355. 10.1038/ng.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Linares J, Jacobs MR, Kristinsson KG, Beall BW, Klugman KP, Parkhill J, Hakenbeck R, Bentley SD, Brueggemann AB. 2013. Pneumococcal capsular switching: a historical perspective. J. Infect. Dis. 207:439–449. 10.1093/infdis/jis703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kononen E, Jousimies-Somer H, Bryk A, Kilp T, Kilian M. 2002. Establishment of streptococci in the upper respiratory tract: longitudinal changes in the mouth and nasopharynx up to 2 years of age. J. Med. Microbiol. 51:723–730 [DOI] [PubMed] [Google Scholar]
- 7.Kreth J, Merritt J, Qi F. 2009. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 28:397–403. 10.1089/dna.2009.0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, Weiden MD. 2013. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1:19. 10.1186/2049-2618-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- 10.Marron A, Carratala J, Gonzalez-Barca E, Fernandez-Sevilla A, Alcaide F, Gudiol F. 2000. Serious complications of bacteremia caused by viridans streptococci in neutropenic patients with cancer. Clin. Infect. Dis. 31:1126–1130. 10.1086/317460 [DOI] [PubMed] [Google Scholar]
- 11.Tunkel AR, Sepkowitz KA. 2002. Infections caused by viridans streptococci in patients with neutropenia. Clin. Infect. Dis. 34:1524–1529. 10.1086/340402 [DOI] [PubMed] [Google Scholar]
- 12.Mitchell J. 2011. Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 26:89–98. 10.1111/j.2041-1014.2010.00601.x [DOI] [PubMed] [Google Scholar]
- 13.Beighton D, Carr AD, Oppenheim BA. 1994. Identification of viridans streptococci associated with bacteremia in neutropenic cancer-patients. J. Med. Microbiol. 40:202–204. 10.1099/00222615-40-3-202 [DOI] [PubMed] [Google Scholar]
- 14.Roos K, Hakansson EG, Holm S. 2001. Effect of recolonisation with “interfering” alpha streptococci on recurrences of acute and secretory otitis media in children: randomised placebo controlled trial. BMJ 322:210–212. 10.1136/bmj.322.7280.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wescombe PA, Hale JD, Heng NC, Tagg JR. 2012. Developing oral probiotics from Streptococcus salivarius. Future Microbiol. 7:1355–1371. 10.2217/fmb.12.113 [DOI] [PubMed] [Google Scholar]
- 16.Kim JO, Romero-Steiner S, Sorensen UBS, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 67:2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301. 10.1038/nrmicro1871 [DOI] [PubMed] [Google Scholar]
- 18.Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 75:83–90. 10.1128/IAI.01475-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilian M, Poulsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, Tettelin H, Sorensen UB. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. 10.1371/journal.pone.0002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rukke HV, Hegna IK, Petersen FC. 2012. Identification of a functional capsule locus in Streptococcus mitis. Mol. Oral Microbiol. 27:95–108. 10.1111/j.2041-1014.2011.00635.x [DOI] [PubMed] [Google Scholar]
- 21.Carvalho MG, Pimenta FC, Moura I, Roundtree A, Gertz RE, Jr, Li Z, Jagero G, Bigogo G, Junghae M, Conklin L, Feikin DR, Breiman RF, Whitney CG, Beall BW. 2013. Non-pneumococcal mitis-group streptococci confound detection of pneumococcal capsular serotype-specific loci in upper respiratory tract. PeerJ 1:e97. 10.7717/peerj.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipsitch M, O'Hagan JJ. 2007. Patterns of antigenic diversity and the mechanisms that maintain them. J. R. Soc. Interface 4:787–802. 10.1098/rsif.2007.0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melin M, Trzcinski K, Meri S, Kayhty H, Vakevainen M. 2010. The capsular serotype of Streptococcus pneumoniae is more important than the genetic background for resistance to complement. Infect. Immun. 78:5262–5270. 10.1128/IAI.00740-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez CJ, Hinojosa CA, Shivshankar P, Hyams C, Camberlein E, Brown JS, Orihuela CJ. 2011. Changes in capsular serotype alter the surface exposure of pneumococcal adhesins and impact virulence. PLoS One 6:e26587. 10.1371/journal.pone.0026587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11:R107. 10.1186/gb-2010-11-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakenbeck R, Konig A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanage WP, Fraser C, Tang J, Connor TR, Corander J. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324:1454–1457. 10.1126/science.1171908 [DOI] [PubMed] [Google Scholar]
- 28.Pearce BJ, Iannelli F, Pozzi G. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153:243–247. 10.1016/S0923-2508(02)01312-8 [DOI] [PubMed] [Google Scholar]
- 29.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205. 10.1016/S0167-7012(01)00369-4 [DOI] [PubMed] [Google Scholar]
- 30.Bricker AL, Camilli A. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131–135. 10.1111/j.1574-6968.1999.tb13460.x [DOI] [PubMed] [Google Scholar]
- 31.Peppoloni S, Ricci S, Orsi CF, Colombari B, De Santi MM, Messino M, Fabio G, Zanardi A, Righi E, Braione V, Tripodi S, Chiavolini D, Cintorino M, Zoli M, Oggioni MR, Blasi E, Pozzi G. 2010. The encapsulated strain TIGR4 of Streptococcus pneumoniae is phagocytosed but is resistant to intracellular killing by mouse microglia. Microbes Infect. 12:990–1001. 10.1016/j.micinf.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 32.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. 2010. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78:704–715. 10.1128/IAI.00881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oggioni M, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew P, Pozzi G. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 61:1196–1210. 10.1111/j.1365-2958.2006.05310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanguinetti L, Toti S, Reguzzi V, Bagnoli F, Donati C. 2012. A novel computational method identifies intra- and inter-species recombination events in Staphylococcus aureus and Streptococcus pneumoniae. PLoS Comput. Biol. 8:e1002668. 10.1371/journal.pcbi.1002668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochud PY, Calandra T, Francioli P. 1994. Bacteremia due to viridans streptococci in neutropenic patients: a review. Am. J. Med. 97:256–264. 10.1016/0002-9343(94)90009-4 [DOI] [PubMed] [Google Scholar]
- 36.Chimalapati S, Cohen J, Camberlein E, Durmort C, Baxendale H, de Vogel C, van Belkum A, Brown JS. 2011. Infection with conditionally virulent Streptococcus pneumoniae Δpab strains induces antibody to conserved protein antigens but does not protect against systemic infection with heterologous strains. Infect. Immun. 79:4965–4976. 10.1128/IAI.05923-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensing BA, Rubens CE, Sullam PM. 2001. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect. Immun. 69:1373–1380. 10.1128/IAI.69.3.1373-1380.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyams CJ. 2010. Ph.D. thesis. University College London, London, United Kingdom [Google Scholar]
- 39.Lammers AJ, De Porto AP, Florquin S, De Boer OJ, Bootsma HJ, Hermans PW, Van der Poll T. 2011. Enhanced vulnerability for Streptococcus pneumoniae sepsis during asplenia is determined by the bacterial capsule. Immunobiology 216:863–870. 10.1016/j.imbio.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 40.Boyer EE. 1919. Studies on the bacterial flora of the mouth and nose of the normal horse. J. Bacteriol. 4:61–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chun J, Kim KY, Lee JH, Choi Y. 2010. The analysis of oral microbial communities of wild-type and Toll-like receptor 2-deficient mice using a 454 GS FLX titanium pyrosequencer. BMC Microbiol. 10:101. 10.1186/1471-2180-10-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergstrom N, Jansson PE, Kilian M, Skov Sorensen UB. 2000. Structures of two cell wall-associated polysaccharides of a Streptococcus mitis biovar 1 strain. A unique teichoic acid-like polysaccharide and the group O antigen which is a C-polysaccharide in common with pneumococci. Eur. J. Biochem. 267:7147–7157. 10.1046/j.1432-1327.2000.01821.x-i2 [DOI] [PubMed] [Google Scholar]
- 43.Denapaite D, Bruckner R, Nuhn M, Reichmann P, Henrich B, Maurer P, Schahle Y, Selbmann P, Zimmermann W, Wambutt R, Hakenbeck R. 2010. The genome of Streptococcus mitis B6—what is a commensal? PLoS One 5:e9426. 10.1371/journal.pone.0009426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjaer TR, Hansen AG, Sorensen UB, Holm AT, Sorensen GL, Jensenius JC, Thiel S. 2013. M-ficolin binds selectively to the capsular polysaccharides of Streptococcus pneumoniae serotypes 19B and 19C and of a Streptococcus mitis strain. Infect. Immun. 81:452–459. 10.1128/IAI.01148-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aaberge IS, Eng J, Lermark G, Lovik M. 1995. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb. Pathog. 18:141–152. 10.1016/S0882-4010(95)90125-6 [DOI] [PubMed] [Google Scholar]
- 46.Ali F, Lee ME, Iannelli F, Pozzi G, Mitchell TJ, Read RC, Dockrell DH. 2003. Streptococcus pneumoniae-associated human macrophage apoptosis after bacterial internalization via complement and Fcgamma receptors correlates with intracellular bacterial load. J. Infect. Dis. 188:1119–1131. 10.1086/378675 [DOI] [PubMed] [Google Scholar]