Abstract
Enterotoxigenic Escherichia coli (ETEC) strains are among the most common causes of diarrheal illness worldwide. These pathogens disproportionately afflict children in developing countries, where they cause substantial morbidity and are responsible for hundreds of thousands of deaths each year. Although these organisms are important targets for enteric vaccines, most development efforts to date have centered on a subset of plasmid-encoded fimbrial adhesins known as colonization factors and heat-labile toxin (LT). Emerging data suggest that ETEC undergoes considerable changes in its surface architecture, sequentially deploying a number of putative adhesins during its interactions with the host. We demonstrate here that one putative highly conserved, chromosomally encoded adhesin, EaeH, engages the surfaces of intestinal epithelial cells and contributes to bacterial adhesion, LT delivery, and colonization of the small intestine.
INTRODUCTION
Infectious diarrhea continues to cause tremendous suffering in developing countries, resulting in an estimated one to two million deaths each year. Enterotoxigenic Escherichia coli (ETEC) contributes significantly to premature deaths from diarrheal illness in young children (1, 2) and causes substantial morbidity in surviving children and adults (3). ETEC strains are perennially the leading etiology of diarrhea in travelers to areas where ETEC strains are endemic (4).
By definition, these organisms secrete heat-labile (LT) and/or heat-stable (ST) enterotoxins that induce host cell production of cyclic nucleotides (cyclic AMP [cAMP] and cGMP, respectively) to activate protein kinases that ultimately result in phosphorylation of the cystic fibrosis transmembrane regulatory channel (CFTR) (5–7) and Na+ ion exchangers (8) on the surfaces of intestinal epithelial cells. Ensuing chloride secretion through CFTR, as well as the commensurate loss of salt and water into the intestinal lumen, results in the cholera-like watery diarrhea characteristic of ETEC infections (9).
In the current paradigm for ETEC pathogenesis, this organism must effectively colonize the small intestine to deliver LT and/or ST efficiently. The majority of pathogenesis (10–12) and molecular epidemiology (13) studies, as well as subsequent vaccine development efforts (14, 15), have focused primarily on plasmid-encoded fimbrial colonization factors (CFs), which are felt to be critical for colonization of the small intestine.
This longstanding but fairly simple view of ETEC pathogenesis in which bacteria adhere via CFs to the small intestine, where these pathogens release their toxin(s), likely underestimates the complexity of these pathogens. More recent investigations have highlighted a number of novel putative virulence factors (16, 17), unique interactions of ETEC with the epithelium (18–20), and an intricate orchestration of multiple pathogen-host events (21) that culminate in successful toxin delivery to epithelial cell targets (22). Collectively, the emerging data suggest that these sophisticated interactions of ETEC strains with their host might be exploited in outlining novel strategies for vaccine development (23).
Unfortunately, despite ETEC's global importance, several obstacles need to be surmounted in order to develop a broadly protective ETEC vaccine (15, 24). One central challenge to ETEC vaccinology is the general plasticity of E. coli genomes (25). Although CFs remain the most extensively studied ETEC vaccine targets (15, 26), they are not universally conserved (27), with at least 26 antigenically distinct structures (15, 28) that vary considerably by time and geography (13). ETEC infections in young children in developing countries appear to provide substantive protection against subsequent diarrheal illness caused by these organisms (2, 29, 30). However, epidemiologic studies (29, 31), as well as recent vaccine trials (32), suggest that other antigens may be involved in protection. Therefore, over time it has become apparent that additional strategies are needed to complement a CF-based approach to ETEC vaccines.
Interestingly, recent studies of ETEC transcriptional modulation following interaction with epithelial cells highlighted a number of genes potentially encoding novel target antigens (21). One gene, eaeH, which was strongly upregulated upon epithelial cell attachment, was originally identified in ETEC by subtractive hybridization with a laboratory strain of E. coli (33). Although our earlier studies suggested that eaeH encodes a surface-expressed antigen that is expressed in the context of epithelial cells (21), its role in the pathogenesis of ETEC has not been explored. The present studies were undertaken to examine the role of this highly conserved antigen in ETEC bacterium-host interaction and toxin delivery.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A complete list of bacterial strains used or generated during the course of these studies is provided in Table 1. Bacteria were grown at 37°C in Luria broth supplemented with antibiotics as appropriate from frozen glycerol stocks maintained at −80°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and/or descriptiona | Source and/or reference |
|---|---|---|
| Strains | ||
| H10407 | ETEC; serotype O78:H11; LT+ STh+ STp+ | Bangladesh, 1973 (10) |
| E24377A | ETEC; serotype O139:H28;CFA/II (CS1/CS3); LT+/ST+ | Egypt (58) |
| Juruá 18/11, 20/10 | ETEC associated with severe cholera-like diarrhea; CS7; LT+ | Amazon (59) |
| 2726950 | ETEC; CS21; ST+; mild, self-limited diarrhea isolate | ICDDR,Bb; 2007 |
| 2785200 | ETEC; CF− LT+; 2007 severe cholera-like disease | ICDDR,B; 2007 |
| 2845650 | ETEC; CS4/CS6 LT+/ST+; mild, self-limited diarrhea | ICDDR,B; 2008 |
| 2860650 | ETEC; CS5+CS6 LT+/ST+; severe cholera-like disease | ICDDR,B; 2008 |
| 2864350 | ETEC; CF− LT+/ST+; severe cholera-like disease | ICDDR,B; 2008 |
| 2875150 | ETEC; CS4/CS6 LT+/ST+; severe cholera-like disease | ICDDR,B; 2008 |
| BCE034_MS14 | ETEC; CFA/I ST+; 2003 asymptomatic colonization | ICDDR,B 2003 |
| P0299917.2 | LT+/ST-H+; severe cholera-like disease | ICDDR,B; 2011 |
| BCE001_MS16 | CF− LT+; asymptomatic colonization | ICDDR,B; 2004 |
| MG1655 | E. coli K-12 (F− λ− ΔilvG rfb-50 rph-1); serotype OR:H48:K− | University of Wisconsin |
| HS | E. coli from healthy laboratory scientist, WRAIR | 60 |
| Nissle 1917 | E. coli O6:K5:H1 commensal strain isolated from a German soldier in 1917 | 61, 62 |
| jf876 | lacZYA::Kmr mutant of H10407 | 49 |
| jf2852 | eaeH::Kmr | This study |
| NiCo21(DE3) | can::CBD fhuA2 [lon] ompT gal (λ DE3) [dcm] arnA::CBD slyD::CBD glmS6 ala ΔhsdS λ DE3 = λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 Δnin-5 | New England Biolabs |
| BL21AI | F− ompT hsdS B (rB− mB−) gal dcm araB::T7RNAP- tetA | 19 |
| Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG λ− | Invitrogen |
| Plasmids | ||
| pDONR221 | Entry cloning plasmid containing ccdB negative selection gene and Cmr cassette between lambda attP sites for recombination with PCR products containing attB regions; Kmr | Invitrogen |
| pET-DEST42 | T7 lac promoter/IPTG-inducible expression plasmid; ccdB, Cmr genes flanked by attR sites for recombination with entry plasmid; C-terminal V5 and His6 tags | Invitrogen |
| pSS001 | 4,254-bp eaeH amplicon cloned into pDONOR221 | This study |
| pSS002 | EaeH expression plasmid derived from LR recombination reaction of pSS001 and pET-DEST42 placing eaeH in frame with V5 and His6 epitope tags | This study |
| pAS004 | 4,257-bp eaeH amplicon with native stop codon cloned into HindIII-BglII sites on pFLAG-CTC | This study |
| pKD46 | Arabinose-inducible λ red recombinase helper plasmid; Ampr; Ts | 36 |
| pKD4 | Template plasmid for mutagenesis using FRT-Kmr cassettes; Kmr Ampr; oriRγ | 36 |
| pFLAG-CTC | 5,348-bp tac promoter expression plasmid for the construction of C-terminal FLAG epitope fusions | Sigma |
Kmr, kanamycin resistant; Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Strr, streptomycin resistant; V5, V5 epitope (GKPIPNPLLGLDST); His6, polyhistidine tag. FLAG indicates the FLAG (DYKDDDDK) epitope tag. Ts, temperature-sensitive replicon; FRT, FLP recombinase\recognition target; oriRγ, pir-dependent origin of replication. WRAIR, Walter Reed Army Institute of Research.
ICDDR,B, International Center for Diarrheal Disease Research, Bangladesh.
Bioinformatic analysis of EaeH.
SignalP (3.0) (34) (http://www.cbs.dtu.dk/services/SignalP/) was used to identify a putative signal peptide region of EaeH, and PSORTb (v3.0 [http://www.psort.org/]) (35) was used to determine the possible subcellular location of the protein. Searches for regions of homology with intimin were performed comparing the intimin (eaeA gene product, GenBank accession number P19809.2) sequence from EPEC against the H10407 genome. Domains within EaeH from H10407 were identified in the Conserved Domain Database using the domain-enhanced lookup time-accelerated BLAST (DELTA-BLAST) algorithm.
Mutagenesis, cloning, and expression of eaeH.
To construct an eaeH deletion mutant, the primers jf062612.9 and jf062612.10 (Table 2) were first used to amplify a kanamycin resistance cassette from pKD4 with 60-bp tails corresponding to the DNA sequence immediately upstream and downstream of eaeH. The resulting amplicon was then introduced into H10407 carrying the pKD46 helper plasmid to affect lambda red-mediated homologous recombination to generate the mutant as previously described (36).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Description |
|---|---|---|
| jf051010.1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTggGAAGGAGATAGAACCATGTCACATTATAAAACAGGT | attB1_forward.Shine-Dalgarno.Kozak.5′ (nt 1 to 21) eaeH |
| jf051010.2 | GGGGACCACTTTGTACAAGAAAGCTGGGTcTGGCATCTCCTCCTCGCCATT | attB2.reverse.eaeH.[nt: 4234–4254]for C-terminal fusion in pDET-DEST42 |
| jf050610.1 | GGAAAAGGAAATCGGGAAA | eaeH locus primer begins 99 bp upstream from eaeH start codon |
| jf050610.2 | CGTAGAAAAGGATGGCAA | eaeH locus primer (reverse) begins 200 bp downstream and 200 bp from the stop codon; 4556 amplicon with jf050610.1 |
| jf062612.9 | CAGACGCCATTATTTGTGTCTGCCTATGTTCGTTAATTCGTTCATCAGGAAATTATCTCAGTGTAGGCTGGAGCTGCTTC | 60-bp homology tail (bases 1 to 60 immediately upstream from eaeH), p1 region of pKD4 |
| jf062612.10 | GGCGTTGAATATTTCAACACCATTATTCTTTATTAGATCGTAACTTTCATACTATTCAAACATATGAATATCCTCCTTA | eaeH H2 primer bases 4219 to 4254/pKD4.p2 60-bp homology tail (bases 1 to 60 immediately downstream from eaeH), p2 region of pKD4 |
| jf020112.3 | ATATCATATGAAGCTAAGCTTATGTCACATTATAAAACAGG | HindIII-eaeH (nt 1 to 18); start codon of eaeH is underlined) |
| jf020112.4 | TGTAGTCGACAGATCAGATCTTTATGGCATCTCCTCCTCGCCA | BglII-eaeH (nt 4237 to 4257; the stop codon of eaeH is underlined) |
The boldfacing and underlining in each sequence are as defined in the final “Description” column. nt, nucleotide(s); attB, bacteriophage lambda recombination sequence.
To construct an eaeH expression plasmid, the primers jf051010.1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTggGAAGGAGATAGAACCATGTCACATTATAAAACAGGT-3′) and jf051010.2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTcTGGCATCTCCTCCTCGCCATT-3′) (Table 2) were first used to amplify a 4,254-bp eaeH gene fragment (lacking only the stop codon) from H10407 genomic DNA. The resulting amplicon, containing 5′ and 3′ terminal attB sites, was then cloned by bacteriophage lambda-mediated recombination with corresponding attP sites on pDONR221, yielding pSS001. After we confirmed the construction of pSS001, this plasmid was recombined with pET-DEST42 placing the eaeH gene in-frame with C-terminal V5 and polyhistidine tags to create the pSS002 expression plasmid. The eaeH gene with its native stop codon was also amplified from H10407 genomic DNA using the primers jf020112.3 and 020112.4 (Table 2), digested with HindIII and BglII, and then directionally cloned into the corresponding sites on pFLAG-CTC yielding pAS004. Genomic DNA was purified by using a Wizard genomic DNA purification kit (Promega, WI).
Recombinant protein production.
The pSS002 expression plasmid encoding the EaeH-V5-6His fusion protein was introduced into BL21AI (Table 1), and recombinants were selected on ampicillin (100 μg/ml). After the induction of BL21AI(pSS002) with IPTG (isopropyl-β-d-thiogalactopyranoside), recombinant polyhistidine-tagged protein was recovered from bacterial lysates (B-PER, bacterial protein extraction reagent; Pierce/Thermo Scientific) by nickel metal affinity chromatography. Western blotting with monoclonal antibody against the V5 epitope (-Gly-Lys-Pro-Ile-Pro-Asn-Pro-Leu-Leu-Gly-Leu-Asp-Ser-Thr-) was then used to confirm expression of the fusion protein.
Outer membrane preparations.
Previously described methods were used to prepare outer membranes from ETEC H10407 or recombinant E. coli expressing eaeH (19, 37). Briefly, clarified bacterial lysates, prepared in a French pressure cell, were layered onto a discontinuous gradient prepared from 2.02, 1.44, and 0.78 M sucrose in HEPES buffer and subjected to ultracentrifugation for 16 h at approximately 80,000 × g at 4°C. Material collected from the 2.02 and 1.44 M interface was diluted 10-fold in HEPES buffer and recentrifuged at approximately 110,000 × g, 4°C for 1 h. The supernatant was discarded, and the resulting outer membrane preparation was stored at −80°C.
Production and purification of polyclonal antibodies.
Polyclonal antisera were produced in rabbits as previously described (16). Briefly, two New Zealand White rabbits were immunized with recombinant V5- and polyhistidine-tagged EaeH. The resulting polyclonal antisera were preabsorbed using an E. coli lysate column (Pierce) and lyophilized strain AAEC191-A (38). Protein A-agarose (Protein A Plus; Thermo Scientific) was used to separate antibodies from serum components. Affinity purification of antibody against recombinant EaeH (rEaeH)-V5-6His immobilized on nitrocellulose was performed as previously described (18, 39).
Flow cytometry.
To identify EaeH on the surface of recombinant E. coli, BL21AI(pSS002) was grown overnight in a 2 ml of Luria broth containing ampicillin (100 μg/ml) at 37°C in a shaking incubator at 250 rpm. The following day, overnight cultures were diluted 1:100 into fresh media, and grown to mid-logarithmic phase (i.e., and optical density at 600 nm of approximately 0.6 to 0.8). IPTG was added to a final concentration of 0.4 mM, and the cultures were aliquoted into separate tubes, followed by incubation for an additional 2 h at three different temperatures (37°C, 30°C, and room temperature) with shaking at 250 rpm. Then, 200 μl of each culture was centrifuged at 10,000 × g and 4°C. Strain H10407 was grown in the same fashion for comparison, omitting the IPTG.
For surface staining, bacterial pellets were washed once with phosphate-buffered saline (PBS) and then incubated with 1% bovine serum albumin (BSA) in PBS for 30 min at room temperature. The bacteria were then incubated with primary antibody (anti-EaeH, rabbit polyclonal antibody or anti-V5 mouse monoclonal antibody) at room temperature for 45 min. After a washing step with PBS, secondary antibody staining was performed at room temperature in the dark with species-specific antibodies conjugated with Alexa Fluor 488 for an additional 45 min. The bacteria were then washed with PBS and resuspended in 500 μl of PBS for acquisition by flow cytometry (FACSCalibur; BD Biosciences). A minimum of 50,000 organism counts were acquired (CellQuest software; Becton Dickinson), and the findings were analyzed using FlowJo (v7.6.3).
Binding of EaeH-coated latex beads to Caco-2 intestinal epithelial cells.
rEaeH-V5-6His was adsorbed on the surface of polystyrene latex beads (1- or 3-μm mean particle size, LB30; Sigma) as described by the manufacturer. Briefly, 100 μl of aqueous bead suspension (∼6.8 × 108 beads) was first prewashed in 25 mM MES [2-(N-morpholino)ethanesulfonic acid] buffer (pH 6.1), and rEaeH-V5-6His was dialyzed against the same buffer. The beads were then mixed with the recombinant protein at a final concentration of 0.4 mg/ml in a total volume of 500 μl containing 0.2% sodium azide overnight at 4°C. Control beads were coated with BSA. After being washed with PBS, the beads were examined by flow cytometry to document successful adsorption of the rEaeH-V5-6His to the bead surfaces. Beads were incubated with polyclonal rabbit anti-EaeH sera (1:1,000), washed and incubated with a 1:200 dilution of fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody (BD Pharmingen), and then washed and resuspended in PBS for examination using a four-color dual-laser flow cytometer (FACSCalibur).
Beads coated with rEaeH or control beads were resuspended in 1 ml of tissue culture medium and added to Caco-2 cells grown on glass coverslips, which were then incubated at 5% CO2 and 37°C. After ∼3 h, the cells were washed with PBS and fixed with an ice-cold solution of 6% paraformaldehyde in PBS. The cells were mounted on glass slides for subsequent visualization by light microscopy (40, 41). Differential interference contrast (DIC) images were obtained on a Zeiss Axiophot microscope by selecting random fields at 20× from both experimental samples and controls. These were imported into ImageJ (v1.45), and the numbers of beads and cells in each image were recorded using the cell counter (ImageJ, menu > Plugins > Analyze > Cell counter).
Confocal microscopy.
To investigate the expression of EaeH in the context of intestinal epithelial cells, ETEC H10407 was grown overnight and diluted 1:100 into fresh Luria broth and added at a multiplicity of infection of ∼1:100 to Caco-2 cells. Caco-2 cells were seeded approximately 48 to 72 h prior to infection onto sterile glass coverslips pretreated with poly-l-lysine. ETEC-infected monolayers were then returned to 37°C and a 5% CO2 atmosphere and removed at various time points for the removal of infected media and washing. After three washes with tissue culture media, the cells were fixed with paraformaldehyde, washed with PBS, and blocked with PBS containing 1% BSA. Affinity-purified anti-EaeH rabbit polyclonal antibody, followed by Alexa Fluor 488-labeled anti-rabbit antibody, was then used to identify organisms that expressed EaeH on the bacterial surface. Intestinal cell membranes were stained as previously described (CellMask, red; Invitrogen) (18). Images were acquired on a Zeiss LSM510 confocal microscope, and files were converted to TIFF image using ImageJ (v1.45).
Scanning electron microscopy (SEM).
Nonconfluent Caco-2 intestinal epithelial cells were seeded onto coverslips pretreated with poly-l-lysine, and grown for 24 to 48 h in a 5% CO2 atmosphere at 37°C. Latex beads (1 or 3 μm in diameter; Sigma) coated with rEaeH were added to the monolayers, followed by incubation for 3 h in tissue culture media at 37°C and 5% CO2. After three gentle washes with PBS, the cells and attached beads were fixed with 0.1% glutaraldehyde in PBS at room temperature for 15 min. Coverslips were washed three times with PBS, followed by subsequent fixation with 3% glutaraldehyde in PBS. The coverslips were then fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.35) for at least 2 h, rinsed (three times for 5 min each time) in 0.1 M cacodylate buffer (pH 7.35), postfixed with 2% osmium tetroxide (pH 7.35; Electron Microscopy Sciences), and rinsed again with 0.1 M cacodylate buffer, followed by rinsing with deionized water. The samples were stained en bloc with 2% aqueous uranyl acetate (Electron Microscopy Sciences), rinsed and dehydrated as previously described in ethyl alcohol (10% through absolute), and dried in a critical-point dryer replacing the ethanol with liquid carbon dioxide. Finally, the samples were sputter coated (EMS 550; Electron Microscopy Sciences) with 60-nm gold palladium. Images were acquired on a scanning electron microscope (Phillips XL30) and saved as tiff files.
Bacterial adherence.
Caco-2 cells were used to examine ETEC-host interactions. As described previously (16), adherence assays were performed using semiconfluent Caco-2 cell monolayers seeded into 96-well tissue culture plates the evening prior to the experiment. Bacteria to be tested were grown overnight in Luria broth with antibiotics as appropriate, diluted 1:100 the morning of the experiment, and then grown to approximately mid-logarithmic phase prior to the addition to the monolayers. For adherence assays, infected monolayers were incubated at 37°C and 5% CO2 for 1 h, washed with tissue culture medium four times, and then lysed in 0.1% Triton X-100 for 5 min, and the bacteria were recovered by plating lysates onto Luria agar.
Heat-labile toxin delivery assays.
To examine the role of eaeH in delivery of heat-labile toxin, ETEC H10407 or mutants were used to infect Caco-2 cells grown to semiconfluence in 96-well plates. After 3 h, the plates were washed with prewarmed tissue culture media, incubated for an additional 2.5 h, and then lysed and processed for cAMP determination by using an enzyme-linked immunosorbent assay (Arbor Assays) as previously described (18, 22).
Intestinal colonization studies.
To investigate the role of EaeH in promoting intestinal colonization, mice were challenged with ETEC as previously described (42). Briefly, after treatment with streptomycin to eliminate competing flora and famotidine to reduce gastric acidity, CD-1 mice (Charles River) were challenged with strain jf876 (lacZYA::Kmr) or strain jf2852 (eaeH::Kmr). To determine the number of bacteria directly attached to the intestinal villi, sections of small intestine were preserved in 10% formalin and embedded in paraffin, and histologic sections were then processed for confocal immunofluorescence microscopy at the Digestive Diseases Research Core Center (DDRCC) at the Washington University School of Medicine. Competition assays were performed as previously described (19). Briefly, mice were challenged with ∼105 CFU of jf876 and an equal number of jf2852 in a final volume of 400 μl. After 24 h, the mice were sacrificed, and saponin lysates of small intestine were plated onto agar plates containing kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The numbers of eaeH mutant bacterial colonies (blue) and eaeH wild-type colonies (white) were used to calculate the competitive index (CI) for each mouse as follows: CI = [mutant (blue)/wild type (white)]output CFU/(mutant/wild type)input CFU, using the input fraction determined at the time of inoculum preparation.
RESULTS
EaeH encodes a surface molecule with features shared by bacterial adhesins and invasins.
The eaeH gene is predicted to encode an outer membrane protein with a putative signal sequence at its amino-terminal end (34, 35), followed by the mature peptide which shares a region of homology with intimin (EaeA) that corresponds to a transmembrane β-barrel (Fig. 1a) (43). Another feature shared with intimin (44), invasin (45), and a diverse superfamily of bacterial virulence proteins (46) is a series of tandem bacterial immunoglobulin-like (BIg) domains similar to those involved in eukaryotic cell surface adhesion proteins such as ICAMs (47). Partial structural characterization of FdeC (48), the EaeH homologue from uropathogenic E. coli, appears to be consistent with this predicted molecular organization.
FIG 1.
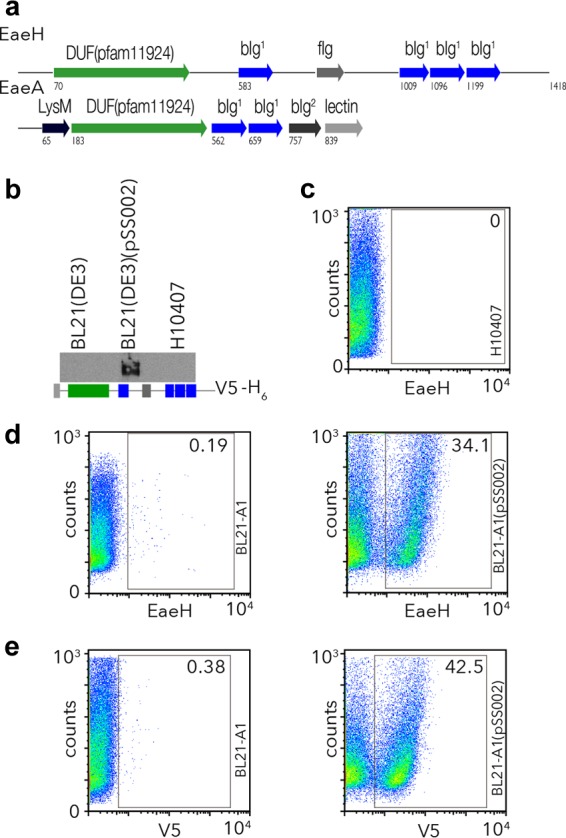
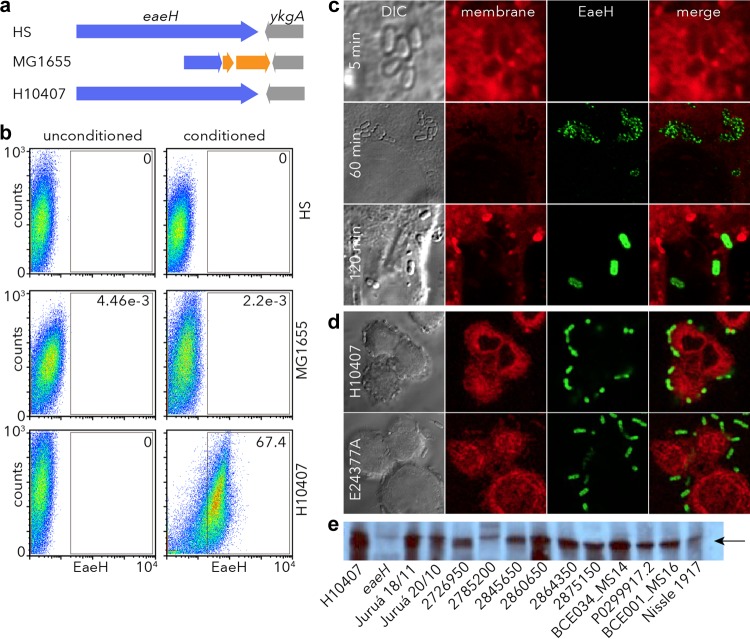
ETEC eaeH encodes a putative surface-expressed protein. (a) Predicted structural features with surface-expressed bacterial virulence molecules. Schematic representation of the EaeH protein from ETEC H10407 (33) (accession GI:71979957 [http://www.ncbi.nlm.nih.gov/protein/AAZ57201.1]) compared to the prototypical intimin (EaeA) molecule (56, 57) from EPEC strain E2348/69 accession GI:229462841 (http://www.ncbi.nlm.nih.gov/protein/229462841) depicts the overall domain structure with multiple bacteria immunoglobulin-like domains (BIg), similar to intimin, as well as an Ig-like fold similar to filamin (fIg). Intimin also contains a carboxy-terminal C-type lectin domain and an amino LysM feature pfam01476 (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam01476), a putative peptidoglycan-binding domain. (b) An anti-EaeH immunoblot is shown for outer membrane preparations from the BL21 expression host strain, BL21AI; the recombinant strain BL21AI(pSS002); and the ETEC H10407 strain grown in Luria broth. The schematic depicts the recombinant EaeH-V5-6His molecule cloned in pSS002 and expressed in BL21AI. (c) EaeH surface expression by ETEC H10407 grown in L broth is minimal by anti-EaeH flow cytometry. (d) EaeH surface expression in recombinant strain compared to control, BL21AI. (e) C-terminal V5 epitope is exposed on the surface of the recombinant, BL21AI(pSS002).
To examine the surface expression of EaeH in ETEC, we cloned the full-length eaeH gene from ETEC H10407 in frame with V5 and His6 polyhistidine tags and expressed it in a recombinant E. coli background (Fig. 1b). Polyclonal antisera raised against EaeH demonstrated the presence of the protein in the outer membrane of the recombinant but not in controls from the expression strain or in ETEC H10407, suggesting that when grown in Luria broth the expression of EaeH by ETEC is poor. Similarly, the recombinant protein could easily be identified by flow cytometry on the surface of BL21AI carrying the EaeH-V5-6His expression plasmid, using either polyclonal anti-EaeH antibody or with V5 monoclonal antibody, but we were not able to identify significant amounts of EaeH on the surface of wild-type ETEC when grown in Luria broth (Fig. 1c to e). Together, these results suggest that whereas eaeH encodes a potential surface-expressed protein, it is poorly expressed when ETEC is grown in Luria broth.
Surface expression of EaeH is induced on pathogen-epithelial cell interactions.
Recent transcriptome analyses of ETEC in contact with host epithelial cells suggested that eaeH expression is activated by pathogen-host interactions. Consequently, we examined the surface expression of the EaeH protein in ETEC and in nonpathogenic E. coli strains (Fig. 2a) after exposure to media conditioned by intestinal epithelial cells. Interestingly, we found that media conditioned by cultured Caco-2 cells was sufficient to induce the surface expression of EaeH in ETEC (Fig. 2b). In contrast, we could not identify EaeH on the surfaces of either strain MG1655 in which the eaeH gene has been interrupted by an insertion sequence or in the commensal E. coli strain HS, in which the gene appears to be intact (Table 3), suggesting that ETEC may be uniquely programmed to respond to host cells by expressing EaeH in the outer membrane.
FIG 2.
Intestinal epithelial cells induce EaeH surface expression in ETEC. (a) eaeH genes in the chromosomes of the commensal E. coli HS strain, the MG1655 E. coli K-12 strain (interrupted by an IS element (orange), and ETEC strain H10407. (b) Flow cytometry data demonstrating surface expression of EaeH in tissue culture media and tissue culture media conditioned by growth of Caco-2 intestinal epithelial cells. (c) Time course demonstrating enhanced EaeH production after H10407 attachment to target epithelial cells. The top panel demonstrates the lack of discernible EaeH expression by ETEC 5 min after attachment to Caco-2 cells, whereas after 60 min (middle row) EaeH is detectable on the bacterial surface, and maximal expression is apparent at 120 min (bottom row). (d) EaeH expression by ETEC strains H10407 and E24377A at 120 min after addition to epithelial cells. Panels from left to right depict DIC images, membrane stains (red), anti-EaeH antibody detected with Alexa Fluor 488 secondary conjugate (green), and merged membrane-EaeH images. (e) EaeH immunoblotting of E. coli bacterial lysates following growth in conditioned media. On the left are shown H10407 and the eaeH mutant, as positive and negative controls, respectively. ETEC strains were selected to represent different phylogenetic groups based on whole-genome sequencing (http://gscid.igs.umaryland.edu/wp.php?wp=comparative_genome_analysis_of_enterotoxigenic_e._coli_isolates_from_infections_of_different_clinical_severity). The Nissle 1917 nonpathogenic commensal isolate is shown on the right. The arrow indicates the predicted migration of EaeH.
TABLE 3.
Comparison of eaeH genes in pathogens and commensal E. colia
| Strain | Pathotype | NCBI protein reference | RAST designation | Gene length (bp) | % Identical | % Similar |
|---|---|---|---|---|---|---|
| H10407 | ETEC | AAZ57201.1 | .peg.385 | 4,257 | ||
| E24377A | ETEC | YP_001461465 | .peg.2865 | 4,254 | 92 | 95 |
| B7A | ETEC | EDV63917.1 | .peg.780 | 4,254 | 92 | 95 |
| MG1655 | K-12 | AAB18025.1 | .peg.302 | 888 | 100* | 100* |
| HS | Commensal | YP_001457122.1 | .peg.327 | 4,254 | 91 | 94 |
| Nissle 1917 | Commensal | CCQ06761 | 4,251 | 94 | 96 | |
| UTI89 | UPEC | YP_539353.1 | .peg.433 | 4,251 | 94 | 96 |
| CFT073 | UPEC | NP_752352.1 | .peg.396 | 4,251 | 94 | 96 |
| TX1999 | EHEC | EGX25301.1 | NA | 4,254 | 99 | 98 |
| EDL933 | EHEC | NP_286024.1 | .peg.332 | 4,251 | 93 | 95 |
RAST, Rapid Annotation using Subsytem Technology (63). NA, not applicable. *, The MG1655 (pseudo)gene is truncated by an IS element; the BLAST-P results are thus based on first 292/296 amino acids.
Earlier transcriptome studies demonstrated that eaeH expression is increased following epithelial cell contact (21). Accordingly, we found that shortly after attachment to host cells (5 min, Fig. 2c) no EaeH could be identified on the surface ETEC strain H10407 attached to the surface of Caco-2 cells, whereas at later time points (60 and 120 min), adherent bacteria were found to express significant amounts of EaeH on their surfaces. Because eaeH appears to be highly conserved in ETEC genomes sequenced to date, we examined whether we could also identify EaeH on the surfaces of ETEC strain E24377A and other ETEC strains either following epithelial cell contact (Fig. 2d) or after growth in conditioned media (Fig. 2e). These studies demonstrated that both eaeH gene and its expression are conserved in ETEC.
EaeH promotes interactions with intestinal epithelial cells.
Based on its structural similarity to known bacterial adhesins and invasins, EaeH might be predicted to function in a similar fashion. After introduction of recombinant EaeH (rEaeH) to cultured intestinal epithelial cells, we observed rEaeH decorating the cellular membrane (Fig. 3a). Likewise, coating latex beads with rEaeH strongly promoted interaction with Caco-2 intestinal epithelial cells compared to control BSA-coated beads (Fig. 3b to e). SEM demonstrated smaller EaeH-coated beads (1 μm) bound to the surfaces of cells, and some appeared to be partially engulfed by the host cell (Fig. 3f), whereas larger (3 μm) beads appeared to be actively engaged by cellular protrusions. Together, these data suggested that EaeH is sufficient to promote epithelial cell interactions.
FIG 3.
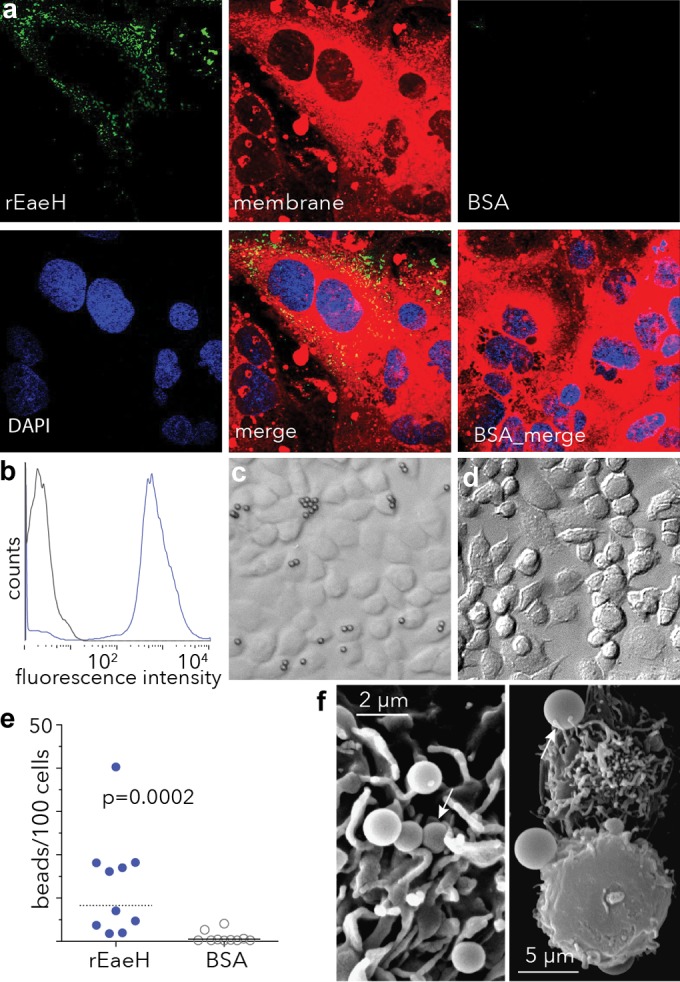
EaeH interacts with the surfaces of intestinal epithelial cells. (a) Binding of rEaeH to the surface of Caco-2 intestinal epithelial cells. Confocal images of rEaeH detected with affinity-purified anti-EaeH polyclonal antibody and secondary AF488 conjugate (green), membrane (CellMask, red), and nuclei stained with DAPI (blue) are shown. Control samples (BSA) processed in an identical fashion are shown on the right. (b) rEaeH-V5-6His bound to the surfaces of latex beads. Beads coated with rEaeH or control beads were analyzed by flow cytometry to confirm binding of EaeH to the bead surface. (c) rEaeH-coated beads in a DIC microscopic image (×20). (d) Control (BSA-coated) beads. (e) Numbers of beads per 100 cells after incubation of Caco-2 cells. The values in panel e were compared using two-tailed nonparametric (Mann-Whitney) testing. (f) SEM image of 1-μm rEaeH-coated latex beads binding to the Caco-2 epithelial cell surface (×10,260) (left panel); SEM image of 3-μm EaeH-coated beads binding to adjacent Caco-2 cells (×3,200) (right panel). Arrows indicate beads either partially engulfed by cells or engaged by cellular processes after attachment.
Similarly, compared to wild-type ETEC, an isogenic eaeH mutant was deficient in adhesion to intestinal epithelial cells in vitro (Fig. 4a). Likewise, affinity-purified anti-EaeH antisera inhibited bacteria adhesion in vitro (Fig. 4b), further suggesting that this highly conserved surface protein plays a role in ETEC pathogen-host interactions. Earlier studies demonstrated that physical contact of ETEC with epithelial cells is required for effective delivery of heat-labile toxin (LT) (49). Theoretically, genes that promote interaction with the intestinal epithelium might also impact the delivery of LT. In accord with other recently described virulence factors that affect epithelial cell adhesion (18, 22), eaeH mutants were impaired in delivery of heat-labile toxin, and antibodies against this surface protein significantly inhibited bacterial activation of cAMP in target epithelial cells (Fig. 4c) in vitro.
FIG 4.
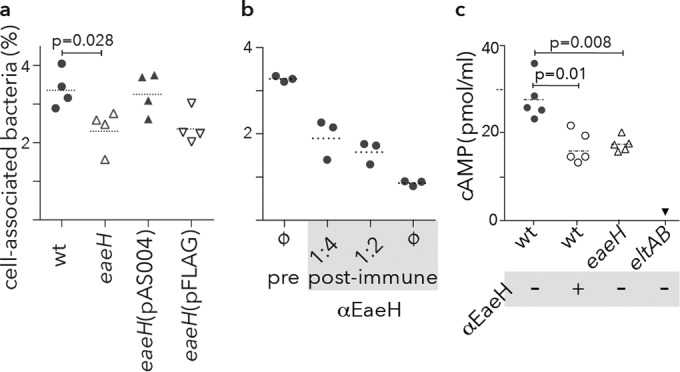
Role of eaeH in bacterial adhesion and toxin delivery. (a) The eaeH gene is required for optimal adhesion to target intestinal epithelial cells in vitro. (b) Antibodies raised against EaeH inhibit adhesion. Protein A-purified polyclonal antibodies from either preimmune rabbit sera (pre) or postimmune antisera (post-immune) raised against recombinant EaeH-V5-6His in the indicated amounts were added to Caco-2 monolayers with ETEC H10407. The percentage of cell-associated bacteria reflects the proportion of bacteria associated with the monolayer at the end of 1 h relative to the inoculum. Dashed horizontal bars represent the geometric mean. (c) EaeH is required for optimal delivery of heat-labile toxin. cAMP production by Caco-2 epithelial cell monolayers after infection with either wild-type (wt) or eaeH mutant bacteria. αEaeH refers to the addition of anti-EaeH antibody. A mutant strain with a deletion of the genes encoding heat-labile toxin (eltAB) is shown as a negative control.
To examine the potential role of EaeH in intestinal colonization, mice were challenged with wild-type and eaeH mutant bacteria. In competition assays, in which the total bacteria residing in lumen of the small intestine were plated onto selective media, we did not observe a phenotypic difference between wild-type bacteria and the eaeH mutant (Fig. 5a). However, immunofluorescence (anti-O78) microscopic examination of intestinal mucosa of mice infected with either wild-type or the eaeH mutant identified significantly more wild-type bacteria attached to the epithelial surface than eaeH mutants (Fig. 5b and c), a finding consistent with the hypothesis that this outer membrane protein is activated by and participates in intimate interactions of ETEC with the intestinal epithelium. Collectively, these data suggest that EaeH could play an important role in the pathogenesis of ETEC infections.
FIG 5.
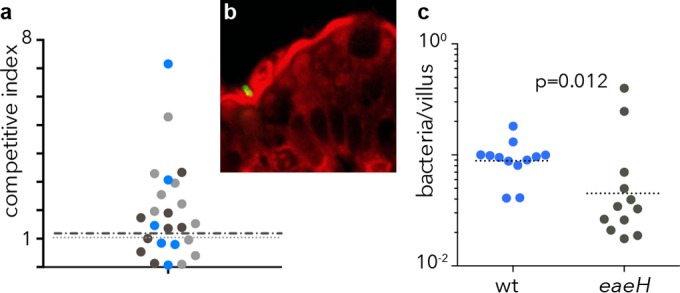
Impact of EaeH on overall colonization and interaction with intestinal epithelial cells in vivo. (a) Competition assay studies between the jf2852 eaeH::Kmr mutant and jf876 (lacZYA::Kmr) wild-type bacteria. The data points represent competitive index values for individual mice from three replicate experiments shown in different colors. The dashed line represents the geometric mean. (b) ETEC H10407 (O78:H11) identified by anti-O78 immunofluorescence staining (green) adherent to the intestinal surface after oral challenge. (c) Wild-type (wt) H10407 and eaeH mutant bacteria attached to the epithelial surface (as shown in panel b). The data represent analyses of three independent sections of the small intestine from four mice in each group (P = 0.012 by two-tailed Mann-Whitney comparison).
DISCUSSION
The enterotoxigenic Escherichia coli pathotype is diverse. At a minimum, these organisms share in the ability to produce and effectively deliver heat-labile and/or heat-stable enterotoxins to their cognate receptors on intestinal epithelial cells. Shortly after the discovery of these organisms in patients presenting with cholera-like diarrhea (50, 51), the first plasmid-encoded fimbrial colonization factors (CFs) were described (10, 52). Over the past 40 years, at least 26 different CFs have been described (13, 28), suggesting that ETEC can use a diverse repertoire of these structures in its colonization of the small intestine.
These antigens constitute the basis for current vaccine strategies that attempt to encompass the most prevalent CFs (14, 15, 26). However, the antigenic diversity of the CFs and the possibility that other molecules may participate in intestinal colonization and toxin delivery have prompted efforts to define additional target antigens that might augment existing approaches. Recent studies provide compelling evidence that the molecular pathogenesis of ETEC is more complex than had been appreciated previously and that a number of novel surface or secreted virulence proteins could serve as viable targets for future development efforts (53).
The eaeH gene encoding one of these molecules was originally identified on the chromosome of ETEC using subtractive hybridization of the prototype H10407 strain and E. coli K-12 (MG1655) (33). More recent attempts to identify novel vaccine candidates by investigation of antigens that are transcriptionally modulated during pathogen-host interactions also highlighted eaeH, since it was strongly upregulated after contact of ETEC with intestinal epithelial cells (21).
The results presented here reaffirm these findings and demonstrate the expression of EaeH by a variety of other ETEC strains. Moreover, we show that this highly conserved antigen plays a role in both epithelial cell adhesion in vitro and intestinal colonization in vivo.
To effectively deliver toxin, ETEC theoretically must find its way to the small intestine, traverse the protective layer of mucin in the intestinal lumen (37), and ultimately engage the epithelial cell. Previous studies suggest that intimate interaction of ETEC with target cells is required for effective uptake of heat-labile toxin and consequent activation of cyclic nucleotides (49). Interestingly, in the present study, eaeH did not appear to impact overall intestinal colonization (which measures both bacteria sequestered in the intestinal lumen and those that are physically attached to the epithelial surface). However, we found significantly fewer eaeH mutant bacteria directly attached to intestinal epithelial cells, suggesting that this gene is required at a later step in ETEC-host interactions. These findings are consistent with in vitro studies suggesting that the activation of eaeH is a late event relative to the activity of other potential adhesins, including the plasmid-encoded CFs, type 1 fimbriae, and the secreted EtpA adhesin (21). Collectively, the emerging data suggest that both bacterial adhesion and intestinal colonization are very complex events that involve interactions through fimbrial structures such as the CFs, secreted proteins such as EtpA, and more intimate connections to epithelial cells via EaeH, a highly conserved E. coli membrane protein.
Although eaeH was originally identified in ETEC, it is shared by other E. coli pathotypes, including strains associated with extraintestinal infections (33, 54, 55), including uropathogenic E. coli (48). Because E. coli is a normal component of the intestinal flora, targeting highly conserved antigens that are shared with some commensal isolates in vaccines could in theory have untoward consequences. Our studies failed to detect EaeH in the commensal HS strain under conditions where expression was robust in ETEC and demonstrated limited production of EaeH in Nissle 1917. Nevertheless, additional study will be required to determine whether this can be generalized to other commensal isolates, preferably those which have not undergone serial passage in multiple laboratories over many years.
ACKNOWLEDGMENTS
We thank Lou Boykins of the Integrated Microscopy Center at the University of Memphis for assistance with the SEM and Michael Whitt of the Department of Microbiology, Immunology, and Biochemistry at the University of Tennessee Health Sciences Center for assistance with the confocal microscopy.
This study was supported by grant 1R01AI089894-01 from the National Institute of Allergy and Infectious Diseases (J.M.F.), Merit Review funding from the Department of Veterans Affairs (J.M.F.), The Bill and Melinda Gates Foundation (OPP1099494) (J.M.F.), and grant P30DK052574 from the National Institute of Diabetes and Digestive and Kidney Disease to support the DDRCC.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health, the Department of Veterans Affairs, or any other funding agencies.
Footnotes
Published ahead of print 16 June 2014
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 2.Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm AM. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect. Immun. 75:3961–3968. 10.1128/IAI.00459-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qadri F, Svennerholm AM, Faruque AS, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465–483. 10.1128/CMR.18.3.465-483.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N, DuPont HL, Ramsey DJ. 2009. Global etiology of travelers' diarrhea: systematic review from 1973 to the present. Am. J. Trop. Med. Hyg. 80:609–614 [PubMed] [Google Scholar]
- 5.Vaandrager AB, Smolenski A, Tilly BC, Houtsmuller AB, Ehlert EM, Bot AG, Edixhoven M, Boomaars WE, Lohmann SM, de Jonge HR. 1998. Membrane targeting of cGMP-dependent protein kinase is required for cystic fibrosis transmembrane conductance regulator Cl− channel activation. Proc. Natl. Acad. Sci. U. S. A. 95:1466–1471. 10.1073/pnas.95.4.1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao AC, de Sauvage FJ, Dong YJ, Wagner JA, Goeddel DV, Gardner P. 1994. Activation of intestinal CFTR Cl− channel by heat-stable enterotoxin and guanylin via cAMP-dependent protein kinase. EMBO J. 13:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. 1991. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 66:1027–1036. 10.1016/0092-8674(91)90446-6 [DOI] [PubMed] [Google Scholar]
- 8.Yun CH, Oh S, Zizak M, Steplock D, Tsao S, Tse CM, Weinman EJ, Donowitz M. 1997. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 94:3010–3015. 10.1073/pnas.94.7.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89–98. 10.1016/j.micinf.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DJ, Jr, Evans DG. 1973. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect. Immun. 8:322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu XQ, Savarino SJ, Bullitt E. 2008. The three-dimensional structure of CFA/I adhesion pili: traveler's diarrhea bacteria hang on by a spring. J. Mol. Biol. 376:614–620. 10.1016/j.jmb.2007.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YF, Poole S, Nishio K, Jang K, Rasulova F, McVeigh A, Savarino SJ, Xia D, Bullitt E. 2009. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:10793–10798. 10.1073/pnas.0812843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isidean SD, Riddle MS, Savarino SJ, Porter CK. 2011. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29:6167–6178. 10.1016/j.vaccine.2011.06.084 [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Sack DA. 2012. Progress and hurdles in the development of vaccines against enterotoxigenic Escherichia coli in humans. Expert review of vaccines 11:677–694. 10.1586/erv.12.37 [DOI] [PubMed] [Google Scholar]
- 15.Svennerholm AM, Lundgren A. 2012. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert review of vaccines 11:495–507. 10.1586/erv.12.12 [DOI] [PubMed] [Google Scholar]
- 16.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245–2258. 10.1128/IAI.74.4.2245-2258.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786–1794. 10.1128/IAI.72.3.1786-1794.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J. Biol. Chem. 286:29771–29779. 10.1074/jbc.M111.251546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy K, Hilliard GM, Hamilton DJ, Luo J, Ostmann MM, Fleckenstein JM. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598. 10.1038/nature07568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76:2106–2112. 10.1128/IAI.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kansal R, Rasko DA, Sahl JW, Munson GP, Roy K, Luo Q, Sheikh A, Kuhne KJ, Fleckenstein JM. 2013. Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect. Immun. 81:259–270. 10.1128/IAI.00919-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy K, Hamilton DJ, Fleckenstein JM. 2012. Cooperative role of antibodies against heat-labile toxin and the EtpA Adhesin in preventing toxin delivery and intestinal colonization by enterotoxigenic Escherichia coli. Clin. Vaccine Immunol. 19:1603–1608. 10.1128/CVI.00351-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608. 10.1016/j.vaccine.2009.05.076 [DOI] [PubMed] [Google Scholar]
- 24.Boedeker EC. 2005. Vaccines for enterotoxigenic Escherichia coli: current status. Curr. Opin. Gastroenterol. 21:15–19 [PubMed] [Google Scholar]
- 25.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmgren J, Bourgeois L, Carlin N, Clements J, Gustafsson B, Lundgren A, Nygren E, Tobias J, Walker R, Svennerholm AM. 2013. Development and preclinical evaluation of safety and immunogenicity of an oral ETEC vaccine containing inactivated Escherichia coli bacteria overexpressing colonization factors CFA/I, CS3, CS5, and CS6 combined with a hybrid LT/CT B subunit antigen, administered alone and together with dmLT adjuvant. Vaccine 31:2457–2464. 10.1016/j.vaccine.2013.03.027 [DOI] [PubMed] [Google Scholar]
- 27.Shaheen HI, Kamal KA, Wasfy MO, El-Ghorab NM, Lowe B, Steffen R, Kodkani N, Amsler L, Waiyaki P, David JC, Khalil SB, Peruski LF., Jr 2003. Phenotypic diversity of enterotoxigenic Escherichia coli (ETEC) isolated from cases of travelers' diarrhea in Kenya. Int. J. Infect. Dis. 7:35–38. 10.1016/S1201-9712(03)90040-3 [DOI] [PubMed] [Google Scholar]
- 28.Del Canto F, Botkin DJ, Valenzuela P, Popov V, Ruiz-Perez F, Nataro JP, Levine MM, Stine OC, Pop M, Torres AG, Vidal R. 2012. Identification of the coli surface antigen 23 (CS23), a novel adhesin of enterotoxigenic Escherichia coli. Infect. Immun. 80:2791–2801. 10.1128/IAI.00263-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinsland H, Valentiner-Branth P, Gjessing HK, Aaby P, Molbak K, Sommerfelt H. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286–291. 10.1016/S0140-6736(03)13971-2 [DOI] [PubMed] [Google Scholar]
- 30.Cravioto A, Reyes RE, Trujillo F, Uribe F, Navarro A, De La Roca JM, Hernandez JM, Perez G, Vazquez V. 1990. Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens. Am. J. Epidemiol. 131:886–904 [DOI] [PubMed] [Google Scholar]
- 31.Steinsland H, Valentiner-Branth P, Aaby P, Molbak K, Sommerfelt H. 2004. Clonal relatedness of enterotoxigenic Escherichia coli strains isolated from a cohort of young children in Guinea-Bissau. J. Clin. Microbiol. 42:3100–3107. 10.1128/JCM.42.7.3100-3107.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darsley MJ, Chakraborty S, Denearing B, Sack DA, Feller A, Buchwaldt C, Bourgeois AL, Walker R, Harro CD. 2012. The oral, live attenuated ETEC vaccine ACE527 reduces the incidence and severity of diarrhea in a human challenge model of diarrheal disease. Clin. Vaccine Immunol. 19:1921–1931. 10.1128/CVI.00364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Savarino SJ, Venkatesan MM. 2006. Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152:1041–1054. 10.1099/mic.0.28648-0 [DOI] [PubMed] [Google Scholar]
- 34.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 35.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6:e1000902. 10.1371/journal.ppat.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blomfield IC, McClain MS, Eisenstein BI. 1991. Type 1 fimbriae mutants of Escherichia coli K-12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439–1445. 10.1111/j.1365-2958.1991.tb00790.x [DOI] [PubMed] [Google Scholar]
- 39.Harlow E, Lane D. 1999. Handling antibodies, p 63–80 Using antibodies: a laboratory manual, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40.Gillen CM, Courtney HS, Schulze K, Rohde M, Wilson MR, Timmer AM, Guzman CA, Nizet V, Chhatwal GS, Walker MJ. 2008. Opacity factor activity and epithelial cell binding by the serum opacity factor protein of Streptococcus pyogenes are functionally discrete. J. Biol. Chem. 283:6359–6366. 10.1074/jbc.M706739200 [DOI] [PubMed] [Google Scholar]
- 41.Dombek PE, Cue D, Sedgewick J, Lam H, Ruschkowski S, Finlay BB, Cleary PP. 1999. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol. Microbiol. 31:859–870. 10.1046/j.1365-2958.1999.01223.x [DOI] [PubMed] [Google Scholar]
- 42.Allen KP, Randolph MM, Fleckenstein JM. 2006. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74:869–875. 10.1128/IAI.74.2.869-875.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Touze T, Hayward RD, Eswaran J, Leong JM, Koronakis V. 2004. Self-association of EPEC intimin mediated by the beta-barrel-containing anchor domain: a role in clustering of the Tir receptor. Mol. Microbiol. 51:73–87. 10.1046/j.1365-2958.2003.03830.x [DOI] [PubMed] [Google Scholar]
- 44.Kelly G, Prasannan S, Daniell S, Fleming K, Frankel G, Dougan G, Connerton I, Matthews S. 1999. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6:313–318. 10.1038/7545 [DOI] [PubMed] [Google Scholar]
- 45.Dersch P, Isberg RR. 2000. An immunoglobulin superfamily-like domain unique to the Yersinia pseudotuberculosis invasin protein is required for stimulation of bacterial uptake via integrin receptors. Infect. Immun. 68:2930–2938. 10.1128/IAI.68.5.2930-2938.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, Ko AI. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929–945. 10.1046/j.1365-2958.2003.03619.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staunton DE, Marlin SD, Stratowa C, Dustin ML, Springer TA. 1988. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell 52:925–933. 10.1016/0092-8674(88)90434-5 [DOI] [PubMed] [Google Scholar]
- 48.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010–12. 10.1128/mBio.00010-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorsey FC, Fischer JF, Fleckenstein JM. 2006. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell Microbiol. 8:1516–1527. 10.1111/j.1462-5822.2006.00736.x [DOI] [PubMed] [Google Scholar]
- 50.Sack RB. 2011. The discovery of cholera-like enterotoxins produced by Escherichia coli causing secretory diarrhoea in humans. Indian J. Med. Res. 133:171–180 [PMC free article] [PubMed] [Google Scholar]
- 51.Sack RB, Gorbach SL, Banwell JG, Jacobs B, Chatterjee BD, Mitra RC. 1971. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J. Infect. Dis. 123:378–385. 10.1093/infdis/123.4.378 [DOI] [PubMed] [Google Scholar]
- 52.Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleckenstein JM, Munson GM, Rasko D. 2013. Enterotoxigenic Escherichia coli: orchestrated host engagement. Gut Microbes 4:392–396. 10.4161/gmic.25861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukiya S, Mizoguchi H, Tobe T, Mori H. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 186:3911–3921. 10.1128/JB.186.12.3911-3921.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077. 10.1073/pnas.0915077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jerse AE, Kaper JB. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843. 10.1073/pnas.87.20.7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements ML, Cheney C. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vicente AC, Teixeira LF, Iniguez-Rojas L, Luna MG, Silva L, Andrade JR, Guth BE. 2005. Outbreaks of cholera-like diarrhoea caused by enterotoxigenic Escherichia coli in the Brazilian Amazon Rainforest. Trans. R. Soc. Trop. Med. Hyg. 99:669–674. 10.1016/j.trstmh.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 60.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 61.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432–5441. 10.1128/JB.186.16.5432-5441.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hancock V, Dahl M, Klemm P. 2010. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J. Med. Microbiol. 59:392–399. 10.1099/jmm.0.008672-0 [DOI] [PubMed] [Google Scholar]
- 63.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]