Abstract
Both interleukin-17A (IL-17A) and IL-17F are proinflammatory cytokines that have an important role in intestinal homeostasis via receptor signaling. These cytokines have been characterized in chickens, but very little is known about their receptors and their functional activity. We provide here the first description of the sequence analysis, bioactivity, and comparative expression analysis of chicken IL-17RA (chIL-17RA) in chickens infected with Salmonella and Eimeria, two major infectious agents of gastrointestinal diseases of poultry of economic importance. A full-length chIL-17RA cDNA with a 2,568-bp coding region was identified from chicken thymus cDNA. chIL-17RA shares ca. 46% identity with mammalian homologues and 29.2 to 31.5% identity with its piscine counterparts. chIL-17RA transcript expression was relatively high in the thymus and in the chicken macrophage cell line HD11. The chIL-17RA-specific small interfering RNA inhibits interleukin-6 (IL-6), IL-8, and IL-1β mRNA expression in chicken embryo fibroblast cells (but not in DF-1 cells) stimulated with chIL-17A or chIL-17F. Interaction between chIL-17RA and chIL-17A was confirmed by coimmunoprecipitation. Downregulation of chIL-17RA occurred in concanavalin A- or lipopolysaccharide-activated splenic lymphocytes but not in poly(I·C)-activated splenic lymphocytes. In Salmonella- and Eimeria-infected chickens, the expression levels of the chIL-17RA transcript were downregulated in intestinal tissues from chickens infected with two Eimeria species, E. tenella or E. maxima, that preferentially infect the cecum and jejunum, respectively. However, chIL-17RA expression was generally unchanged in Salmonella infection. These results suggest that chIL-17RA has an important role in mucosal immunity to intestinal intracellular parasite infections such as Eimeria infection.
INTRODUCTION
The interleukin-17 (IL-17) cytokine family consists of six similar members (IL-17A, IL-17B, IL-17C, IL-17D, IL-17E [also called IL-25], and IL-17F), which vary in amino acid sequence homology and biological function. IL-17 cytokines as homodimers or heterodimers generally function through engagement of the IL-17 receptor (IL-17R) family, which includes five molecules (IL-17RA, IL-17RB, IL-17RC, IL-17RD, and IL-17RE) (1–3). The heterodimer of IL-17RA and IL-17RB is a receptor for IL-17E. The heterodimer composed of IL-17RA and IL-17RC acts as a receptor for homodimers or heterodimers of IL-17A and IL-17F (3, 4). These single transmembrane domain-containing receptors possess certain conserved structural characteristics, including two extracellular fibronectin III-like domains and a cytoplasmic SEF (similar expression to FGF)/IL-17R (SEFIR) domain. The SEFIR domain of IL-17R interacts with Act1 (NF-κB activator 1). This complex allows the incorporation of TRAF6 to induce the activation of NF-κB, C/EBP, or AP-1 and the production of proinflammatory cytokines (1, 5–7).
IL-17RA was initially identified in the murine EL4 cDNA expression library, which encodes a type 1 transmembrane protein of 864 amino acids, resulting in a 97-kDa transmembrane receptor (8). IL-17RA mRNA is widely expressed in the brain, spleen, lung, liver, and kidney, with particularly high levels in lymphoid tissues (8). IL-17RA mRNA has also been detected in epithelial cells, fibroblasts, B and T lymphocytes, myelomonocytic cells, mesenchymal stem cells, and macrophages (8–11), which suggests that it is involved in a variety of biological activities. IL-17RA expression increases in the lungs of C3H/HeN mice infected with Chlamydia muridarum (12) and in the bladders of bacillus Calmette-Guerin (BCG)-stimulated mice (13). IL-17RA knockout mice have a reduced survival rate after Toxoplasma gondii infection (14) and Klebsiella pneumoniae inoculation (15, 16). In contrast, IL-17RA-deficient mice have an increased survival rate after infections with Toxoplasma gondii (17) or Trypanosoma cruzi (18).
The growing literature on the importance of the IL-17/IL-17R axis in host defense includes many studies that describe the critical role of the IL-17 family of cytokines in avian immune responses (19–22). Three cytokines—IL-17A, IL-17D, and IL-17F—have been identified in an expressed sequence tag (EST) cDNA library prepared from intestinal intraepithelial lymphocytes (IELs) of Eimeria-infected chickens (23), from a testis cDNA library prepared from Korean native chickens (24), and from concanavalin A (ConA)-activated chicken splenic lymphocytes (19), respectively. However, additional IL-17 receptors remain to be identified in this economically important species. In the present study, we described a full-length cDNA encoding a chicken IL-17RA (chIL-17RA) protein, together with expression and bioactivity studies, to gain better insight into its function in chickens. Real-time reverse transcription-PCR (RT-PCR) was used to evaluate expression profiles of chIL-17RA in chickens infected with two distinct gastrointestinal infectious organisms, Salmonella and Eimeria.
MATERIALS AND METHODS
Animals and infections.
Male ROSS chicks (Samhwa, Chungnam, South Korea) were given unlimited access to anticoccidial/antibiotic-free feed and water. Constant light was provided for the duration of the experiments. Fifty 10-day-old chicks were infected via the oral route with 104 sporulated Eimeriatenella (Korean isolate 291-7) (25) or E. maxima (Korean isolate 291-3) (26) oocysts and transferred to disposable cages (27). Eimeria spp. present in feces were cleaned by flotation on 5.25% sodium hypochlorite and washed three times with phosphate-buffered saline (PBS). Samples of intestine were collected from five chicks at 0, 1, 4, 7, and 10 days postinfection. Forty 14-day-old chicks were infected via the oral route with 2 × 108 CFU of S. gallinarum or S. Typhimurium. Salmonellae spp. that were grown overnight to stationary phase in Luria-Bertani broth at 37°C with aeration were used for the infection. Samples of liver and spleen were collected from five chicks at 0, 1, 4, and 7 days postinfection. Control chicks received the same volume of PBS. All animal experiments were approved by the Institutional Animal Care and Use Committee at Gyeongsang National University, Jinju, Republic of Korea.
Cloning of chicken IL-17RA.
Total RNA was extracted from thymus using RiboEx reagent (Geneall, South Korea) and was treated with RNase-free DNase I (Fermentas, Canada). Single-stranded cDNA was synthesized from total RNA using oligo(dT) primers and a Transcriptor first-strand cDNA synthesis kit (Roche Applied Science, Germany). Based on a predicted cDNA sequence (accession number XM_416389), 5′/3′-rapid amplification of cDNA ends (RACE) was performed using chIL-17RA-specific primers (for 5′ RACE, 5′-CCTCCATGGGTTAAAGCTCA-3′; for 3′ RACE, 5′-GCAGAGCTGATGAACATGA-3′) with thymus cDNA, a high-fidelity DNA polymerase (Bioneer, South Korea), and a 5′/3′ RACE kit (5′/3′ RACE Second Generation; Roche Applied Science) according to the manufacturer's instructions. PCR products were cloned into TA vectors (RBC, Taiwan) and then sequenced (Macrogen, South Korea). A DNA Engine thermocycler (Bio-Rad, USA) was used for the PCR with the following cycles: 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C, with a final 5-min extension at 72°C. The cDNA sequence was submitted to GenBank (accession number KF743557).
Sequence analysis.
Protein identification was performed using the Expert Protein Analysis System (ExPASy [www.expasy.org/tools/]). The signal peptide sequence was predicted using the SignalP 4.1 software program (http://www.cbs.dtu.dk/services/SignalP). Domain structures were predicted using SMART6 (http://smart.embl-heidelberg.de) (28) and DomPred (http://bioinf.cs.ucl.ac.uk/dompred) software (29). Amino acid multiple alignments were generated using CLUSTAL Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo/). Prediction of disulfide bond partners was performed using DiANNA 1.1 software (30). Homology analysis was performed using MatGat software (31).
Cell culture.
Primary splenic lymphocytes from 3-week-old chickens and established chicken cell lines, HD11 macrophages (32), DF-1 fibroblast cells (33), DT40 B cells (34), and CU205 and CU91 reticuloendotheliosis virus (REV)-transformed lymphoblast cell lines (35) were cultured in Dulbecco modified Eagle medium (DMEM; HyClone, USA) supplemented with 10% fetal bovine serum and penicillin-streptomycin (10,000 U/ml; HyClone) at 41°C in 5% CO2. Splenic lymphocytes were resuspended to 5 × 106 cells/ml and stimulated with 25 μg/ml poly(I·C) and 10 μg/ml lipopolysaccharide (LPS from E. coli O111:B4; Sigma-Aldrich) or 10 μg/ml ConA (Amersham Bioscience, Sweden) for 0, 4, 8, 24, 48, or 72 h. Primary chicken embryonic fibroblasts (CEF) were isolated from 11-day-old embryos and cultured as described above.
Plasmid construction and cell transfection.
Full-length chIL-17RA cDNA harboring myc (chIL-17RA-myc) was amplified from single-stranded cDNA harvested from thymus by PCR using the following specific primers (5′-GATCAAGCTTGCTATGGCGGGTGCGGGGCGGCCGGG-3′ and 5′-GATCGAATTCCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCACAGTCCTCTGGAGATGGGCTCAT-3′). Primers contained HindIII and EcoRI restriction enzyme sites (single underline) and myc sequences (boldface). PCR products were digested with HindIII and EcoRI and cloned into the corresponding restriction sites of pcDNA3.1 (Invitrogen, USA). The cells were transiently transfected into COS-7 cells with 10 μg of constructs or empty vector (negative control) using Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Growth media were replaced with serum-free media 5 h posttransfection. Cells were incubated for an additional 24 or 48 h in serum-free DMEM at 37°C in 5% CO2 and, when necessary, treated with peptide-N-glycosidase F (PNGase F; New England BioLabs, USA).
Immunoprecipitation.
COS-7 cells were transfected with chIL-17RA-myc or with chIL-17A (19) using Lipofectamine reagent according to the manufacturer's instructions. Lysates from chIL-17RA-myc-transfected COS-7 were incubated with concentrated supernatant from chIL-17A for 3 h. Anti-myc rabbit antibody (Cell Signaling, USA) and protein A-agarose beads (Cell Signaling) were added, followed by incubation at 4°C overnight. After incubation, the beads were washed five times with nondenaturing lysis buffer, boiled with sample buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Western blot analysis.
The samples were mixed with equal volumes of sample buffer (0.125 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.004% bromophenol blue), heated for 5 min at 95°C, resolved on 10% SDS-polyacrylamide gels, and electroblotted onto polyvinyl difluoride membranes (Bio-Rad). Membranes were blocked with PBS containing 1% nonfat dry milk for 16 h at 4°C. The membranes were then incubated overnight at 4°C with chIL-17A monoclonal antibody (1G8) (27) or anti-myc rabbit antibody (Cell Signaling). After three washes with PBS containing 0.05% Tween 20 (PBS-T), the membranes were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Promega, USA) in PBS containing 1% nonfat dry milk for 40 min at room temperature. Membranes were washed five times with PBS-T, followed by five washes with distilled water, visualized using an enhanced chemiluminescence kit and Western blotting detection reagents (GE Healthcare Life Sciences, USA). They were detected using the ChemiDoc imaging system (Bio-Rad, USA).
RNA interference.
CEF cells were transfected with 100 nM chIL-17RA-specific small interfering RNA (siIL-17RA; data not shown) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Briefly, CEF cells were seeded at a density of 5 × 105 cells per well in six-well plates and transfected with siIL-17RA-Lipofectamine complexes. After 24 h posttransfection, the cells were stimulated with the supernatant of COS-7 cells transfected with chIL-17A or chIL-17F for another 24 h. Total RNA was extracted from the CEF cells using RiboEx reagent (Geneall, South Korea), treated with RNase-free DNase I (Fermentas, Canada) and used for analysis of IL-6, IL-8, IL-1β, and transforming growth factor 1β (TGF-1β) by real-time quantitative PCR. A nontargeting siRNA (siNC) was used as a negative control for non-sequence-specific effects (Bioneer, South Korea; data not shown).
Quantitative real-time RT-PCR.
Normal tissues, mitogen-activated splenic lymphocytes, and tissue samples pooled from five chickens infected with Eimeria and Salmonella spp. were subjected to real-time RT-PCR analysis in triplicate. Random hexamer primers were used for the cDNA synthesis. The real-time RT-PCR was performed using a CFX96 real-time PCR system (Bio-Rad) with SYBR green (Bioneer) and the primers (data not shown). A melting curve was obtained at the end of each run to verify the presence of a single amplification product without primer dimers. Standard curves were generated using serial, 5-fold dilutions of thymus cDNA. The relative expression levels of individual transcripts were quantified with β-actin as a reference for normalization using Bio-Rad CFX software (19).
Statistical analysis.
Data were analyzed using the Student t test or a one-way analysis of variance (ANOVA), followed by Dunnett's multiple-comparison test (InStat software; GraphPad, San Diego, CA). Differences were considered significant at a P value of <0.05. The data were expressed as means ± the standard errors (SE) of triplicate trials from two or three independent experiments.
RESULTS
Cloning and characterization of chIL-17RA cDNA.
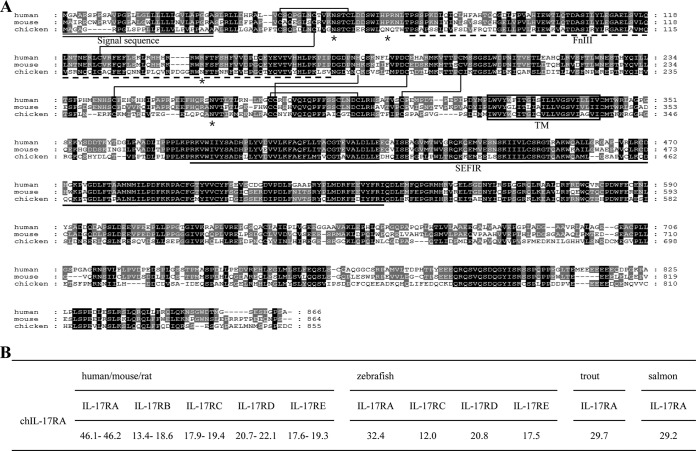
Full-length cDNA of chIL-17RA was first cloned from chicken thymus cDNA. The chIL-17RA cDNA was ∼3.3 kb and contained a 2,568-bp open reading frame predicted to encode a putative 855-amino-acid protein with a calculated molecular mass of 96.58 kDa (nonglycosylated) and an isoelectric point of 4.98. The chIL-17RA amino acid sequence contained a signal sequence (amino acids 1 to 27), an extracellular region (amino acids 28 to 313), a transmembrane region (amino acids 314 to 336), and an extremely long cytoplasmic region (amino acids 337 to 855) (Fig. 1A). Amino acid sequence comparison using MatGat software (31) indicated that chIL-17RA shared ca. 46% identity with its human, mouse, and rat counterparts and 29.2 to 31.5% identity with trout, salmon, and zebrafish. The chIL-17RA shared ca. 12.0 to 22.1% identity with other IL-17R family members.
FIG 1.
Molecular features and amino acid identities of chicken IL-17RA. (A) Multiple alignment of chicken and mammalian IL-17RA deduced amino acid sequences. Sequences were aligned using CLUSTAL W2 software (www.ebi.ac.uk/Tools/msa/clustalw2/). The conserved amino acid sequences are shaded to indicate homology. The predicted signal sequence is indicated with a double underline. The putative transmembrane region is boxed. The putative FnIII domain amino acid and SEFIR domain are indicated with a dashed underline and a bold underline, respectively. The 12 cysteine residues that form six potential disulfide bonds are indicated with a single line. Asterisks (*) indicate four potential N-linked glycosylation sites in the extracellular region. (B) Percent amino acid identities between chicken IL-17RA and mammalian or piscine IL-17Rs. The GenBank accession numbers used in the comparison were KF743557 (chicken), NP_055154 (human IL-17RA), NP_032385 (mouse IL-17RA), NP_001101353 (rat IL-17RA), NP_001117885 (trout IL-17RA), NP_001158836 (salmon IL-17RA), NP_001093473 (zebrafish IL-17RA), NP_061195 (human IL-17RB), NP_062529 (mouse IL-17RB), NP_001100760 (rat IL-17RB), NP_703190 (human IL-17RC), NP_849273 (mouse IL-17RC), NP_001164036 (rat IL-17RC), XP_005169166 (zebrafish IL-17RC-like), NP_060033 (human IL-17RD), NP_602319 (mouse IL-17RD), NP_001178866 (rat IL-17RD), NP_705946 (zebrafish IL-17RD), NP_705613 (human IL-17RE), NP_001004091 (mouse IL-17RE), and XP_002666936 (zebrafish IL-17RE-like).
chIL-17RA contained a putative extracellular fibronectin III-like domain (FnIII) and an intracellular SEFIR domain (Fig. 1A). A multiple alignment with human and mouse revealed conservation of 17 cysteine residues. Twelve of these residues were predicted to be involved in the formation of disulfide bonds by DiANNA software (30). The translated chIL-17RA sequence also contained four potential N-linked glycosylation sites (Asn-X-Ser/Thr) in the extracellular region (Fig. 1A).
Distribution of chIL-17RA mRNA in normal tissues, cell lines, and mitogen-stimulated splenic lymphocytes.
Quantitative RT-PCR analysis was used to examine the expression of chIL-17RA transcripts in various normal tissues and six chicken cell lines (Fig. 2A and B). The expression of chIL-17RA transcripts was detected in all normal tissues and cell lines, including relatively high expression in thymus, HD11, CU205, and CEF. The muscle and CU91 expressed very weak levels of chIL-17RA transcripts. For ConA-, poly(I·C)-, or LPS-stimulated splenic lymphocytes (Fig. 2C), chIL-17RA mRNA transcript was significantly downregulated at all time points in ConA- and LPS-stimulated splenic lymphocytes compared to unactivated and cultured splenic lymphocytes but not in poly(I·C)-stimulated samples.
FIG 2.
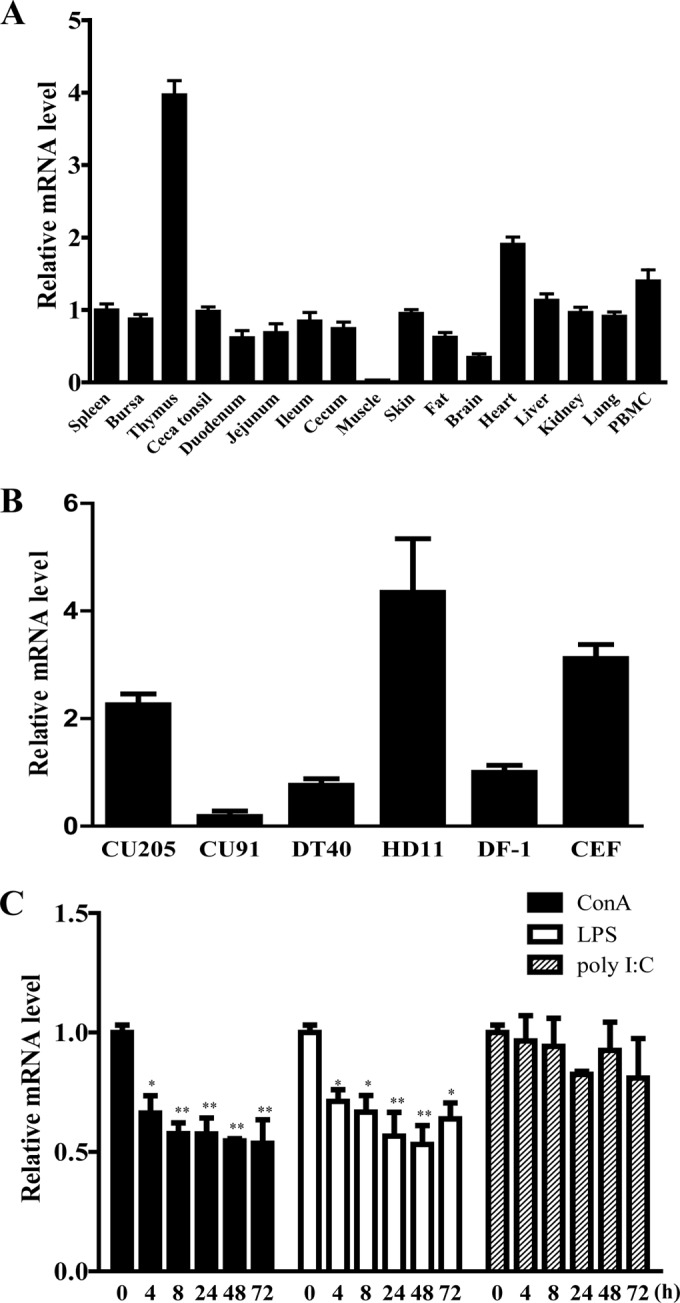
Expression of chIL-17RA mRNA in normal tissues, cell lines, and activated-splenic lymphocytes. (A) Total RNA was extracted from various tissues from 2-week-old healthy chickens. Tissue samples were pooled from five chickens and subjected to quantitative real-time PCR analysis. Gene expression levels were normalized with β-actin and calibrated with spleen tissue expression. (B) Total RNA was extracted from various chicken cell lines and chicken embryonic fibroblasts. CU205 and CU91, T cell lines; DF-1, fibroblast cell line; DT40, B cell line; HD11, macrophage cell line; CEF, chicken embryonic fibroblast. Gene expression levels were normalized with β-actin and calibrated with DF-1 cell expression. (C) Splenic lymphocytes were isolated from 3-week-old chickens, and activated with 10 μg/ml ConA, 10 μg/ml LPS, or 25 μg/ml poly(I·C) for the indicated times. The y axis represents the fold change in expression of chIL-17RA from activated lymphocytes, compared to unactivated and cultured splenic lymphocytes. Expression levels were normalized to β-actin. All data represent the means ± the SE of triplicate trials from two or three independent experiments. P < 0.05 (*) or P < 0.01 (**) were considered to be statistically significant.
Molecular mass of IL-17RA.
Compared to seven or eight N-glycosylation sites in mammalian IL-17RA genes (e.g., human and mouse), the chIL-17RA gene included four N-glycosylation sites in the extracellular region. Thus, molecular masses of chIL-17RA were identified in PNGase F-treated COS-7 cells transfected with a chIL-17RA-myc construct. An 89.7-kDa protein (lower arrow in Fig. 3A; calculated molecular mass of 96.58 kDa) is likely the backbone of chIL-17RA, and the 96.6-kDa protein (upper arrow) represents an N-linked glycosylated form of the protein. The band was confirmed as the chIL-17RA molecule using matrix-assisted laser desorption ionization–time of flight mass spectrometry and analysis of the peptide sequence (see Fig. S1 in the supplemental material).
FIG 3.
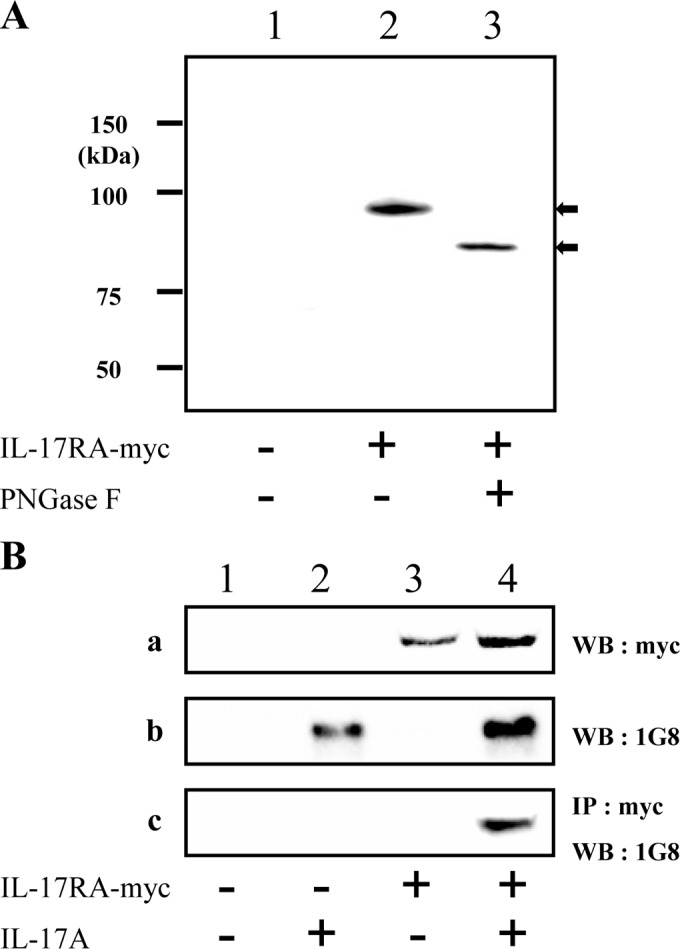
Molecular mass and coimmunoprecipitation of chIL-17RA. (A) Whole-cell lysates of COS-7 cells were collected 48 h (lanes 1 and 2) after transient transfection with chIL-17RA-myc construct (lane 2) or empty pcDNA3.1 (lane 1). To determine the size of the chIL-17RA backbone, cell lysate was incubated with 500 U of PNGase F (lane 3) at 37°C for 1 h. Cell lysates from COS-7 cells were separated using SDS-PAGE under reducing conditions, followed by Western blotting with mouse anti-myc antibody. The data represent two independent experiments with similar pattern results. (B) Interaction between myc-tagged chIL-17RA and chIL-17A. Lysate of COS-7 cells transfected with chIL-17RA-myc construct was incubated with the concentrated supernatant of chIL-17A transfected COS-7, followed by incubation with anti-myc rabbit antibody. The complex was precipitated using protein A-agarose. The precipitate was separated using SDS-PAGE under reducing conditions and immunoblotted with 1G8, a specific antibody for chIL-17A (27) (row c). The nonimmunoprecipitated samples were blotted as loading controls (rows a and b). The data represent three independent experiments with similar results.
Interaction between chIL-17RA and chIL-17A.
We used coimmunoprecipitation experiments to determine whether chIL-17RA interacts with chIL-17A. Lysates of chIL-17RA-myc transfected COS-7 were incubated with the supernatants of chIL-17A-transfected COS-7 and analyzed by Western blotting. The observed interaction did not appear to be due to nonspecific binding to protein A-beads, myc proteins, and 1G8 antibody (Fig. 3B, lanes 1, 2, and 3), whereas chIL-17RA eluted together with chIL-17A (Fig. 3B, lane 4), suggesting that chIL-17RA directly interacts with chIL-17A.
Inhibition of cytokine production by chIL-17RA-specific siRNA in CEF.
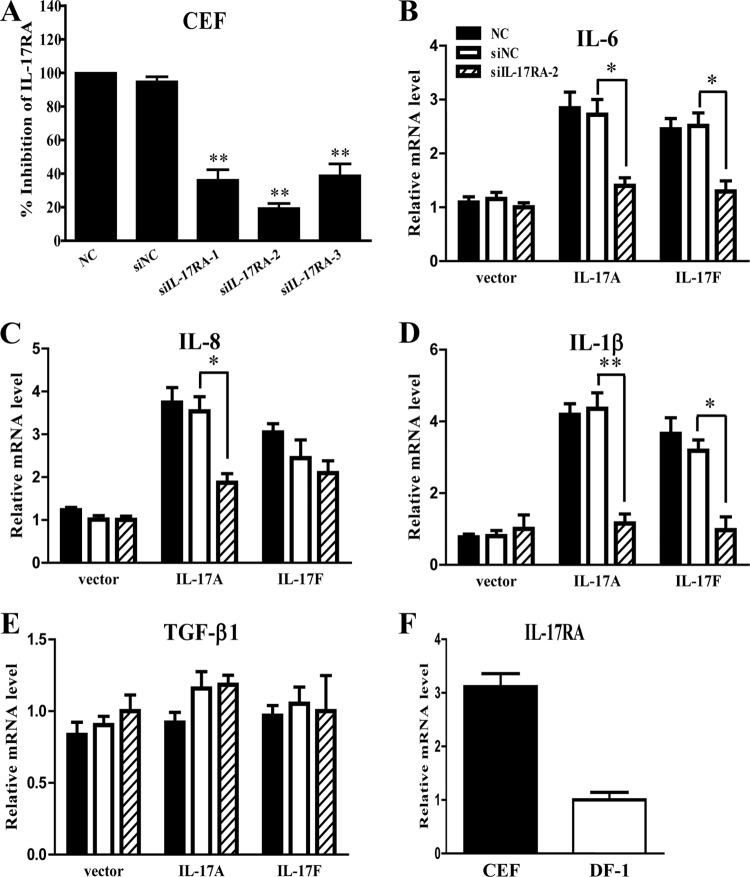
Three small interfering RNA (siRNA) sequences targeting genes within the intracellular region (siIL-17RA-1 and -2) and extracellular region (siIL-17RA-3) of chIL-17RA were synthesized. All siRNA sequences were evaluated for their capacity to inhibit expression of chIL-17RA transcript in primary CEF transfected with siIL-17RA for 24 h by quantitative RT-PCR analysis (Fig. 4A). All siRNA sequences induced a significant reduction of chIL-17RA mRNA expression compared to the nonsense siRNA (siNC) and the nontreated cells (NC) used as a negative control. Inhibition was most efficient with the siIL-17RA-2 molecule, reducing chIL-17RA mRNA expression by up to 80.6% (Fig. 4A).
FIG 4.
Inhibition of cytokine production by chIL-17RA-specific siRNA in CEF. (A) Inhibition of chIL-17RA mRNA expression by chIL-17RA-specific siRNA (siIL-17RA) in CEF cells. CEF cells were transfected with three siIL-17RA or siNC for 24 h and subjected to quantitative real-time PCR analysis. P < 0.01 (**) was considered to be statistically significant (nonsense siRNA [siNC] used as a negative control). (B to E) After chIL-17RA inhibition by siIL-17RA-2 or siNC, CEF cells were stimulated with supernatants of COS-7 cells transfected with vector only, chIL-17A, or chIL-17F plasmid. To measure the biological activity of chIL-17RA, proinflammatory cytokines, IL-6 (B), IL-8 (C), IL-1β (D), and TGF-β1 (E) were analyzed by quantitative real-time PCR. P < 0.05 (*) or P < 0.01 (**) were considered to be statistically significant (nonsense siRNA [siNC] used as a negative control). NC, nontreated CEF cells. Gene expression levels were normalized with β-actin and calibrated with siIL-17RA-2 in vector group. (F) Expression levels of chIL-17RA mRNA in CEF cells and DF-1 cells. All data represent the means ± the SE of triplicate trials from two independent experiments.
To determine the inhibitory effect of the siIL-17RA-2 sequence on the production of IL-6, IL-8 IL-1β, and TGF-β1 (also designated TGF-β4) (36, 37) cytokines, CEF cells transfected with siIL-17RA-2 were stimulated with chIL-17A or chIL-17F for 24 h. Compared to cells transfected with the nonsense siRNA (siNC) used as a negative control, the siIL-17RA-2 in cells stimulated with chIL-17A or chIL-17F significantly inhibited the expression levels of IL-6 and IL-1β mRNA. IL-1β mRNA expression was reduced by 69.6 and 73.6% in chIL-17F- and chIL-17A-stimulated samples, respectively. IL-8 mRNA expression was significantly reduced only in the chIL-17A sample (Fig. 4B to D). No significant difference in expression levels of TGF-β1 mRNA (also designated TGF-β4) was observed both in chIL-17F and chIL-17A samples (Fig. 4E).
The fibroblast cell line DF-1 transfected with siIL-17RA-2 was stimulated with chIL-17A or chIL-17F for 24 h. However, no inhibition was observed in expression levels of IL-6, IL-8 IL-1β, or TGF-β1 mRNA. However, all siRNA sequences induced a significant reduction of chIL-17RA mRNA expression in comparison with siNC or NC used as a negative control (see Fig. S2 in the supplemental material). Therefore, we investigated whether chIL-17RA transcript is differentially expressed between CEF and DF-1 cells. It is interesting that chIL-17RA transcript expression was ∼3.1-fold higher in CEF cells compared to DF-1 cells (Fig. 4F).
Quantitative analysis of chIL-17RA mRNA expression in Salmonella- and Eimeria-infected chickens.
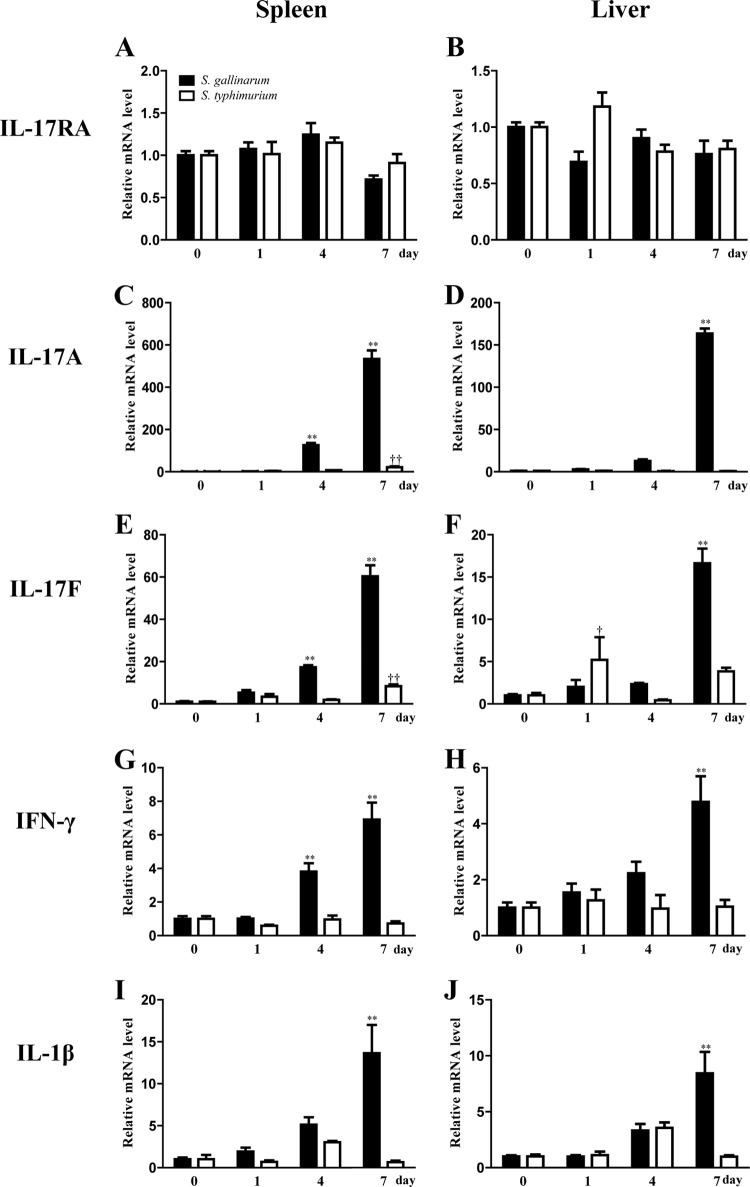
Using quantitative real-time PCR, the expression profiles of chIL-17RA, IL-17A, IL-17F, IFN-γ, and IL-1β were determined in the livers and spleens of chickens infected with S. gallinarum or S. Typhimurium (Fig. 5). Expression profiles were also generated from intestinal tissue samples from chickens infected with two Eimeria species, E. tenella and E. maxima, which preferentially infect the cecum and jejunum, respectively (Fig. 6). Compared to levels in normal spleen and liver, the expression levels of chIL-17RA mRNA were unchanged in the spleens and livers of chickens infected with S. gallinarum or S. Typhimurium. (Fig. 5A and B). Expression levels of the IL-17A, IL-17F, IFN-γ, and IL-1β were generally elevated on day 4 and 7 in the spleen and liver of chickens infected with S. gallinarum, but not S. Typhimurium, which displayed an increased IL-17A and IL-17F expression in the spleen on day 7 postinfection (Fig. 5C to J).
FIG 5.
mRNA expression profiles of chIL-17RA and related cytokines in Salmonella-infected chickens. Fourteen-day-old chicks were infected via the oral route with 2 × 108 CFU of S. gallinarum or S. Typhimurium. The liver and spleen of Salmonellae-infected chickens were collected on days 0, 1, 4, and 7 postinfection. Tissue samples were pooled from five chickens. Expression levels of chIL-17RA and related cytokines were analyzed using quantitative real-time PCR. Gene expression levels were normalized with β-actin and calibrated with expression of uninfected group. All data represent the means ± the SE of triplicate trials from two independent experiments. **, P < 0.01 (S. gallinarum-infected chickens compared to uninfected chickens); ††, P < 0.01 (S. Typhimurium-infected chickens compared to uninfected chickens).
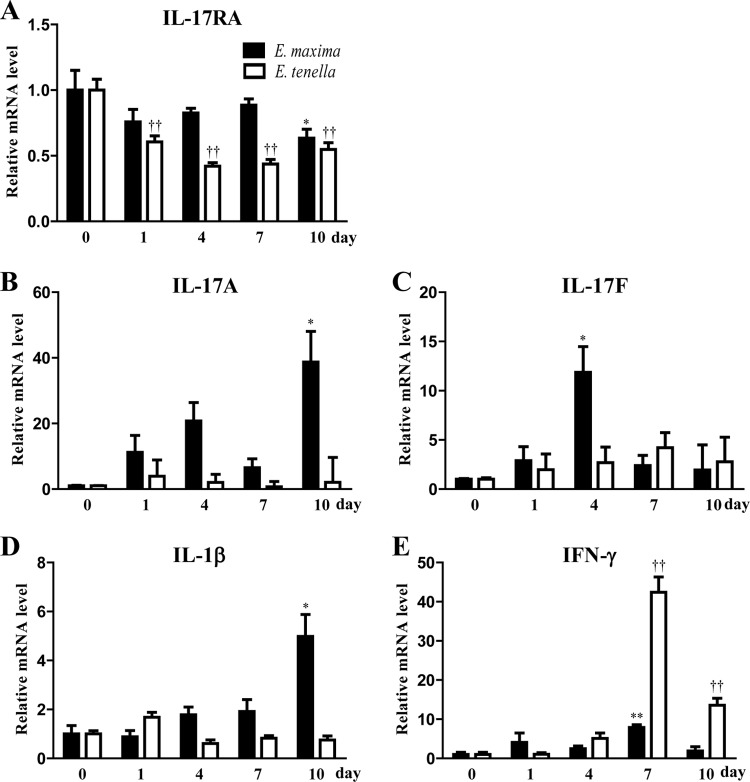
FIG 6.
mRNA expression profiles of chIL-17RA and related cytokines in Eimeria-infected chickens. Ten-day-old chicks were infected via the oral route with 104 sporulated E. tenella or E. maxima oocysts. Intestinal tissues (jejunum for E. maxima and cecum for E. tenella) were collected on 0, 1, 4, 7, and 10 days after infection. Tissue samples were pooled from five chickens. The expression levels of chIL-17RA and related cytokines were analyzed using quantitative real-time PCR. Gene expression levels were normalized with β-actin and calibrated with expression of uninfected group. All data represent the means ± the SE of triplicate trials from two independent experiments. P < 0.05 (*) or P < 0.01 (**), E. maxima-infected chickens compared to uninfected chickens; P < 0.01 (††), E. tenella-infected chickens compared to uninfected chickens.
Compared to uninfected chickens, the expression levels of chIL-17RA transcript were generally downregulated in the jejuna and ceca of E. maxima- or E. tenella-infected chickens (Fig. 6A). The expression levels of chIL-17F and chIL-17A transcripts were highly expressed on days 4 and 10 after E. maxima infection, respectively. However, the expression levels of chIL-17A and chIL-17F transcripts were unchanged in E. tenella infection (Fig. 6B and C). Eimeria species are intracellular protozoan parasites that typically invade intestinal epithelial cells and induce increased levels of cytokine genes (e.g., IL-1β and IFN-γ) (25, 38, 39). Therefore, IL-1β and IFN-γ transcript levels were monitored postinfection. Compared to control chickens, IL-1β transcript expression was ∼5.0-fold higher on day 10 only in E. maxima-infected chickens (Fig. 6D). The IFN-γ transcript expression levels were approximately 42.4- and 13.5-fold higher in E. tenella-infected chickens on days 7 and 10, respectively. In E. maxima-infected chickens, the IFN-γ transcript expression was ∼7.9-fold higher on day 7 (Fig. 6E). Given that the infections with Eimeria were successful and that the samples were prepared properly, the reduction in chIL-17RA mRNA may be caused by either of the intracellular protozoan parasites, E. maxima or E. tenella.
DISCUSSION
IL-17RA functions as the signal-transducing component for homodimers and heterodimers of IL-17A and IL-17F and for homodimers of IL-17E (3). Our previous studies have demonstrated that chIL-17A and chIL-17F of CEF induced the expression of inflammatory cytokines such as IL-1β, IL-6, or IL-8 (19, 22, 23). We have described here full-length cDNA encoding chicken IL-17RA and demonstrated specific siRNA-mediated inhibition of inflammatory cytokine transcripts in CEF cells stimulated with chIL-17A or chIL-17F.
ChIL-17RA is a transmembrane protein composed of a 286-amino-acid extracellular region, a 23-amino-acid transmembrane region, and a 519-amino-acid intracellular region. ChIL-17RA revealed a putative extracellular FnIII domain and an intracellular SEFIR domain, which are common structural characteristics of the IL-17R family (3, 7, 40). Multiple alignment of chIL-17RA between mammalian counterparts revealed a high degree of conservation within an intracellular SEFIR domain which shared a 68 to 72% identity. This result suggests that signaling pathways engaged by IL-17RA are similar between chickens and mammals. Generally, similar to chIL-17A and chIL-17F, chIL-17RA (19, 23) shared higher identity with mammalian IL-17RA than did piscine IL-17RA. Twelve of the many conserved cysteine residues that are potentially involved in the formation of disulfide bonds (e.g., in mammalian IL-17RA) were also present in chIL-17RA. In rainbow trout IL-17RA, eight of the conserved cysteines were predicted to be involved in the formation of four disulfide bridges (40). The length of chIL-17RA amino acid sequence is shorter than vertebrate IL-17RA, except in the zebrafish (accession no. NP_001093473), which encodes 782-amino-acid sequences.
Distribution analysis of chIL-17RA transcript revealed a relatively high expression in the normal thymus and heart. These observations agree with the high expression of IL-17RA mRNA in the thymus of rainbow trout (40) and wild-type mice (11). Mouse IL-17RA mRNA is also preferentially expressed in immune cells such as T cells and macrophage cell lines (11). Generally, upregulation of Th17 cytokines and IL-17RA expression has been implicated with myocarditis, hypertrophy, myocardial injury, and myocardial infarction (41–43), although the reason why there is high expression of chIL-17RA in chicken heart tissues compared to muscles remains to be investigated. In our study, a relatively large amount of IL-17RA transcript was detected in two chicken cell lines, HD11 macrophage and CU205 REV-transformed lymphoblast cell lines. Interestingly, high levels of IL-17A and IL-17F transcripts were detected in CU205 cells (19, 23) that express T cell receptor 3 (TCR3), but not CD3, CD4, CD8, TCR1, or TCR2 (35), and carry a NK cell marker as identified by 28-4 monoclonal antibody (23, 44). To investigate whether the mRNA expression profiles of chIL-17RA could be modulated, primary CEF cells were stimulated in vitro (Fig. 2C). The expression levels of chIL-17RA were unchanged or decreased significantly in splenic lymphocytes stimulated with ConA, LPS, or poly(I·C). Similarly, after LPS stimulation, mouse IL-17RA is significantly reduced in bone marrow-derived macrophages, but the expression levels are unchanged in poly(I·C) (45). In contrast, rainbow trout IL-17RA transcript increases in head kidney primary leukocytes activated with poly(I·C) but do not change with LPS stimulation (40). These findings suggest that in vitro expression profiles of IL-17RA may differ between birds and fish.
A heterodimer composed of IL-17RA and IL-17RC acts as a receptor for IL-17A and/or IL-17F (3, 4). A suppressive role of RNA interference was not apparent in the DF-1 fibroblast cell line stimulated with chIL-17A and chIL-17F (see Fig. S2 in the supplemental material). This observation can be partly explained by less expression of chIL-17RA transcript in DF-1 cells than in primary CEF. The availability of chicken IL-17RC that remains to be identified will enhance future study of the role of chIL-17RA complexes.
Salmonella species are important zoonotic pathogens that cause distinct gastrointestinal diseases in humans and animals, including chickens. This pathogen enters the small intestine and is carried to various organs, including the spleen and liver (46–48). Expression of chicken IL-17 transcript increases in the cecum during Salmonella enterica serovar Enteritidis infection (20, 49). In the present study, chIL-17RA, the receptor for chIL-17A and chIL-17F, was largely unresponsive in the liver and spleen during S. gallinarum or S. Typhimurium infection. However, chIL-17A and chIL-17F transcripts were upregulated in the liver and spleen during S. gallinarum infection. Consistently, the chIL-17A protein was mainly detected in the liver and spleen in S. gallinarum-infected chickens (see Fig. S3 in the supplemental material). IL-17RA expression increases in the lungs of Chlamydia-susceptible C3H/HeN mice infected with Chlamydia muridarum, but not in Chlamydia-resistant C57BL/6 mice (12). On the other hand, IL-17RA expression increases in the bladders of BCG-stimulated C57BL/6 mice (13). Compared to wild-type C57BL/6 mice, IL-17RA-deficient C57BL/6 mice exhibit higher Helicobacter pylori colonization density (50), elevated bacterial burdens following Staphylococcus aureus infection (51), and increased mortality following Klebsiella pneumoniae inoculation (15, 16). It is noteworthy that expression levels of IL-17RA transcript are upregulated or downregulated depending on which rainbow trout organs are infected intraperitoneally with Yersinia ruckeri (40).
Eimeria species, etiological agents of distinct gastrointestinal diseases, cause the most costly diseases that affect the poultry industry worldwide (21, 52). The chIL-17A expression increases in the duodenum and jejunum of E. acervulina- and E. maxima-infected chickens, respectively (53, 54). Expression levels of chIL-17A transcript are upregulated or downregulated in the cecum of E. tenella-infected chickens (19, 22, 53). In the present study, when chickens were infected with E. tenella (cecum) or E. maxima (jejunum), the chIL-17A expression increased during E. maxima infection but was unchanged during E. tenella infection. IFN-γ transcript, but not IL-17A and IL-17F, was highly expressed on days 7 and 10 in E. tenella-infected chickens (Fig. 6). IFN-γ can function as a negative regulator of IL-17 synthesis (55, 56). We also found that expression levels of chIL-17RA transcript were generally downregulated in chickens infected with E. maxima or E. tenella. Similarly, expression levels of IL-17RA are significantly reduced in kidney of rainbow trout infected with the parasite Tetracapsuloides bryosalmonae (40), and IL-17RA knockout mice challenged with Toxoplasma gondii displayed reduced mucosal damage (14). However, discrepancies in survival rates occur in IL-17RA knockout mice infected orally with 30 cysts of the 76K strain of T. gondii, an intracellular protozoan parasite (14, 17).
The avian immune system provides an important model for the study of comparative immunology. We cloned chIL-17RA among several chicken IL-17 receptors and investigated expression patterns and functions. In expression profiles of chIL-17RA transcript in chickens infected with two distinct gastrointestinal diseases caused by Salmonella or Eimeria, the chIL-17RA was significantly decreased in E. tenella and in E. maxima infection. This result indicates that chIL-17RA-mediated immunoregulation has an important role in local defenses against intracellular parasitism. Further studies using IL-17RA knockout chickens or other Eimeria species will be required to understand the specific contribution of the chIL-17RA molecule during Eimeria infection.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program (NRF-2013R1A1A4A01006646) and by the Cooperative Research Program for Agriculture Science and Technology Development (PJ008527032013), RDA, Republic of Korea.
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02141-14.
REFERENCES
- 1.Gaffen SL. 2009. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9:556–567. 10.1038/nri2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reynolds JM, Angkasekwinai P, Dong C. 2010. IL-17 family member cytokines: regulation and function in innate immunity. Cytokine Growth Factor Rev. 21:413–423. 10.1016/j.cytogfr.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwakura Y, Ishigame H, Saijo S, Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity 34:149–162. 10.1016/j.immuni.2011.02.012 [DOI] [PubMed] [Google Scholar]
- 4.Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, Tocker J, Peschon J. 2006. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J. Immunol. 177:36–39. 10.4049/jimmunol.177.1.36 [DOI] [PubMed] [Google Scholar]
- 5.Chang SH, Park H, Dong C. 2006. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J. Biol. Chem. 281:35603–35607. 10.1074/jbc.C600256200 [DOI] [PubMed] [Google Scholar]
- 6.Gu C, Wu L, Li X. 2013. IL-17 family: cytokines, receptors and signaling. Cytokine 64:477–485. 10.1016/j.cyto.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang W, Kolls JK, Zheng Y. 2008. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity 28:454–467. 10.1016/j.immuni.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. 1995. Herpesvirus saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 3:811–821. 10.1016/1074-7613(95)90070-5 [DOI] [PubMed] [Google Scholar]
- 9.Yao Z, Spriggs MK, Derry JM, Strockbine L, Park LS, VandenBos T, Zappone JD, Painter SL, Armitage RJ. 1997. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine 9:794–800. 10.1006/cyto.1997.0240 [DOI] [PubMed] [Google Scholar]
- 10.Silva WA, Jr, Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JL, Zanette DL, Santos AR, Zago MA. 2003. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 21:661–669. 10.1634/stemcells.21-6-661 [DOI] [PubMed] [Google Scholar]
- 11.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. 2009. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30:108–119. 10.1016/j.immuni.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Chen Q, Moore J, Kolls JK, Halperin S, Wang J. 2009. Critical role of the interleukin-17/interleukin-17 receptor axis in regulating host susceptibility to respiratory infection with Chlamydia species. Infect. Immun. 77:5059–5070. 10.1128/IAI.00403-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saban MR, Simpson C, Davis C, Wallis G, Knowlton N, Frank MB, Centola M, Gallucci RM, Saban R. 2007. Discriminators of mouse bladder response to intravesical bacillus Calmette-Guerin (BCG). BMC Immunol. 8:6. 10.1186/1471-2172-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly MN, Kolls JK, Happel K, Schwartzman JD, Schwarzenberger P, Combe C, Moretto M, Khan IA. 2005. Interleukin-17/interleukin-17 receptor-mediated signaling is important for generation of an optimal polymorphonuclear response against Toxoplasma gondii infection. Infect. Immun. 73:617–621. 10.1128/IAI.73.1.617-621.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194:519–527. 10.1084/jem.194.4.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Happel KI, Dubin PJ, Zheng M, Ghilardi N, Lockhart C, Quinton LJ, Odden AR, Shellito JE, Bagby GJ, Nelson S, Kolls JK. 2005. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202:761–769. 10.1084/jem.20050193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guiton R, Vasseur V, Charron S, Arias MT, Van Langendonck N, Buzoni-Gatel D, Ryffel B, Dimier-Poisson I. 2010. Interleukin 17 receptor signaling is deleterious during Toxoplasma gondii infection in susceptible BL6 mice. J. Infect. Dis. 202:427–435. 10.1086/653738 [DOI] [PubMed] [Google Scholar]
- 18.Tosello Boari J, Amezcua Vesely MC, Bermejo DA, Ramello MC, Montes CL, Cejas H, Gruppi A, Acosta Rodriguez EV. 2012. IL-17RA signaling reduces inflammation and mortality during Trypanosoma cruzi infection by recruiting suppressive IL-10-producing neutrophils. PLoS Pathog. 8:e1002658. 10.1371/journal.ppat.1002658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim WH, Jeong J, Park AR, Yim D, Kim YH, Kim KD, Chang HH, Lillehoj HS, Lee BH, Min W. 2012. Chicken IL-17F: identification and comparative expression analysis in Eimeria-infected chickens. Dev. Comp. Immunol. 38:401–409. 10.1016/j.dci.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 20.Matulova M, Varmuzova K, Sisak F, Havlickova H, Babak V, Stejskal K, Zdrahal Z, Rychlik I. 2013. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet. Res. 44:37. 10.1186/1297-9716-44-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min W, Kim WH, Lillehoj EP, Lillehoj HS. 2013. Recent progress in host immunity to avian coccidiosis: IL-17 family cytokines as sentinels of the intestinal mucosa. Dev. Comp. Immunol. 41:418–428. 10.1016/j.dci.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Liu R, Song M, Hu Y, Pan B, Cai J, Wang M. 2013. Eimeria tenella: interleukin 17 contributes to host immunopathology in the gut during experimental infection. Exp. Parasitol. 133:121–130. 10.1016/j.exppara.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 23.Min W, Lillehoj HS. 2002. Isolation and characterization of chicken interleukin-17 cDNA. J. Interferon Cytokine Res. 22:1123–1128. 10.1089/10799900260442548 [DOI] [PubMed] [Google Scholar]
- 24.Hong YH, Lillehoj HS, Park DW, Lee SH, Han JY, Shin JH, Park MS, Kim JK. 2008. Cloning and functional characterization of chicken interleukin-17D. Vet. Immunol. Immunopathol. 126:1–8. 10.1016/j.vetimm.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 25.Jeong J, Kim WH, Yoo J, Lee C, Kim S, Cho JH, Jang HK, Kim DW, Lillehoj HS, Min W. 2012. Identification and comparative expression analysis of interleukin 2/15 receptor β chain in chickens infected with E. tenella. PLoS One 7:e37704. 10.1371/journal.pone.0037704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yim D, Kang SS, Kim DW, Kim SH, Lillehoj HS, Min W. 2011. Protective effects of aloe vera-based diets in Eimeria maxima-infected broiler chickens. Exp. Parasitol. 127:322–325. 10.1016/j.exppara.2010.08.010 [DOI] [PubMed] [Google Scholar]
- 27.Yoo J, Chang HH, Bae YH, Seong CN, Choe NH, Lillehoj HS, Park JH, Min W. 2008. Monoclonal antibodies reactive with chicken interleukin-17. Vet. Immunol. Immunopathol. 121:359–363. 10.1016/j.vetimm.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryson K, Cozzetto D, Jones DT. 2007. Computer-assisted protein domain boundary prediction using the DomPred server. Curr. Protein Peptide Sci. 8:181–188. 10.2174/138920307780363415 [DOI] [PubMed] [Google Scholar]
- 30.Ferre F, Clote P. 2005. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33:W230–W232. 10.1093/nar/gki412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. 10.1186/1471-2105-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390. 10.1016/0092-8674(79)90057-6 [DOI] [PubMed] [Google Scholar]
- 33.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295–304. 10.1006/viro.1998.9290 [DOI] [PubMed] [Google Scholar]
- 34.Baba TW, Humphries EH. 1985. Formation of a transformed follicle is necessary but not sufficient for development of an avian leukosis virus-induced lymphoma. Proc. Natl. Acad. Sci. U. S. A. 82:213–216. 10.1073/pnas.82.1.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schat KA, Pratt WD, Morgan R, Weinstock D, Calnek BW. 1992. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 36:432–439. 10.2307/1591524 [DOI] [PubMed] [Google Scholar]
- 36.Pan H, Halper J. 2003. Cloning, expression, and characterization of chicken transforming growth factor beta 4. Biochem. Biophys. Res. Commun. 303:24–30. 10.1016/S0006-291X(03)00300-0 [DOI] [PubMed] [Google Scholar]
- 37.Halper J, Burt DW, Romanov MN. 2004. On reassessment of the chicken TGFβ4 gene as TGFβ1. Growth Factors 22:121–122. 10.1080/08977190410001712878 [DOI] [PubMed] [Google Scholar]
- 38.Dalloul RA, Bliss TW, Hong YH, Ben-Chouikha I, Park DW, Keeler CL, Lillehoj HS. 2007. Unique responses of the avian macrophage to different species of Eimeria. Mol. Immunol. 44:558–566. 10.1016/j.molimm.2006.02.004 [DOI] [PubMed] [Google Scholar]
- 39.Rothwell L, Young JR, Zoorob R, Whittaker CA, Hesketh P, Archer A, Smith AL, Kaiser P. 2004. Cloning and characterization of chicken IL-10 and its role in the immune response to Eimeria maxima. J. Immunol. 173:2675–2682. 10.4049/jimmunol.173.4.2675 [DOI] [PubMed] [Google Scholar]
- 40.Monte MM, Wang T, Holland JW, Zou J, Secombes CJ. 2013. Cloning and characterization of rainbow trout interleukin-17A/F2 (IL-17A/F2) and IL-17 receptor A: expression during infection and bioactivity of recombinant IL-17A/F2. Infect. Immun. 81:340–353. 10.1128/IAI.00599-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avalos AM, Apablaza FA, Quiroz M, Toledo V, Peña JP, Michea L, Irarrázabal CE, Carrión FA, Figueroa FE. 2012. IL-17A levels increase in the infarcted region of the left ventricle in a rat model of myocardial infarction. Biol. Res. 45:193–200. 10.4067/S0716-97602012000200012 [DOI] [PubMed] [Google Scholar]
- 42.Rose NR. 2011. Critical cytokine pathways to cardiac inflammation. J. Interferon Cytokine Res. 31:705–710. 10.1089/jir.2011.0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barry SP, Ounzain S, McCormick J, Scarabelli TM, Chen-Scarabelli C, Saravolatz LI, Faggian G, Mazzucco A, Suzuki H, Thiemermann C, Knight RA, Latchman DS, Stephanou A. 2013. Enhanced IL-17 signalling following myocardial ischaemia/reperfusion injury. Int. J. Cardiol. 163:326–334. 10.1016/j.ijcard.2011.08.849 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Gobel TW, Kaspers B, Stangassinger M. 2001. NK and T cells constitute two major, functionally distinct intestinal epithelial lymphocyte subsets in the chicken. Int. Immunol. 13:757–762. 10.1093/intimm/13.6.757 [DOI] [PubMed] [Google Scholar]
- 45.Barin JG, Baldeviano GC, Talor MV, Wu L, Ong S, Quader F, Chen P, Zheng D, Caturegli P, Rose NR, Cihakova D. 2012. Macrophages participate in IL-17-mediated inflammation. Eur. J. Immunol. 42:726–736. 10.1002/eji.201141737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lillehoj HS, Kim CH, Keeler CL, Jr, Zhang S. 2007. Immunogenomic approaches to study host immunity to enteric pathogens. Poult. Sci. 86:1491–1500. 10.1093/ps/86.7.1491 [DOI] [PubMed] [Google Scholar]
- 47.Parsons BN, Humphrey S, Salisbury AM, Mikoleit J, Hinton JC, Gordon MA, Wigley P. 2013. Invasive non-typhoidal Salmonella typhimurium ST313 are not host-restricted and have an invasive phenotype in experimentally infected chickens. PLoS Negl. Trop. Dis. 7:e2487. 10.1371/journal.pntd.0002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuda K, Chaudhari AA, Kim SW, Lee KM, Lee JH. 2010. Physiology, pathogenicity and immunogenicity of lon and/or cpxR deleted mutants of Salmonella gallinarum as vaccine candidates for fowl typhoid. Vet. Res. 41:59. 10.1051/vetres/2010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crhanova M, Hradecka H, Faldynova M, Matulova M, Havlickova H, Sisak F, Rychlik I. 2011. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar Enteritidis infection. Infect. Immun. 79:2755–2763. 10.1128/IAI.01375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Algood HM, Allen SS, Washington MK, Peek RM, Jr, Miller GG, Cover TL. 2009. Regulation of gastric B cell recruitment is dependent on IL-17 receptor A signaling in a model of chronic bacterial infection. J. Immunol. 183:5837–5846. 10.4049/jimmunol.0901206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidlak D, Kielian T. 2012. Differential effects of interleukin-17 receptor signaling on innate and adaptive immunity during central nervous system bacterial infection. J. Neuroinflammation 9:128,2094-9-128. 10.1186/1742-2094-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald V, Shirley MW. 2009. Past and future: vaccination against Eimeria. Parasitology 136:1477–1489. 10.1017/S0031182009006349 [DOI] [PubMed] [Google Scholar]
- 53.Hong YH, Lillehoj HS, Lee SH, Dalloul RA, Lillehoj EP. 2006. Analysis of chicken cytokine and chemokine gene expression following Eimeria acervulina and Eimeria tenella infections. Vet. Immunol. Immunopathol. 114:209–223. 10.1016/j.vetimm.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 54.Hong YH, Lillehoj HS, Lillehoj EP, Lee SH. 2006. Changes in immune-related gene expression and intestinal lymphocyte subpopulations following Eimeria maxima infection of chickens. Vet. Immunol. Immunopathol. 114:259–272. 10.1016/j.vetimm.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 55.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141. 10.1038/ni1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molesworth-Kenyon SJ, Yin R, Oakes JE, Lausch RN. 2008. IL-17 receptor signaling influences virus-induced corneal inflammation. J. Leukoc. Biol. 83:401–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.