Abstract
Porphyromonas gingivalis is associated with chronic periodontitis, an inflammatory disease of the tooth's supporting tissues. Macrophages are important in chronic inflammatory conditions, infiltrating tissue and becoming polarized to an M1 or M2 phenotype. As responses to stimuli differ between these phenotypes, we investigated the effect of P. gingivalis lipopolysaccharide (LPS) on M1 and M2 macrophages. M1 and M2 polarized macrophages were produced from murine bone marrow macrophages (BMMϕ) primed with gamma interferon (IFN-γ) or interleukin-4 (IL-4), respectively, and incubated with a low or high dose of P. gingivalis LPS or control TLR2 and TLR4 ligands. In M1-Mϕ, the high dose of P. gingivalis LPS (10 μg/ml) significantly increased the expression of CD40, CD86, inducible nitric oxide synthase, and nitric oxide secretion. The low dose of P. gingivalis LPS (10 ng/ml) did not induce costimulatory or antibacterial molecules but did increase the secretion of IL-1α, IL-6, IL-12p40, IL-12p70, and tumor necrosis factor alpha (TNF-α). P. gingivalis LPS marginally increased the expression of CD206 and YM-1, but it did enhance arginase expression by M2-Mϕ. Furthermore, the secretion of the chemokines KC, RANTES, eotaxin, and MCP-1 from M1, M2, and nonpolarized Mϕ was enhanced by P. gingivalis LPS. TLR2/4 knockout macrophages combined with the TLR activation assays indicated that TLR2 is the main activating receptor for P. gingivalis LPS and whole cells. In conclusion, although P. gingivalis LPS weakly activated M1-Mϕ or M2-Mϕ compared to control TLR ligands, it induced the secretion of inflammatory cytokines, particularly TNF-α from M1-Mϕ and IL-10 from M2-Mϕ, as well as chemotactic chemokines from polarized macrophages.
INTRODUCTION
Chronic periodontitis is a chronic inflammatory disease associated with specific bacteria in a biofilm (subgingival plaque) and is characterized by resorption of the alveolar bone and other supporting tissues of the teeth (1, 2). Typically, chronic periodontitis is characterized by a dense inflammatory cell infiltrate of the gingival tissue, including macrophages (3). In the mucosal tissues, macrophages often are the first immune cell to encounter immunostimulatory compounds derived from invading pathogens. Ligation of Toll-like receptors (TLRs) on the macrophage surface by bacterial pathogen-associated molecular patterns, such as lipopolysaccharide (LPS), leads to macrophage activation (4).
Although chronic periodontitis is associated with a polymicrobial biofilm (subgingival plaque), one species of the biofilm, Porphyromonas gingivalis, is recognized as a keystone pathogen linked to disease onset and progression (5, 6). Previous investigations into the effect of P. gingivalis LPS on nonpolarized macrophages have shown that the induced immune responses is varied and that many cytokines were only transiently expressed compared to Escherichia coli LPS and other Gram-negative pathogens (7–9). Furthermore, P. gingivalis LPS is atypical in that it is structurally different from the canonical enterobacterial LPS and has been reported to stimulate both TLR4 and TLR2 (10–12). The stimulation of TLR4 has been linked to penta-acylated lipid A structures (13–15); however, the molecular entity for stimulation of TLR2, even in highly purified LPS samples, has not yet been identified (12). It has been suggested that TLR2 stimulation is due to the presence of novel lipoprotein contaminants that copurify with the P. gingivalis LPS (12).
The exposure of macrophages to cytokines prior to TLR ligation is a process that more closely resembles in vivo macrophage activation, especially during a chronic infection where naive monocytes/macrophages would be recruited from the bloodstream to an already inflamed site via a cytokine/chemokine gradient. However, no investigation has utilized cytokine priming to induce an M1 or M2 macrophage phenotype to study the effect P. gingivalis LPS has on these polarized macrophages.
Macrophages display a remarkable amount of plasticity in their physiological responses, and the cytokine environment at the time of TLR ligation has a profound effect on the phenotype of the activated macrophage (16). Gamma interferon (IFN-γ) polarizes murine macrophages toward an M1 phenotype (pre-M1-Mϕ) and, when exposed to E. coli LPS, they mature into a classically activated macrophage, designated M1 macrophages (M1-Mϕ) (17). M1-Mϕ exhibit high levels of phagocytosis and nitric oxide production and upregulate the expression of costimulatory molecules on the cell surface (17, 18). M1-Mϕ play a critical role in the resolution of bacterial infections through phagocytosis and killing of pathogens, the initiation and maintenance of inflammation, and the recruitment of adaptive immunity effector cells such as T lymphocytes (19).
Alternative pathways of macrophage activation exist depending on the stimulus applied to the macrophage. Interleukin-4 (IL-4) priming results in the generation of alternatively activated macrophages, designated M2 macrophages (M2-Mϕ) (20). M2-Mϕ have been associated with fibrosis and are characterized by arginase production, which breaks down arginine into urea and l-ornithine, a precursor of collagen formation (20–23). M2-Mϕ express high levels of CD206, FIZZ1, and YM-1, low levels of costimulatory molecules, such as CD40 and CD86, and low levels of nitric oxide (18). Despite the indication that M2-Mϕ have an important role in limiting host tissue destruction in chronic infections (24) and the presence of fibrosis in chronically diseased gingiva (25), alternatively activated macrophages in chronic periodontitis have received limited attention.
As chronic periodontitis is characterized by inflammation and alveolar bone resorption and macrophages, M1 macrophages in particular have an important role in chronic inflammatory diseases (26). Investigating the activation of macrophage phenotypes in response to a periodontal pathogen may provide insights into important host-pathogen interactions. We have previously shown that macrophages in the gingival tissue of mice exhibiting alveolar bone resorption through infection with P. gingivalis expressed high levels of CD86 and lower levels of CD206, suggesting M1 macrophage polarization (27). The aim of this study was to investigate the response P. gingivalis LPS induces in pre-M1-Mϕ and pre-M2-Mϕ and the subsequent maturation of these macrophages.
MATERIALS AND METHODS
Ethics statement.
All animal experimental procedures were carried out in strict accordance with the recommendations in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (66). The protocols for the experiments were approved by The University of Melbourne Ethics Committee for Animal Experimentation (approval number 1212363).
Generation of macrophage phenotypes.
All cell culture reagents were obtained from Sigma-Aldrich Pty Ltd. (NSW, Australia) unless specified otherwise. Mammalian cells were routinely grown in complete minimal essential media (MEM), consisting of MEM supplemented with 10% (vol/vol) fetal bovine serum (FBS), 20 mM l-glutamine, 10 mM sodium pyruvate, and 100 U/100 μg penicillin-streptomycin. Immortalized macrophages (iMACs) were the gift of Eicke Latz (University of Bonn, Germany). The mouse fibroblast cell line L929 (obtained from the cell culture collection at the Melbourne Dental School) was grown in complete MEM supplemented with 1:100 dilutions of 100× MEM nonessential amino acid solution. To make CSF-1-containing L929 supernatant, cells were seeded at 1 × 106 cells/ml and allowed to grow for 7 days. Supernatant then was sterile filtered (0.22 μM) and stored at −70°C. All cells were grown at 37°C in 5% CO2 atmosphere.
To generate bone marrow macrophages (BMMϕ), mouse femurs aseptically harvested and sterilized in 70% (vol/vol) ethanol for 5 min then were washed twice in Dulbecco's phosphate-buffered saline (PBS). The epiphyses were removed using a sterile scalpel, and the contents of the femurs were flushed out with complete MEM using a 25-guage needle. Bone marrow cells were seeded at 1 × 106 cells/ml in complete MEM supplemented with 5% (vol/vol) L929 supernatant and grown for 5 days in cell culture petri dishes (Sigma-Aldrich). Spent media was removed, and fresh complete MEM supplemented with 5% (vol/vol) L929 conditioned media was added on day 3.
To prime macrophages, cells were incubated overnight in complete MEM supplemented with either 20 U/ml of IFN-γ (ABD Serotec, Oxford, United Kingdom) or 150 U/ml of IL-4 (ABD Serotec) to generate pre-M1-Mϕ and pre-M2-Mϕ, respectively. To activate the macrophages, the pre-M1-Mϕ and pre-M2-Mϕ or nonpolarized M0-Mϕ were incubated with either 10 ng/ml Escherichia coli LPS (Sigma-Aldrich), 10 ng/ml Pam3CSK4 (Invivogen, CA, USA), or 10 ng/ml or 10 μg/ml ultrapure P. gingivalis LPS (Invivogen) and incubated overnight in complete MEM.
Analysis of LPS.
The ultrapure P. gingivalis LPS (Invivogen) was prepared using a modification of the methods of Westphal and Jann (28) and Hirschfeld et al. (29) and contained penta-acylated and tetra-acylated lipid A structures (Invivogen, personal communication). For analysis, it was diluted in Novex Tricine SDS-PAGE loading buffer and heated to 85°C for 10 min, and 5 mg was loaded onto a 10 to 20% gradient tricine gel and subjected to SDS-PAGE at 125 V for 90 min (Life Technologies, NSW, Australia). O-LPS was stained using ProQ Emerald 300 lipopolysaccharide stain (Life Technologies) according to the manufacturer's instructions. Protein was stained using Coomassie SimplyBlue safe protein stain (Life Technologies) according to the manufacturer's instructions. Stained SDS-PAGE gels were visualized using the Fujifilm LAS3000 imaging system (Fujifilm, Tokyo, Japan). LPS separated by SDS-PAGE was transferred to a nitrocellulose membrane using the X-cell II transblot module (Life Technologies) at 30 V for 60 min. The membrane then was blocked with 5% (wt/vol) skim milk powder in PBS for 1 h at room temperature and probed with the detection antibodies anti-A-LPS (30, 31) and anti-RgpA/B (30, 32) in 1% (wt/vol) skim milk powder in PBS overnight at 4°C. The anti-A-LPS antibody (monoclonal antibody [MAb] 1B5) was a gift from M. A. Curtis (31). The membrane was washed 3× with PBS, and bound antibody was detected by incubation with a 1:2,000 dilution of a goat anti-mouse IgG antibody in 1% (wt/vol) skim milk powder in PBS (Southern Biotech, Birmingham, AL) for 1 h at room temperature. Development of the blot was accomplished after incubation in a 1:2,000 dilution of swine anti-goat horseradish peroxidase (HRP) conjugate (Southern Biotech) for 1 h at room temperature. Bands were visualized by the addition of 20 ml 3,3′,5,5′-tetramethylbenzidine (TMB) substrate for 10 min at room temperature (Mabtech, Nacka, Sweden).
Amino acid analysis of the P. gingivalis LPS preparation.
A sample of ultrapure P. gingivalis LPS (Invivogen) was prepared as a 20 μg/ml solution in MilliQ water and hydrolyzed at 150°C for 1 h. The sample was dried and derivatized using the AccQ-Tag method according to the manufacturer's protocol (Waters Pty Ltd., NSW, Australia). The resultant hydrolyzed components were separated using an Agilent 1200 series liquid chromatograph instrument (Agilent, NSW, Australia) equipped with a UV detector (model G1316A) and an AccQ-Tag column (3.9 by 150 mm). Amino acid compounds were detected at 150-nm absorbance using an acetate-phosphate solution (eluent A; sodium acetate trihydrate-phosphoric acid-triethylamine at 19:6:1 [% wt/wt] in MilliQ water) and 60% (vol/vol) acetonitrile in MilliQ water as elution buffers at a flow rate of 1.0 ml/min and norleucine as an internal standard. Chemstation software 03.01-SR1.1 (Agilent Technologies) was used for peak integration and amino acid identification.
TLR activation assay.
Human embryonic kidney 293 cells (HEK293) stably transfected with TLR4/MD2 or TLR2 (cell lines were a kind gift from A. Mansell and P. Hertzog, Monash University) were grown in complete Dulbecco's MEM (DMEM) supplemented with 150 μg/ml Geneticin (Life technologies). To assay for activity, 2 × 104 cells were seeded into 96-well plates (Sigma-Aldrich) and the following day were transiently transfected with 10 ng of pNF-κB-firefly luciferase (Stratagene, CA, USA) and 5 ng pTK-renilla luciferase (Promega, WI, USA) per well using Fugene HD transfection reagent according to the manufacturer's instructions (Promega). pTK-renilla luciferase was used as a transfection control. For HEK293 cells expressing TLR4/MD2, 1 ng of a plasmid encoding CD14 was added to the transfection mix. After 24 h at 37°C, cells were incubated with serial dilutions (50 μg/ml to 0.5 ng/ml) of P. gingivalis LPS, E. coli LPS, or PAM3CSK for 4 h before lysis with 100 μl of passive lysis buffer (Promega). Half the lysate was added to 50 μl of luciferase assay substrate (Promega), and half was added to 50 μl of 2 μg/ml coelenterazine (Promega). The assays were read on a Victor3 1420 multilabel counter (PerkinElmer, MA, USA) and expressed as the luminescence of firefly luciferase relative to that of renilla luciferase.
Nitric oxide assay.
Nitric oxide was measured using the Greiss reagent kit (Life Technologies) according to the manufacturer's instructions. Briefly, the day after macrophage activation with TLR ligand, 150 μl of the BMMϕ supernatant was combined with 130 μl of distilled water (dH2O), 10 μl of N-(1-naphthyl) ethylenediamine dihydrochloride (1 mg/ml), and 10 μl of sulfanilic acid (1.0 mM). A standard curve was generated using 2-fold serial dilutions of a 100 μM nitrite standard solution (100 μM to 1.56 μM). The reaction was allowed to proceed for 30 min at room temperature, and then the absorbance was measured at 550 nm on a Victor3 1420 multilabel counter (PerkinElmer).
Arginase assay.
TLR ligand-activated M0-Mϕ, M1-Mϕ, and M2-Mϕ were lysed with the addition of 100 μl PBS containing 0.1% (vol/vol) Triton X-100. After the addition of 100 μl of 25 mM Tris, 1 mM MnCl2, the collected cell lysate was heated to 55°C for 10 min. Once cooled, 200 μl of 0.5 M arginine in PBS (Sigma-Aldrich) was added to the cell lysate solution, which then was incubated at 37°C for 1 h. The arginase reaction was stopped by the addition of 900 μl of 44.6N H2PO4, 36N H2SO4. The cell lysate reaction solution then was incubated for 30 min at 100°C after the addition of 40 μl of 9% (vol/vol) isonitrosopropiophenone in ethanol (Sigma-Aldrich). Absorbance was measured at 550 nm on a Victor3 1420 multilabel counter (PerkinElmer). A standard curve generated using 2-fold serial dilutions of 200 mM urea (200 mM to 3.12 mM) was used to quantify the assay (Sigma-Aldrich).
Flow cytometry analysis of surface marker expression.
Cells from TLR activation assays were washed twice in situ in 2 ml PBS containing 0.1% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich) (PBS-BSA buffer), removed from the petri dish using a 23-gauge syringe, and then incubated with anti-mouse CD16/CD32 antibody (clone 2.4G2; BD Biosciences, Franklin Lakes, NJ) for 20 min on ice. Cells were washed in 5 ml PBS-BSA buffer a further two times prior to incubation with various fluorochrome-conjugated antibodies against molecules of interest for 30 min on ice. Detection antibodies used for phenotyping and activation analysis were anti-CD206-fluorescein isothiocyanate (FITC) (clone MR5D3; AbD Serotec), anti-CD40-allophycocyanin (APC) (clone 3/23), anti-CD86-phycoerythrin (PE)-Cy7 (clone GL1), anti-CD11c-FITC (clone HL3), anti-CD11b-APC (clone M1/70), and anti-CD11a-PE (clone 2D7). Unless otherwise stated, all antibodies were purchased from BD Biosciences. All cells were washed twice in 5 ml PBS-BSA buffer and then analyzed on a Beckman Coulter FC500 flow cytometer (Beckman Coulter Pty Ltd., NSW, Australia). The FC500 was equipped with an argon ion laser operating at an excitation wavelength of 488 nm and a red solid-state diode laser operating at 635 nm. The fluorescence from FITC was measured with a 525-nm filter (FL1), PE was measured through a 575-nm filter (FL2), APC was measured through a 660-nm filter (FL4), and PE-Cy7 was measured through a 755-nm filter (FL5). The data were analyzed using FlowJo software, V7.0 (Tree Star, OR, USA). Forward and side scatter properties were used to acquire a total of 10,000 cells and to gate out the cell debris.
Cytokine bead array analysis of cell culture supernatant.
Cell culture supernatant from TLR activation assays was analyzed for cytokines using the Bioplex Pro mouse cytokine 23-plex assay (Bio-Rad, NSW, Australia). The 23-plex assay measures IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-17A, eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, KC, MCP-1, MIP-1α, MIP-1β, RANTES, and tumor necrosis factor alpha (TNF-α). The assay was performed according to the manufacturer's instructions. Briefly, 50 μl of beads was added to the assay plate and washed 2 times with 100 μl wash buffer. The samples, standards, blanks, and controls then were added in a volume of 50 μl and incubated with the beads for 1 h at room temperature, no light, and constant mixing at 300 rpm on a MX4 micromixer (FINEPCR, Seoul, South Korea). Beads then were washed 3 times with 100 μl of wash buffer using a Bioplex ProII wash station (Bio-Rad), and 25 μl/well of biotinylated anticytokine detection antibody was added. Plates then were incubated at room temperature with no light and constant mixing at 300 rpm for 1 h. Wells then were washed 3 times with 100 μl of wash buffer using a Bioplex ProII wash station (Bio-Rad) before bound biotin-labeled anti-cytokine antibody was detected by the addition of 25 μl of streptavidin-PE. Plates were incubated at room temperature with no light and constant mixing at 300 rpm for 10 min. Beads then were washed 3 times with 100 μl of wash buffer using a Bioplex ProII wash station (Bio-Rad), and the beads were resuspended in 125 μl of assay buffer before reading the assay on a Bio-plex 200 system (Bio-Rad).
RNA extraction and reverse transcriptase PCR.
BMMϕ from the TLR activation assays were collected and incubated with 0.5 ml of RNA Protect reagent and stored at 4°C until RNA extraction (Qiagen, NSW, Australia). RNA was extracted from BMMϕ using the RNeasy plus RNA extraction kit by following the manufacturer's instructions (Qiagen). RNA was quantified by absorbance at 260 nm using a NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). First-strand cDNA synthesis was performed using ImProm II reverse transcriptase (Promega). RNA (0.5 mg) was added to 0.5 μg of oligo(dT) primer and incubated at 70°C for 5 min before being chilled on ice. The RNA-primer mix then was added to 5 μl of 5× buffer, 2 μl of MgCl2, 1.2 μl of deoxynucleoside triphosphate (dNTP), 0.5 μl of RNasin, and 1 μl of ImProm II reverse transcriptase enzyme and mixed with up to 30 μl with nuclease-free water. The reaction mix was incubated at 42°C for 1 h and then at 70°C for 15 min. PCR was performed to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH), inducible nitric oxide synthase (iNOS), YM-1, and Arg-1 mRNA using Taq DNA polymerase (M0267S; New England BioLabs, MA, USA). PCRs were set up in a total volume of 25 μl, consisting of 2.5 μl of 10× buffer, 0.2 μM forward primer, 0.2 μM reverse primer, 0.5 μl of dNTP, 20 ng of cDNA, and 0.2 U of Taq polymerase, and it was mixed with up to 25 μl with nuclease-free water. PCR was performed on a G-storm thermocycler (G-storm, Somerset, United Kingdom) with an initial denaturing step of 95°C for 30 s; 30 cycles of 95°C for 30 s; an annealing temperature of either 60°C for iNOS, 55°C for Arg-1, 55°C for YM-1, or 55°C for GAPDH for 30 s; and extension at 68°C for 1 min. A final extension of 68°C for 5 min then was performed before storage of the amplified gene products at 4°C. Primers for iNOS, Arg-1, and GAPDH were from reference 18, and YM-1 was from reference 33. The primers used in this study were iNOS forward, CCCTTCCGAAGTTTCTGGCAGCAGC; iNOS reverse, GGCTGTCAGAGCCTCGTGGCTTTGG; Arg-1 forward, CAGAAGAATGGAAGAGTCAG; Arg-1 reverse, CAGATATGCAGGGAGTCACC; YM-1 forward, GGGCATACCTTTATCCTGAG; YM-1 reverse, CCACTGAAGTCATCCATGTC; GAPDH forward, GCACTTGGCAAAATGGAGAT; and GAPDH reverse, CCAGCATCACCCCATTAGAT. A 2% (wt/vol) agarose gel in 1× Tris-acetate-EDTA (TAE) buffer was prepared with Sybrsafe directly incorporated to stain PCR products (Life Technologies). PCR products were separated by electrophoresis at 70 V for 1 h in 1× TAE buffer. The PCR products then were visualized using a Fujifilm LAS3000 imaging system (Fujifilm, Tokyo, Japan). Quantification of the density of PCR products was performed using the Multi Gauge V3.0 software suite (Fujifilm). The densities of YM-1, Arg-1, and iNOS were normalized against the expression of GAPDH and displayed relative to unstimulated BMMϕ.
Statistical analysis.
Data were analyzed by two-way analysis of variance (ANOVA) with Bonferroni posttest and are presented as means ± standard deviations (SD) (GraphPad Prism V5.0). Statistical significance was considered at P < 0.05. The data presented are representative of at least three biological replicates.
RESULTS
Characterization of the P. gingivalis LPS preparation.
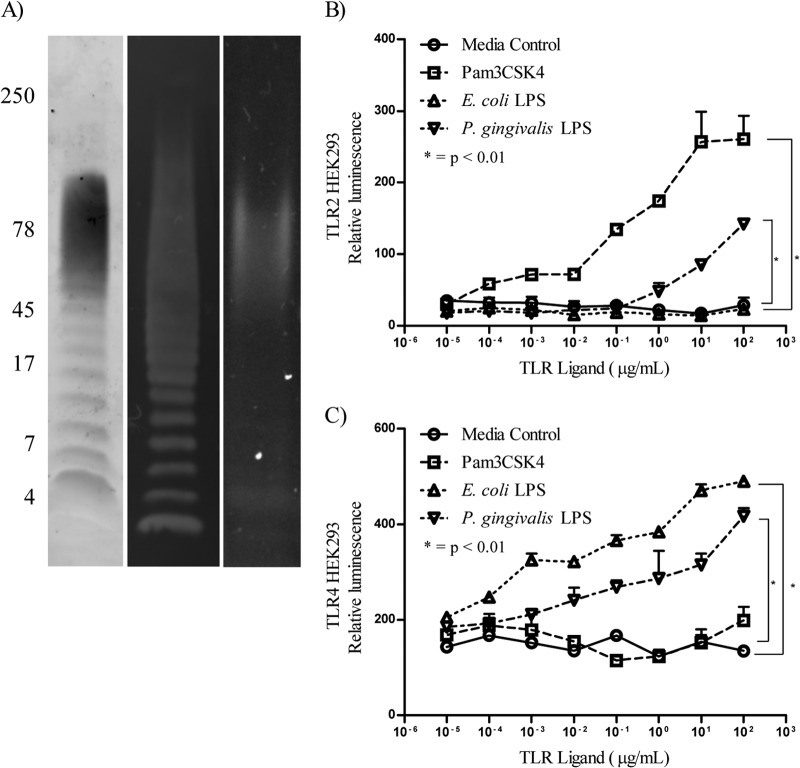
The commercially available ultrapure P. gingivalis LPS from Invivogen initially was subjected to Tricine SDS-PAGE and immunoblotting and stained with ProQ Emerald 300 and Coomassie SimplyBlue safe stain or probed with the P. gingivalis A-LPS-specific MAb 1B5. The P. gingivalis LPS formed a characteristic O-LPS ladder, and probing the immunoblot with MAb 1B5 confirmed the presence of A-LPS in the preparation (Fig. 1A). Protein was detected by Coomassie stain and found to be present as a diffuse 60- to 80-kDa band (Fig. 1A). We further characterized P. gingivalis LPS by amino acid analysis after acid hydrolysis. Amino acid compositional analysis confirmed the presence of amino acids in the LPS preparation (Table 1). The total amount of amino acid in the sample was 5.6% by weight. No amino acids were detected without acid hydrolysis, indicating that the amino acids were present as proteins/peptides. As A-LPS or a glycolipid reactive with MAb 1B5 is recognized as the covalently attached anchor of a class of secreted proteins referred to as CTD proteins, which include the abundant gingipains (RgpA/B and Kgp) (30, 32), we probed the LPS preparation for the presence of RgpA/B using antibodies raised to RgpB (30, 32). The presence of the modified form of RgpB as a diffuse band at 60 to 80 kDa was confirmed in the LPS preparation, whereas the discrete (unconjugated) form of the protease, which runs as a sharp band at 50 kDa, was not detected. These results indicate that the P. gingivalis LPS preparation contained O-LPS, A-LPS, and CTD protein conjugate.
FIG 1.
Characterization of P. gingivalis LPS preparation. (A) The P. gingivalis LPS used in this study first was analyzed by SDS-PAGE and stained with ProQ emerald to detect LPS (lane 1). An immunoblot was probed with MAb 1B5 (lane 2) to show the presence of O-LPS and A-LPS, respectively, and stained with Coomassie SimplyBlue safe stain to detect protein (lane 3). Molecular mass markers are indicated in kilodaltons. We then performed TLR activation assays using HEK293 cells stably transfected with TLR2 (B) or TLR4/MD2 (C), transiently transfected with plasmids encoding CD14, NK-κB-luciferase, and TK-renilla luciferase, and incubated with serial dilutions of the P. gingivalis LPS-p and control TLR ligands.
TABLE 1.
Amino acid analysis of the P. gingivalis LPS preparation
| Amino acid | % (wt/wt) amino acid residue |
|---|---|
| Asp/Asn | 0.22 ± 0.03 |
| Ser | 0.22 ± 0.08 |
| Glu/Gln | 1.10 ± 0.17 |
| Gly | 0.28 ± 0.04 |
| His | |
| Arg | |
| Thr | |
| Ala | 1.42 ± 0.18 |
| Pro | 1.07 ± 0.07 |
| Tyr | 0.21 ± 0.02 |
| Val | 0.18 ± 0.03 |
| Met | |
| Lys | 0.16 ± 0.01 |
| Ile | 0.09 ± 0.01 |
| Leu | 0.64 ± 0.08 |
| Phe | 0.03 ± 0.02 |
| Total | 5.62 ± 0.74 |
As P. gingivalis LPS previously has been thought to activate TLR2, the ability of the P. gingivalis LPS to stimulate cells via TLR2 and/or TLR4 was investigated. HEK293 cells expressing TLR2 or TLR4/MD2/CD14 were transiently transfected with pNF-κB-firefly luciferase and pTK-renilla luciferase and then incubated with serial dilutions of P. gingivalis LPS, a TLR2 ligand (Pam3CSK4), and a TLR4 ligand (E. coli LPS), and TLR activation was monitored by a luciferase reporter assay. Pam3CSK4 and the P. gingivalis LPS, but not E. coli LPS, significantly (P < 0.0001) activated the TLR2-expressing HEK293 cells compared to medium-alone controls (Fig. 1B). For TLR4-expressing HEK293 cells, E. coli LPS and the P. gingivalis LPS, but not Pam3CSK4, induced significant (P < 0.0001) activation compared to medium-alone controls (Fig. 1C). Although the P. gingivalis LPS induced activation via TLR2 and TLR4, the level of activation was significantly (P < 0.05) less than that of the defined TLR ligands of E. coli LPS and Pam3CSK4 (Fig. 1B and C). As P. gingivalis LPS contains both TLR2- and TLR4-stimulating components, we compared the ability of the P. gingivalis LPS to activate and induce cytokine secretion in bone marrow-derived macrophages with the response induced by E. coli LPS and Pam3CSK4 in all of the subsequent assays.
Effect of P. gingivalis LPS on IFN-γ-primed macrophages.
The ability of P. gingivalis LPS to induce a response and activate murine M1 macrophages was investigated. Bone marrow cells were removed from the tibia of C57BL6 mice and differentiated into macrophages for 5 days in the presence of CSF-1 derived from L929 fibroblasts, termed M0 cells, and used as control cells throughout. After this period, M0 cells were found to be CD11b and F4/80 positive and expressed low levels of CD86 and CD206, indicating a nonpolarized macrophage phenotype (Fig. 2A to C). BMMϕ (M0 cells) then were incubated overnight with IFN-γ to prime an M1 phenotype (pre-M1-Mϕ, CD86+, and CD206−) (Fig. 2D) before activation with two concentrations of P. gingivalis LPS, E. coli LPS, or Pam3CSK4 to generate an M1 phenotype (M1-Mϕ). A representative flow cytometry plot is shown in Fig. 2E.
FIG 2.
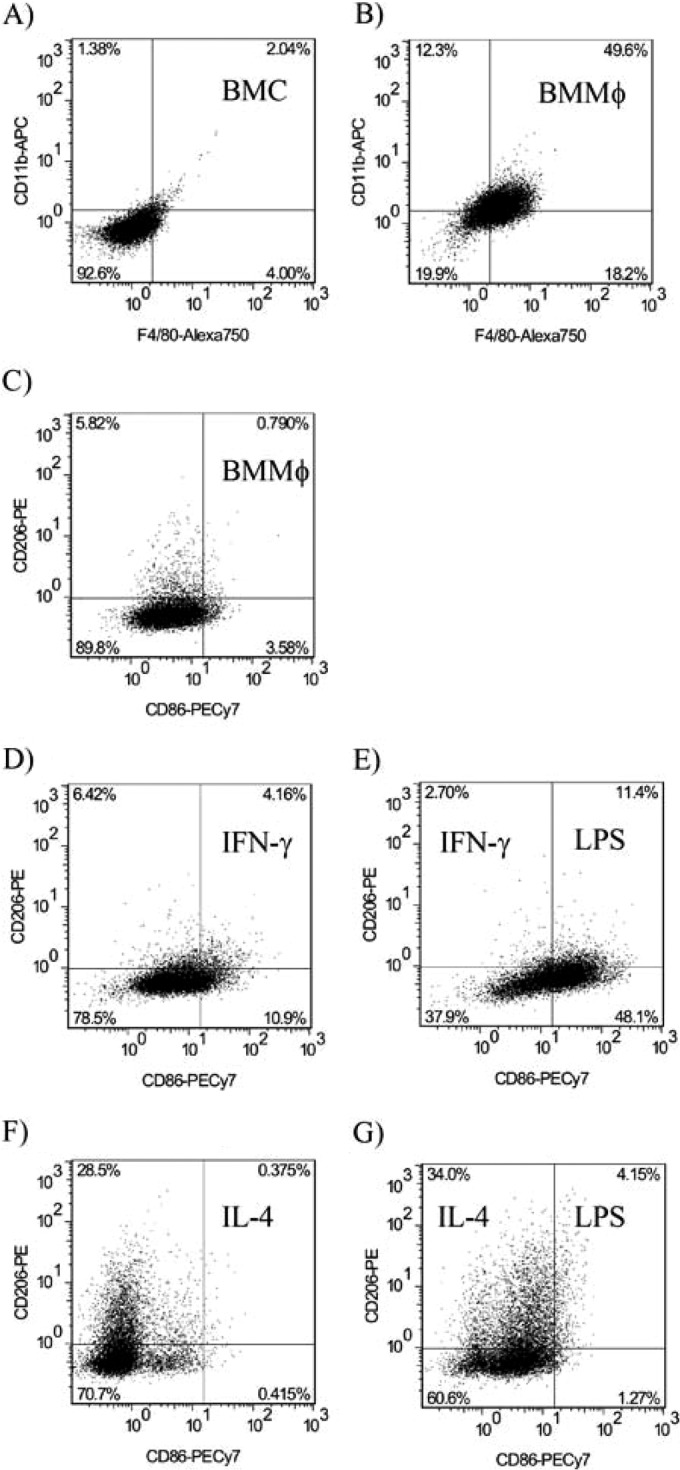
Characterization of bone marrow-derived M0, M1, and M2 macrophages. (A) Bone marrow-derived cells were grown in DMEM supplemented with L929-derived CSF-1 for 5 days and were found to be CD11b positive and F4/80 positive. (B) After that period, the cells were CD11b+ F4/80+ and were termed bone marrow-derived macrophages (M0-Mϕ). (C) M0-Mϕ expressed low levels of CD86 and CD206. Priming with IFN-γ increased the expression of CD86 (D), and TLR ligation of primed cells further boosted the CD86 expression (E). Priming with IL-4 increased the expression of CD206 (F); however, TLR ligation showed no further increase in CD206 expression (G).
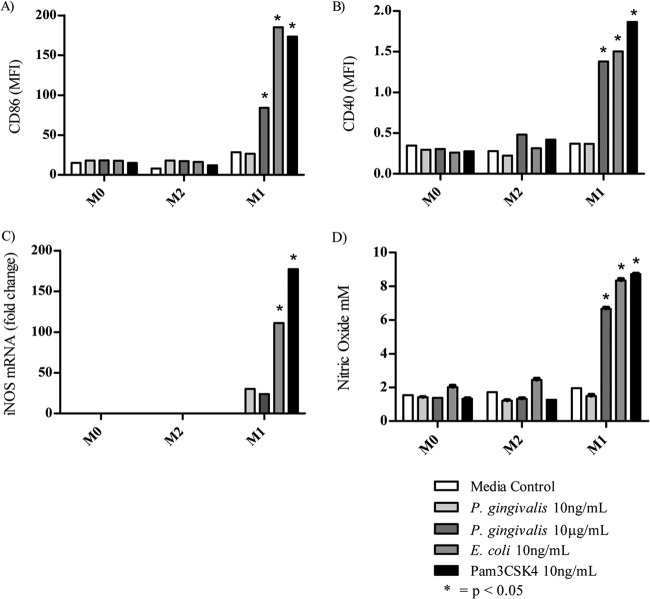
Initial experiments using 10 ng/ml P. gingivalis LPS showed no increase in expression of M1 macrophage costimulatory markers CD40 or CD86 or nitric oxide secretion (data not shown). Further experiments were performed using higher concentrations of P. gingivalis LPS; however, significant upregulation of costimulatory markers CD40 (P < 0.05) and CD86 (P < 0.0001) compared to control cells were not observed until 10 μg/ml P. gingivalis LPS was added to pre-M1-Mϕ (Fig. 3A and B). The addition of 10 ng/ml E. coli LPS or Pam3CSK4 resulted in significant upregulation of CD40 (P < 0.01) and CD86 (P < 0.0001). The concentration of P. gingivalis LPS required to significantly upregulate CD40 and CD86 was 1,000-fold higher than the concentration required by E. coli LPS or Pam3CSK4.
FIG 3.
Expression of M1 markers on BMMϕ after TLR ligation. M0-Mϕ were incubated with IFN-γ to generate pre-M1-Mϕ before activation with various TLR ligands. The M1-Mϕ generated were analyzed for their expression of CD86 (A) and CD40 (B) by flow cytometry after approximately 24 h. MFI, mean fluorescence intensity. (C) The expression of inducible nitric oxide synthase mRNA was measured by semiquantitative reverse transcription-PCR. (D) The production of nitric oxide was measured by the Griess reaction. M1 markers were not expressed after the addition of IL-4. Data are presented as means ± SD from three biological replicates.
An important differentiating factor between M1 and M2 macrophages is the difference in arginine metabolism: M1-Mϕ convert arginine via iNOS to nitric oxide and citrulline, and M2-Mϕ convert arginine via arginase-1 to polyamine and urea. Therefore, we investigated the metabolism of arginine in M1 macrophages, as iNOS mRNA expression and nitric oxide (NO) secretion were induced by P. gingivalis LPS. High levels of iNOS mRNA were observed in M1-Mϕ activated with E. coli LPS (P < 0.0001) and Pam3CSK4 (P < 0.0001); however, although both the low and high doses of P. gingivalis LPS produced significantly (P < 0.01) higher levels of iNOS mRNA than did pre-M1-Mϕ cells, the level was significantly (P < 0.01) less than that induced by the other TLR ligands (Fig. 3C). Despite the lower levels of iNOS mRNA detected in the M1-Mϕ activated with the high dose of P. gingivalis LPS, these macrophages produced significant (P < 0.0001) and similar amounts of NO compared to that induced by E. coli LPS and Pam3CSK4 (Fig. 3D). We were unable to detect CD40, CD86, or iNOS mRNA or NO production in M0 and M2-Mϕ activated with any of the TLR ligands (Fig. 3A to D).
Effect of P. gingivalis LPS on IL-4-primed macrophages.
The ability of P. gingivalis LPS to induce an immune response and activate murine BMMϕ IL-4-primed macrophages (pre-M2-Mϕ) was investigated. BMMϕ were generated as described above and incubated overnight with IL-4 to produce a pre-M2-Mϕ phenotype (CD86− and CD206+) (Fig. 2F) before stimulation with two concentrations of P. gingivalis LPS, E. coli LPS, or Pam3CSK4 (Fig. 2G) to generate an M2 phenotype (M2-Mϕ).
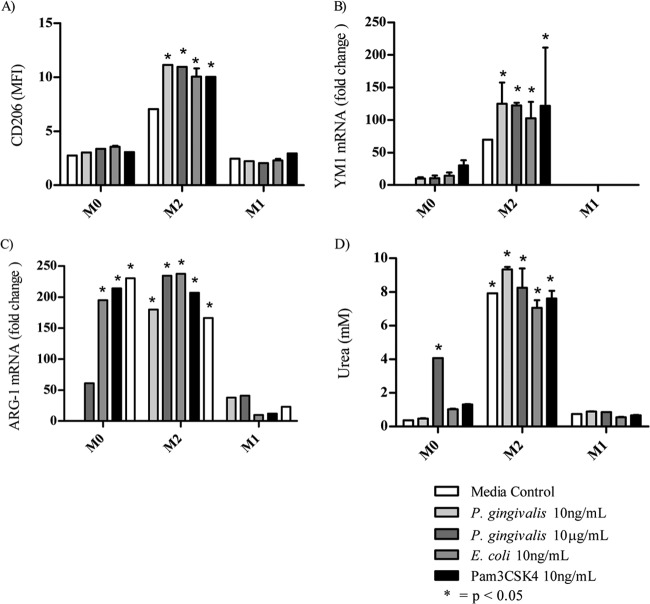
Prior to TLR ligand activation, the priming of M0-Mϕ to a pre-M2-Mϕ phenotype resulted in the significantly (P < 0.05) increased expression of M2-Mϕ costimulatory markers CD206 and YM-1 mRNA as well as the increased expression of Arg-1 mRNA (Fig. 4A to C). Despite the upregulation of Arg-1 mRNA in the M0-Mϕ after TLR ligation, only incubation with P. gingivalis LPS (10 μg/ml) resulted in urea secretion (Fig. 4D). Interestingly, all of the TLR ligands increased the expression of YM-1 mRNA and Arg-1 mRNA but not CD206 in the nonpolarized M0-Mϕ (Fig. 4A to C).
FIG 4.
Expression of M2 markers on BMMϕ after TLR ligation. M0-Mϕ were incubated with IL-4 to generate pre-M2-Mϕ before activation with various TLR ligands. (A) The M2-Mϕ generated were analyzed for their expression of CD206 by flow cytometry after approximately 24 h. The expression of YM-1 (B) and Arg-1 (C) mRNA was measured by semiquantitative reverse transcription-PCR. (D) The production of arginase was measured by analyzing the breakdown of l-arginine into urea. M1 markers were not expressed after the addition of IL-4. Data are presented as means ± SD from three biological replicates.
Incubation of pre-M2-Mϕ with P. gingivalis LPS resulted in a small but significant (P < 0.05) increase in CD206 and YM-1 mRNA expression comparable to that induced by E. coli LPS and Pam3CSK4 (Fig. 4A and B). Of the TLR ligands, only P. gingivalis LPS induced a small but significant increase in Arg-1 mRNA expression and urea secretion in M2-Mϕ (Fig. 4C and D). We were unable to detect significant levels of M2 markers pre- or postincubation with TLR ligands in pre-M1-Mϕ.
The production of cytokines and chemokines by M0, M1, and M2 polarized macrophages after polarization with P. gingivalis LPS preparation.
After demonstrating the increased expression of macrophage activation markers in response to P. gingivalis LPS, we investigated the production of cytokines and chemokines by M0, M1, and M2 polarized macrophages incubated with the different TLR ligands by cytokine bead array. Generally, inflammatory cytokines are produced by M1-Mϕ, whereas M2-Mϕ produce low levels of inflammatory cytokines. BMMϕ were produced, primed, and activated with the different TLR ligands as described above, and after 48 h the culture supernatants were collected and analyzed by cytokine bead (Bioplex) array assay.
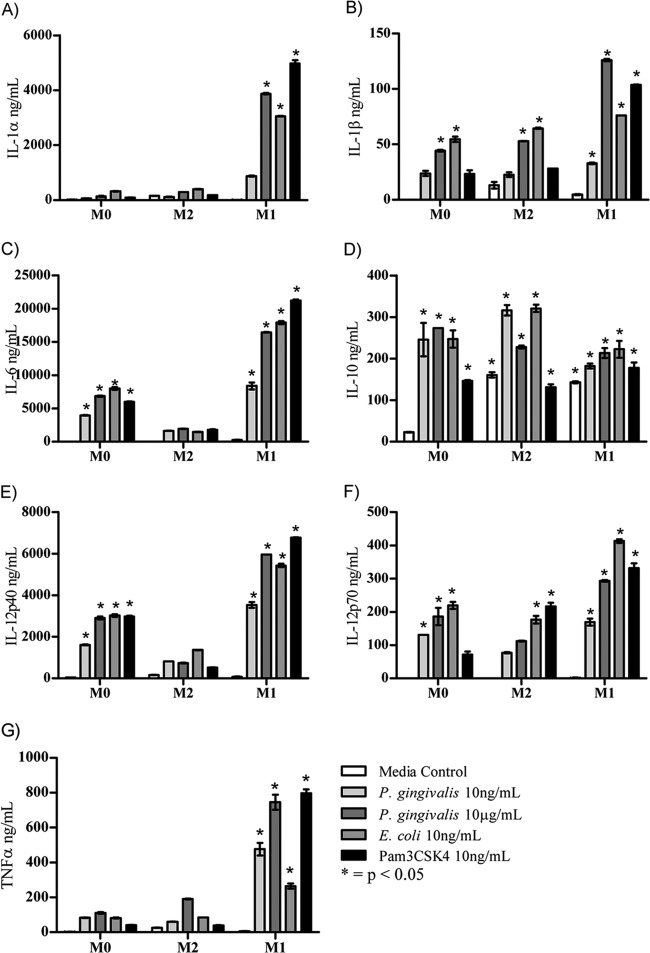
Of the 24 cytokines and chemokines analyzed in the culture supernatant, only twelve, i.e., IL-1α, IL-1β, IL-6, IL-10, IL-12p40, IL-12p70, TNF-α, KC, RANTES, eotaxin, MCP-1, and MIP-1α, were consistently secreted by either M0, M1, or M2 polarized Mϕ after incubation with P. gingivalis LPS, E. coli LPS, or Pam3CSK4 (Fig. 5 and 6). None of the TLR ligands induced the secretion of IL-2, IL-3, IL-4, IL-5, IL-9, IL-13, IL-17A, IFN-γ, G-CSF, GM-CSF, or MIP-1β by M0, M1, or M2-Mϕ (data not shown).
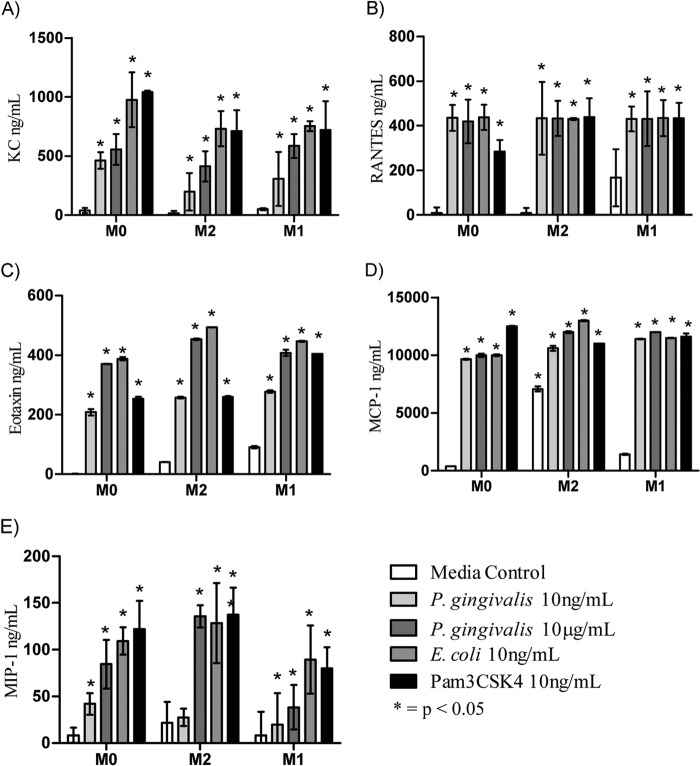
FIG 5.
Cytokine expression by bone marrow-derived macrophages. BMMϕ were primed with either IL-4 or IFN-γ overnight and activated with various concentrations of TLR ligand. Culture supernatant was removed after 48 h and analyzed using a 23-plex Bioplex assay. (A) IL-1α; (B) IL-1β; (C) IL-6; (D) IL-10; (E) IL-12p40; (F) IL-12p70; (G) TNF-α. Expression of IL-2, IL-3, IL-4, IL-5, IL-9, IL-13, IL-17A, or IFN-γ was not detected.
FIG 6.
Chemokine expression by bone marrow-derived macrophages. BMMϕ were primed with either IL-4 or IFN-γ overnight and activated with various concentrations of TLR ligand. Culture supernatant was removed 48 h later and analyzed using a 23-plex Bioplex assay. (A) KC, (B) RANTES, (C) eotaxin, (D) MCP-1, and (E) MIP-1α. Expression of G-CSF, GM-CSF, and MIP-1β was not detected.
The addition of the low-dose P. gingivalis LPS (10 ng/ml) to M0-Mϕ resulted in significant (P < 0.01) expression of IL-1β, IL-6, IL-10, IL-12 p40, and IL-12 p70 compared to that of medium-alone controls (Fig. 5B to F). The exposure of pre-M1-Mϕ to the low dose of P. gingivalis LPS gave significantly higher (P < 0.0001) expression of all of the cytokines mentioned above, as well as inducing the expression of IL-1α and TNF-α (Fig. 5A and G). Although P. gingivalis LPS at both concentrations induced the above-mentioned cytokines in pre-M1-Mϕ, to induce a similar level of cytokine as that induced by E. coli LPS and Pam3CSK4, a 1,000-fold higher concentration of P. gingivalis LPS was required. However, P. gingivalis LPS at the same concentration (10 ng/ml) as that of E. coli LPS induced significantly (P < 0.05) larger amounts of TNF-α (Fig. 5G). TLR ligation of pre-M1-Mϕ significantly (P < 0.05) increased the levels of cytokine production compared to M0-Mϕ regardless of the TLR ligand used, indicating that M1 polarized Mϕ have a much higher sensitivity to P. gingivalis LPS, even at low doses.
Incubation of pre-M2-Mϕ with P. gingivalis LPS resulted in the secretion of IL-1β, IL-12p70, and IL-10 at levels similar to that induced by either E. coli LPS and/or Pam3CSK4 and significantly (P < 0.05) higher than that of the medium-alone M2 polarized Mϕ control (Fig. 5B, D, and F). However, the levels of proinflammatory cytokines (IL-1β and IL-12p70) produced by M2-Mϕ always were lower than those produced by M1-Mϕ.
The production of chemokines KC, eotaxin, RANTES, MCP-1, and MIP-1α were dependent on TLR ligation and not on priming with cytokines, as there were no distinct differences in the levels of chemokines induced by the TLR ligands in M0, M1, and M2 polarized Mϕ (Fig. 6). RANTES and MCP-1 in particular were produced at similar concentrations after the addition of all TLR ligands tested (Fig. 6B and D). KC, MIP-1α, and eotaxin responded to P. gingivalis LPS in a dose-dependent manner and were produced at lower levels than those of E. coli LPS or Pam3CSK4 (Fig. 6A, C, and E).
The effect of TLR2 and TLR4 deficiency on macrophage activation by P. gingivalis LPS.
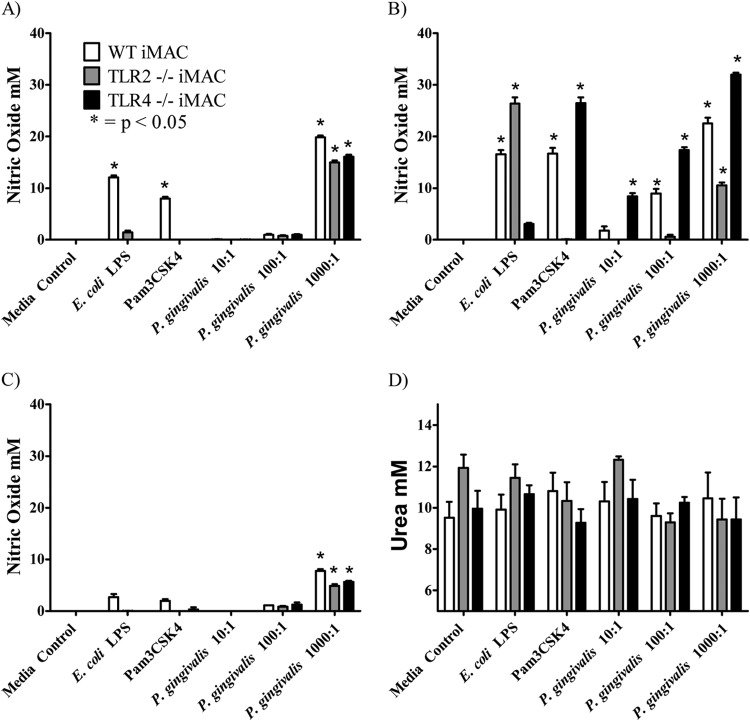
As the M1 and M2 macrophages responded differently to the TLR2 and TLR4 ligands (Pam3CSK4 and E. coli LPS, respectively) as well as to P. gingivalis LPS, the effect of removing one TLR signaling pathway on macrophage activation was investigated. Immortalized bone marrow-derived macrophages (iMACs), either from C57BL/6 mice (wild type [WT]) or TLR2- or TLR4-deficient mice (TLR2−/− and TLR4−/−), were primed with cytokines (IFN-γ and IL-4), and the resultant pre-M1 and pre-M2 macrophages then were exposed to P. gingivalis LPS (10 μg/ml) or control ligands. Nitric oxide secretion and arginase activity were measured as markers of mature M1 or M2 activation, respectively.
Unprimed M0 macrophages, as well as IL-4-primed M2 macrophages, produced negligible amounts of nitric oxide in response to E. coli LPS and no nitric oxide in response to Pam3CSK4 or P. gingivalis LPS (data not shown). Pre-M1 (IFN-γ-primed) macrophages derived from WT mice responded to all of the ligands tested in a manner similar to that previously observed with BMMϕ (Fig. 7A). Pre-M1 TLR2−/− macrophages were not activated by Pam3CSK4 but did respond to E. coli LPS, albeit at reduced levels compared to the pre-M1 WT macrophages. However, TLR2 deficiency in these pre-M1 macrophages completely eliminated the production of nitric oxide in response to P. gingivalis LPS (Fig. 7A). TLR4 deficiency removed the ability of pre-M1 to produce nitric oxide in response to E. coli LPS and had a reduced response to P. gingivalis LPS and Pam3CSK4 compared to the pre-M1 WT macrophages (Fig. 7A).
FIG 7.

Polarization of macrophages lacking TLR2 or TLR4. Immortalized macrophages (wild type, TLR2−/−, or TLR4−/−) were left unprimed or were primed overnight with IFN-γ (A) or IL-4 (B) and then activated with E. coli LPS (10 ng/ml), Pam3CSK4 (10 ng/ml), or P. gingivalis LPS (10 μg/ml). (A) Nitric oxide expression was used as a marker for M1 polarization. Results were normalized against the medium control. (B) Arginase activity (urea production) was used as a marker for M2 polarization.
Arginase was used as a marker for M2 macrophage polarization in this study, and as observed previously, arginase activity was induced by the addition of IL-4 alone. The additional exposure to TLR ligands had no additive effect on the level of arginase produced by these cell types (Fig. 7B).
M1 and M2 macrophage activation by P. gingivalis whole cells.
We have shown that P. gingivalis LPS weakly activates M1 macrophages. Therefore, experiments were performed with whole P. gingivalis cells in order to determine the polarization of macrophages to intact bacteria. Immortalized bone marrow-derived macrophages were primed with cytokine (IFN-γ and IL-4), and the resultant pre-M1 and pre-M2 macrophages then were exposed to various multiplicities of infection (MOI) of P. gingivalis whole cells. The next day, nitric oxide or arginase activity levels were measured as a marker of mature M1 or M2 activation.
Unprimed (M0) WT macrophages produced low levels of nitric oxide in response to both E. coli LPS and Pam3CSK4 as well as the high (1,000:1 MOI) dose of P. gingivalis whole cells (Fig. 8A). The TLR2−/− and TLR4−/− M0 macrophages produced nitric oxide in response to the high dose of P. gingivalis but at a lower level than that of the M0 WT macrophages. A similar activation pattern was observed for the IL-4-primed macrophages and the M0 (unprimed) macrophages (Fig. 8C).
FIG 8.
Polarization of macrophages by P. gingivalis whole cells. Immortalized macrophages (wild type, TLR2−/−, or TLR4−/−) were left unprimed (A), primed overnight with IFN-γ (B) or IL-4 (C and D), and then activated with E. coli LPS (10 ng/ml), Pam3CSK4 (10 ng/ml), or P. gingivalis at various MOIs (10:1, 100:1, or 1,000:1). (A, B, and C) Nitric oxide expression was used as a marker for M1 polarization. (D) Arginase activity (urea production) was used as a marker for M2 polarization. Results shown in panels A, B, and C were normalized against the medium control.
The M1 (IFN-γ-primed) WT macrophages produced significant amounts of nitric oxide in response to E. coli LPS and Pam3CSK4 and a dose-dependent response to P. gingivalis whole cells (Fig. 8B). The M1 TLR2−/− macrophages did not respond to Pam3CSK4 but did produce significant amounts of nitric oxide in response to E. coli LPS. M1 TLR2−/− macrophages produced nitric oxide in response to P. gingivalis whole cells only at the highest MOI of 1,000:1 and then at significantly (P < 0.05) lower levels than that produced by the M1 WT macrophages, indicating that the TLR2 ligands of P. gingivalis whole cells are the major stimulatory ligands. Confirming this, the M1 (IFN-γ-primed) TLR4−/− macrophages produced significant quantities of nitric oxide only in response to E. coli LPS, not Pam3CSK4, and were not compromised in their ability to respond to P. gingivalis whole cells. Moreover, they produce higher quantities of nitric oxide than the M1 WT macrophages.
Arginase was used as a marker for M2 macrophage polarization in this study, and as observed previously, arginase activity was induced by the addition of IL-4 alone. The additional exposure to TLR ligands had no effect on the level of arginase produced by these cell types (Fig. 8D).
DISCUSSION
The innate immune system is the first line of defense against pathogenic organisms. Macrophages are a critical component of this innate immune response and are integral for initiating and sustaining the adaptive immune response (19). We investigated the effect of priming macrophages with IFN-γ or IL-4 to produce a primed M1 or M2 macrophage phenotype, respectively, on their activation and immune response to P. gingivalis LPS. There are a number of studies that have investigated the cytokines present in diseased tissue during chronic periodontitis, reviewed in Gemmell et al. (34) and Pathirana et al. (35), and it is widely acknowledged that the levels of various proinflammatory cytokines are increased. This inflammatory milieu is the first environment that monocytes recruited from the bloodstream will encounter; therefore, a naive macrophage will be exposed to various cytokines and stimuli shortly after exiting the bloodstream. We have used exposure to two different polarizing cytokines and TLR ligands to mimic this exposure.
P. gingivalis LPS previously has been thought to be a TLR2 agonist based on its ability to activate cells from C3H/HeJ mice (7, 36). We found that the LPS preparation contained O-LPS, A-LPS, and CTD protein conjugate. The presence of a second type of LPS in P. gingivalis was first demonstrated by Rangarajan et al. (37), who showed that the O antigen of O-LPS was replaced with a phosphorylated-branch mannan repeating unit. The authors also demonstrated differences in the lipid A structures of the two types of LPS, with the A-LPS containing nonphosphorylated tetra- and penta-acylated isoforms which exhibited reduced proinflammatory activity compared with total LPS, containing both O-LPS and A-LPS (37). Recently, Chen et al. (30) suggested that A-LPS or a glycolipid containing the phosphorylated mannan epitope that reacts with MAb 1B5 was the cell surface anchor for a unique range of proteins secreted through the outer membrane and covalently attached to the surface of P. gingivalis. These secreted proteins contain a C-terminal signal sequence (CTD) which is cleaved from the protein at the surface and then is covalently attached to the membrane anchor (30, 32). The most abundant CTD proteins are the gingipains (RgpA/B and Kgp), and the RgpB conjugate is displayed on an SDS-PAGE gel as a diffuse 60- to 80-kDa band, whereas the unconjugated form of the protein is a sharp 50-kDa band (30, 32). The P. gingivalis LPS preparation used in this study contained the diffuse 60- to 80-kDa RgpB-conjugated band but not the 50-kDa unconjugated enzyme, indicating that the preparation contained the CTD protein conjugate.
We analyzed the activity of the P. gingivalis LPS preparation using a TLR2 or TLR4 HEK293 cell luciferase reporter assay system and found that both TLR2- and TLR4-expressing HEK293 cells were stimulated, indicating that the preparation was able to ligate TLR2 and TLR4. The TLR4 ligand of P. gingivalis LPS has been shown to be penta-acylated forms of lipid A, with the diphosphorylated form being the most potent (15, 38, 39). Although P. gingivalis TLR2 signaling has been attributed to a Pg1828 lipopeptide contaminant of the LPS preparation (10), Jain et al. (12) recently showed that a P. gingivalis mutant not producing PG1828 still stimulated TLR2 and suggested that P. gingivalis expresses a novel class of lipoproteins which contaminate P. gingivalis LPS preparations that stimulate TLR2. At this stage, due to the complex and atypical nature of P. gingivalis LPS, the molecular structure of the TLR2 agonist has not been identified (10–12, 38, 40–44). However, it is possible that the novel contaminant responsible for TLR2 stimulation is associated with the CTD protein conjugate, which is present even in ultrapure LPS preparations.
Several previous studies into the effects of P. gingivalis LPS on macrophages, both murine and human, have been reported. It has been found that P. gingivalis LPS fails to induce sustained cytokine responses in thioglycolate-induced peritoneal macrophages. In particular, IL-12p35 and TNF-α mRNA were quickly induced, but levels decreased within 10 h (36). The transient production of cytokines by human macrophages in response to P. gingivalis LPS also has been observed (45). Other groups, however, found that high levels of TNF-α and IL-12, along with many other macrophage-derived cytokines and chemokines, still were produced by mouse macrophages at 24 h (46). In this investigation, nonpolarized (M0) macrophages were found to produce significant levels of IL-1β, IL-6, IL-12, and TNF-α 48 h after TLR stimulation. Differences in LPS preparations may explain the variation in cytokine expression observed between reports, as the complex and atypical nature of P. gingivalis LPS has only recently been discovered. In the current study, we did find that several of the cytokines previous groups tested were induced by P. gingivalis LPS even at low doses, and exposure to IFN-γ or IL-4, producing M1 or M2 polarized Mϕ, induces significantly higher proinflammatory cytokine responses than M0-Mϕ.
The results of this study indicate that P. gingivalis LPS, when used at concentrations similar to those used for E. coli LPS or Pam3CSK4, is unable to cause the maturation of macrophages primed with IFN-γ into M1 macrophages, as measured by upregulation of CD40 and CD86 and iNOS expression. However, the cytokine expression profile of these cells suggests that low doses of P. gingivalis LPS are able to produce significant levels of cytokines despite not being able to induce high levels of costimulatory marker expression or antimicrobial compounds. The ability to induce cytokine expression but not upregulate surface marker expression may be an example of the ability of P. gingivalis to dysregulate the immune response (47). The ability to promote inflammation by IL-1β, IL-6, TNF-α, and IL-12 production and recruit other immune cells through chemokine production but not activate the adaptive immune system through costimulatory marker expression may result in a chronic inflammatory condition without clearance of the pathogen or pathogen products (e.g., outer membrane vesicles) released from the biofilm into the subjacent gingival tissue (48).
We further demonstrated that to obtain levels of costimulatory marker activation at a level similar to that of E. coli LPS or Pam3CSK4, 1,000-fold higher levels of P. gingivalis LPS are required. P. gingivalis is known to produce monophosphorylated penta- or tetra-acylated lipid A molecules depending on the environmental stimulus (49, 50). A number of studies have shown that monophosphorylated penta- or tetra-acylated lipid A molecules have significantly reduced biological activity compared to diphosphorylated hexa-acylated lipid A molecules typically produced by E. coli (39, 51, 52). Despite P. gingivalis producing lipid A moieties of differing structures depending on environmental stimuli, only penta-acylated monophosphorylated forms or forms with fewer acyl chains and, hence, even less stimulatory activity have been observed (38, 49). This gives a plausible explanation for the low level of costimulatory marker activation by P. gingivalis LPS even after macrophage priming with IFN-γ.
The P. gingivalis LPS preparation, even at low concentrations, induced TNF-α at levels higher than that of E. coli LPS and comparable to that of Pam3CSK4. TNF-α, as well as other proinflammatory cytokines, such as IL-1β and IL-6, plays an important role in the pathogenesis of chronic periodontitis and is implicated in the systemic complications arising from the disease (53, 54). The higher TNF-α production by macrophages in response to P. gingivalis LPS observed in this study indicates that macrophages are a significant source of this inflammatory cytokine and play a central role in driving the inflammation observed in chronic periodontitis. Indeed, it has been shown recently that the absence of bone loss in TLR2 knockout mice can be reconstituted by the adoptive transfer of TLR2-expressing macrophages (55), emphasizing their importance in chronic periodontitis. However, the lack of costimulatory molecule expression and the inability to produce significant quantities of nitric oxide at biologically relevant doses indicates that they are not fully effective at clearing P. gingivalis. These results must be interpreted in the context of the whole bacterium and, indeed, the whole polymicrobial biofilm in which P. gingivalis exists. Other TLR ligands released into the host tissue may be able to compensate for the lack of immunological activity of the P. gingivalis LPS. Interestingly, where P. gingivalis has emerged as a major constituent of the biofilm against the periodontal pocket epithelium, its LPS may subvert innate responses to the other bacteria, as it has been shown to interfere with TLR4 signaling by LPS from other bacteria (14).
The results of the TLR knockout experiments, combined with the TLR activation assays, indicate that TLR2 is the main activating receptor for the P. gingivalis LPS preparation. The lack of TLR2 completely eliminated nitric oxide production by macrophages in response to P. gingivalis LPS, whereas the lack of TLR4 only lowered the response. The knockout macrophage cell lines also were exposed to whole P. gingivalis cells to investigate the ability of other TLR ligands present on the cell to compensate for the lack of TLR2 stimulation. Unprimed M0 and IL-4-primed M2 macrophages were able to produce nitric oxide at the highest MOI tested, 1,000:1, in the latter case overcoming the IL-4 effect. This indicates that a high enough exposure to TLR ligands can alter the phenotype of the macrophage regardless of the cytokine priming.
Macrophages derived from C57BL6 mice exhibited a dose-dependent response to P. gingivalis whole cells, as did the cells lacking TLR4. The elimination of TLR2 signaling resulted in no nitric oxide production by M1 macrophages until the highest MOI (1,000:1) where nitric oxide was produced, although at lower levels than that produced by WT. These results also suggest that TLR2 is the main signaling pathway that macrophages use to detect P. gingivalis.
The current study also investigated the ability of P. gingivalis LPS to stimulate alternatively activated macrophages, such as M2 polarized Mϕ. This was measured by a lack of inflammatory cytokine production, the production of arginase-1, and the expression of YM-1 (among other markers) (56). Our results demonstrated that low levels of YM-1 and high levels of arginase-1 were able to be induced by the addition of TLR ligand alone without the need to prime macrophages with IL-4. It has been shown previously that, in addition to IFN-γ and IL-4 cytokines producing M1 and M2 polarized macrophage phenotypes, respectively, the addition of GM-CSF and CSF-1 polarizes murine macrophages (57, 58). The use of CSF-1 in this study to generate a M0 macrophage phenotype from bone marrow cells may have predisposed the M0-Mϕ to the generation of M2 markers. However, this M2-Mϕ predisposition did not prevent the expression of M1 macrophages markers after the addition of IFN-γ, indicating a certain amount of plasticity in the phenotypes (26). Furthermore, our results demonstrate that IL-4 alone was able to induce the M2 phenotype without the addition of further TLR ligands, although the addition of TLR ligand did increase the expression of certain M2 markers. M2 macrophages have been shown play a role in tissue remodelling by increasing the production of effectors that both synthesize and degrade collagen (59) and are becoming the focus of research in fibrotic diseases (60, 61). Fibrotic gingival tissue is commonly observed in chronic periodontitis (25), and gingival fibroblasts have been thought to be largely responsible for collagen remodelling during disease (62). However, an M2 macrophage marker, CD163 (RM3/1), found associated with healthy gingival tissue, also has been found in chronically inflamed periodontal tissue (63, 64). It has been shown recently that P. gingivalis tolerates M2 macrophages but not M1 macrophages (65), potentially suppressing the anti-inflammatory and tissue repair functions of M2 macrophages while the proinflammatory M1 macrophages remain unaffected. Considering the role of M2 macrophages in differentiation of fibrocytes and fibroblasts in liver fibrosis (60), M2 macrophages require more attention in the context of periodontal disease, particularly as better analytical techniques and more reliable biomarkers become available.
The alternative activation of macrophages during this study was found to be largely independent of TLR ligation. However, nonpolarized (M0) macrophages upregulated the M2 marker arginase-1 after exposure to P. gingivalis LPS, which may have been due to their culture with CSF-1 and warrants further investigation. The significant levels of cytokines produced by M1 macrophages in this study, combined with the inability to induce nitric oxide and costimulatory marker expression at similar levels of TLR ligand, may contribute to the immune dysregulation observed during chronic periodontitis. This dysregulation may lead to the survival and persistence of P. gingivalis in subgingival plaque and its products in gingival tissues, resulting in chronic inflammation.
ACKNOWLEDGMENT
This work was supported by National Health and Medical Research Council Australia grant APP1029878.
Footnotes
Published ahead of print 21 July 2014
REFERENCES
- 1. Oliver RC, Brown LJ. 1993. Periodontal diseases and tooth loss. Periodontol. 2000 2:117–127. 10.1111/j.1600-0757.1993.tb00224.x [DOI] [PubMed] [Google Scholar]
- 2. Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- 3. Moskow BS, Polson AM. 1991. Histologic studies on the extension of the inflammatory infiltrate in human periodontitis. J. Clin. Periodontol. 18:534–542. 10.1111/j.1600-051X.1991.tb00086.x [DOI] [PubMed] [Google Scholar]
- 4. Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499–511. 10.1038/nri1391 [DOI] [PubMed] [Google Scholar]
- 5. Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10:717–725. 10.1038/nrmicro2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- 7. Bainbridge BW, Coats SR, Darveau RP. 2002. Porphyromonas gingivalis lipopolysaccharide displays functionally diverse interactions with the innate host defense system. Ann. Periodontol. 7:29–37. 10.1902/annals.2002.7.1.29 [DOI] [PubMed] [Google Scholar]
- 8. Gordon S, Taylor PR. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964. 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- 9. Kocgozlu L, Elkaim R, Tenenbaum H, Werner S. 2009. Variable cell responses to P. gingivalis lipopolysaccharide. J. Dent. Res. 88:741–745. 10.1177/0022034509341166 [DOI] [PubMed] [Google Scholar]
- 10. Asai Y, Hashimoto M, Fletcher HM. 2005. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated. Infect. Immun. 73:2157–2157. 10.1128/IAI.73.4.2157-2163.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimoto M, Asai Y, Ogawa T. 2004. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int. Immunol. 16:1431–1437. 10.1093/intimm/dxh146 [DOI] [PubMed] [Google Scholar]
- 12. Jain S, Coats SR, Chang AM, Darveau RP. 2013. A novel class of lipoprotein lipase-sensitive molecules mediates TLR2 activation by Porphyromonas gingivalis. Infect. Immun. 81:1277–1286. 10.1128/IAI.01036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogawa T, Asai Y, Hashimoto M, Takeuchi O, Kurita T, Yoshikai Y, Miyake K, Akira S. 2002. Cell activation by Porphyromonas gingivalis lipid A molecule through Toll-like receptor 4- and myeloid differentiation factor 88-dependent signaling pathway. Int. Immunol. 14:1325–1332. 10.1093/intimm/dxf097 [DOI] [PubMed] [Google Scholar]
- 14. Coats SR, Reife RA, Bainbridge BW, Pham TT, Darveau RP. 2003. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infect. Immun. 71:6799–6807. 10.1128/IAI.71.12.6799-6807.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herath TD, Darveau RP, Seneviratne CJ, Wang CY, Wang Y, Jin L. 2013. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One 8:e58496. 10.1371/journal.pone.0058496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benoit M, Desnues B, Mege J-L. 2008. Macrophage polarization in bacterial infections. J. Immunol. 181:3733–3739. 10.4049/jimmunol.181.6.3733 [DOI] [PubMed] [Google Scholar]
- 17. Dalton D, Pitts-Meek S, Keshav S, Figari I, Bradley A, Stewart T. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259:1739–1742. 10.1126/science.8456300 [DOI] [PubMed] [Google Scholar]
- 18. Edwards JP, Zhang X, Frauwirth KA, Mosser DM. 2006. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 80:1298–1298. 10.1189/jlb.0406249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. 2005. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 23:901–944. 10.1146/annurev.immunol.23.021704.115816 [DOI] [PubMed] [Google Scholar]
- 20. Stein M, Keshav S, Harris N, Gordon S. 1992. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 176:287–292. 10.1084/jem.176.1.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doyle AG, Herbein G, Montaner LJ, Mintyo AJ, Caputo D, Gordon S, Minty AJ, Caput D, Ferrara P. 1994. Interleukin-13 alters the activation state of murine macrophages in vitro: comparison with interleukin-4 and interferon-γ. Eur. J. Immunol. 24:1441–1445. 10.1002/eji.1830240630 [DOI] [PubMed] [Google Scholar]
- 22. Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. 2001. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J. Immunol. 167:6533–6544. 10.4049/jimmunol.167.11.6533 [DOI] [PubMed] [Google Scholar]
- 23. Gharib SA, Johnston LK, Huizar I, Birkland TP, Hanson J, Wang Y, Parks WC, Manicone AM. 2014. MMP28 promotes macrophage polarization toward M2 cells and augments pulmonary fibrosis. J. Leukoc. Biol. 95:9–18. 10.1189/jlb.1112587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho VW, Sly LM. 2009. Derivation and characterization of murine alternatively activated (M2) macrophages. Methods Mol. Biol. 531:173–185. 10.1007/978-1-59745-396-7_12 [DOI] [PubMed] [Google Scholar]
- 25. Chavrier C, Couble ML, Hartmann D, Grimaud JA, Magloire H. 1987. Immunohistochemical study of types I, III and IV collagen in fibrosis of diseased gingiva during chronic periodontitis: a light and electron microscopic study. J. Periodont. Res. 22:29–36. 10.1111/j.1600-0765.1987.tb01536.x [DOI] [PubMed] [Google Scholar]
- 26. Galli SJ, Borregaard N, Wynn TA. 2011. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat. Rev. Immunol. 12:1035–1044. 10.1038/ni.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lam RS, O'Brien-Simpson NM, Lenzo JC, Holden JA, Brammar GC, Walsh KA, McNaughtan JE, Rowler DK, Van Rooijen N, Reynolds EC. 28 July 2014. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J. Immunol. 10.4049/jimmunol.1400853 [DOI] [PubMed] [Google Scholar]
- 28. Westphal O, Jann K. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further application to the procedure p 83 In Whistler RL. (ed), Methods in carbohydrate chemistry, vol 5 Academic Press, New York, NY [Google Scholar]
- 29. Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 165:618–622. 10.4049/jimmunol.165.2.618 [DOI] [PubMed] [Google Scholar]
- 30. Chen Y-Y, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 79:1380–1401. 10.1111/j.1365-2958.2010.07530.x [DOI] [PubMed] [Google Scholar]
- 31. Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 287:24605–24617. 10.1074/jbc.M112.369223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh G. 2002. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J. Leukoc. Biol. 71:597–597. 10.1371/journal.pone.0031205 [DOI] [PubMed] [Google Scholar]
- 34. Gemmell E, Carter CL, Hart DNJ, Drysdale KE, Seymour GJ. 2002. Antigen-presenting cells in human periodontal disease tissues. Oral Microbiol. Immunol. 17:388–393. 10.1034/j.1399-302X.2002.170609.x [DOI] [PubMed] [Google Scholar]
- 35. Pathirana RD, O'Brien-Simpson NM, Reynolds EC. 2010. Host immune responses to Porphyromonas gingivalis antigens. Periodontol. 2000 52:218–237. 10.1111/j.1600-0757.2009.00330.x [DOI] [PubMed] [Google Scholar]
- 36. Hirschfeld M, Weis JJ, Toshchakov V, Salkowski CA, Cody MJ, Ward DC, Qureshi N, Michalek SM, Vogel SN. 2001. Signaling by toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477–1477. 10.1128/IAI.69.3.1477-1482.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920–2932. 10.1128/JB.01868-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell. Microbiol. 11:1587–1599. 10.1111/j.1462-5822.2009.01349.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, Darveau RP. 2014. Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infect. Immun. 82:650–659. 10.1128/IAI.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72:5041–5051. 10.1128/IAI.72.9.5041-5051.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kumada H, Haishima Y, Watanabe K, Hasegawa C, Tsuchiya T, Tanamoto K, Umemoto T. 2008. Biological properties of the native and synthetic lipid A of Porphyromonas gingivalis lipopolysaccharide. Oral Microbiol. Immunol. 23:60–69. 10.1111/j.1399-302X.2007.00392.x [DOI] [PubMed] [Google Scholar]
- 42. Ogawa T, Yagi T. 2010. Bioactive mechanism of Porphyromonas gingivalis lipid A. Periodontol. 2000 54:71–77. 10.1111/j.1600-0757.2009.00343.x [DOI] [PubMed] [Google Scholar]
- 43. Ogawa T, Asai Y, Makimura Y, Tamai R. 2007. Chemical structure and immunobiological activity of Porphyromonas gingivalis lipid A. Front. Biosci. 12:3795–3812. 10.2741/2353 [DOI] [PubMed] [Google Scholar]
- 44. Sawada N, Ogawa T, Asai Y, Makimura Y, Sugiyama A. 2007. Toll-like receptor 4-dependent recognition of structurally different forms of chemically synthesized lipid As of Porphyromonas gingivalis. Clin. Exp. Immunol. 148:529–536. 10.1111/j.1365-2249.2007.03346.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barksby HE, Nile CJ, Jaedicke KM, Taylor JJ, Preshaw PM. 2009. Differential expression of immunoregulatory genes in monocytes in response to Porphyromonas gingivalis and Escherichia coli lipopolysaccharide. Clin. Exp. Immunol. 156:479–487. 10.1111/j.1365-2249.2009.03920.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou Q, Desta T, Fenton M, Graves DT, Amar S. 2005. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect. Immun. 73:935–943. 10.1128/IAI.73.2.935-943.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tam V, O'Brien-Simpson NM, Chen Y-Y, Sanderson CJ, Kinnear B, Reynolds EC. 2009. The RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis inactivate the Th2 cytokines interleukin-4 and interleukin-5. Infect. Immun. 77:1451–1458. 10.1128/IAI.01377-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. 2009. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect. Immun. 77:1246–1261. 10.1128/IAI.01038-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. 2006. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 74:4474–4485. 10.1128/IAI.01924-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, Tarelli E, Marsh PD. 2011. Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infect. Immun. 79:1187–1193. 10.1128/IAI.00900-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bäckhed F, Normark S, Schweda EKH, Oscarson S, Richter-Dahlfors A. 2003. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microb. Infect. 5:1057–1063. 10.1016/S1286-4579(03)00207-7 [DOI] [PubMed] [Google Scholar]
- 52. Flad H-D, Loppnow H, Rietschel ET, Ulmer AJ. 1993. Agonists and antagonists for lipopolysaccharide-induced cytokines. Immunobiology 187:303–316. 10.1016/S0171-2985(11)80346-3 [DOI] [PubMed] [Google Scholar]
- 53. Diya Z, Lili C, Shenglai L, Zhiyuan G, Jie Y. 2008. Lipopolysaccharide (LPS) of Porphyromonas gingivalis induces IL-1beta, TNF-alpha and IL-6 production by THP-1 cells in a way different from that of Escherichia coli LPS. Innate Immun. 14:99–107. 10.1177/1753425907088244 [DOI] [PubMed] [Google Scholar]
- 54. Nishimura F, Iwamoto Y, Mineshiba J, Shimizu A, Soga Y, Murayama Y. 2003. Periodontal disease and diabetes mellitus: the role of tumor necrosis factor-α in a 2-way relationship. J. Periodontol. 74:97–102. 10.1902/jop.2003.74.1.97 [DOI] [PubMed] [Google Scholar]
- 55. Papadopoulos G, Weinberg EO, Massari P, Gibson FC, Wetzler LM, Morgan EF, Genco CA. 2013. Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J. Immunol. 190:1148–1157. 10.4049/jimmunol.1202511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. 2005. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol. 174:6561–6562. 10.4049/jimmunol.174.11.6561 [DOI] [PubMed] [Google Scholar]
- 57. Fleetwood AJAJ, Lawrence T, Hamilton JA, Cook AD. 2007. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178:5245–5252. 10.4049/jimmunol.178.8.5245 [DOI] [PubMed] [Google Scholar]
- 58. Sierra-Filardi E, Vega MA, Sa P, Corbı AL, Sánchez-Mateos P, Corbí AL, Puig-Kröger A. 2010. Heme oxygenase-1 expression in M-CSF-polarized M2 macrophages contributes to LPS-induced IL-10 release. Immunobiology 215:788–795. 10.1016/j.imbio.2010.05.020 [DOI] [PubMed] [Google Scholar]
- 59. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25:677–686. 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 60. Heymann F, Trautwein C, Tacke F. 2009. Monocytes and macrophages as cellular targets in liver fibrosis. Inflamm. Allergy Drug Targets 8:307–318. 10.2174/187152809789352230 [DOI] [PubMed] [Google Scholar]
- 61. Wynn TA. 2004. Fibrotic disease and the TH1/TH2 paradigm. Nat. Rev. Immunol. 4:583–594. 10.1038/nri1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Larjava H, Sandberg M, Vuorio E. 1989. Altered distribution of type I collagen mRNA in periodontal disease. J. Periodont. Res. 24:171–177. 10.1111/j.1600-0765.1989.tb02002.x [DOI] [PubMed] [Google Scholar]
- 63. Chapple CC, Srivastava M, Hunter N. 1998. Failure of macrophage activation in destructive periodontal disease. J. Pathol. 186:281–286 [DOI] [PubMed] [Google Scholar]
- 64. Schlegel Gómez R, Langer P, Pelka M, Driesch P, Johannessen AC, Simon M., Jr 1995. Variational expression of functionally different macrophage markers (27E10, 25F9, RM3/1) in normal gingiva and inflammatory periodontal disease. J. Clin. Periodontol. 22:341–346. 10.1111/j.1600-051X.1995.tb00159.x [DOI] [PubMed] [Google Scholar]
- 65. Foey AD, Crean S. 2013. Macrophage subset sensitivity to endotoxin tolerisation by Porphyromonas gingivalis. PLoS One 8:e67955–e67955. 10.1371/journal.pone.0067955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. National Health and Medical Research Council. 2013. Australian code for the care and use of animals for scientific purposes, 8th ed. National Health and Medical Research Council, Canberra, Australia [Google Scholar]