Abstract
Ehrlichia chaffeensis is an obligately intracellular Gram-negative bacterium that selectively infects mononuclear phagocytes. We recently reported that E. chaffeensis utilizes a type 1 secretion (T1S) system to export tandem repeat protein (TRP) effectors and demonstrated that these effectors interact with a functionally diverse array of host proteins. By way of these interactions, TRP effectors modulate host cell functions; however, the molecular basis of these interactions and their roles in ehrlichial pathobiology are not well defined. In this study, we describe the first bacterial protein posttranslational modification (PTM) by the small ubiquitin-like modifier (SUMO). The E. chaffeensis T1S effector TRP120 is conjugated to SUMO at a carboxy-terminal canonical consensus SUMO conjugation motif in vitro and in human cells. In human cells, TRP120 was selectively conjugated with SUMO2/3 isoforms. Disruption of TRP120 SUMOylation perturbed interactions with known host proteins, through predicted SUMO interaction motif-dependent and -independent mechanisms. E. chaffeensis infection did not result in dramatic changes in the global host SUMOylated protein profile, but a robust colocalization of predominately SUMO1 with ehrlichial inclusions was observed. Inhibiting the SUMO pathway with a small-molecule inhibitor had a significant impact on E. chaffeensis replication and recruitment of the TRP120-interacting protein polycomb group ring finger protein 5 (PCGF5) to the inclusion, indicating that the SUMO pathway is critical for intracellular survival. This study reveals the novel exploitation of the SUMO pathway by Ehrlichia, which facilitates effector-eukaryote interactions necessary to usurp the host and create a permissive intracellular niche.
INTRODUCTION
Ehrlichia chaffeensis, the etiologic agent of the life-threatening tick-borne zoonosis human monocytotropic ehrlichiosis (HME), is an obligately intracellular Gram-negative bacterium that selectively infects mononuclear phagocytes and replicates in cytoplasmic vacuoles resembling endosomes (1–3). The mechanisms through which E. chaffeensis directs internalization, establishes intracellular infection, and avoids innate and adaptive host defenses are not well understood. However, we identified a group of type 1 secretion (T1S) system ehrlichial tandem repeat protein (TRP) effectors, similar to the repeats-in-toxin family of exoproteins, that are involved in novel molecular interactions with a large group of functionally diverse host cell proteins and host cell DNA (4–9).
E. chaffeensis TRP120 is a major immunoreactive protein that is found on the surfaces of infectious dense-cored ehrlichiae and is expressed in both arthropod and mammalian cells (10, 11). A single major linear epitope (22 amino acids) in the tandem repeat region of TRP120 has been identified that elicits protective antibodies (12). Following T1S, TRP120 crosses the ehrlichial vacuole membrane through an unknown mechanism, similar to the Chlamydia trachomatis protein effector CPAF (13), and is present in the host cell cytosol, where it is involved in numerous interactions with functionally important eukaryotic proteins, some of which exhibit multifunctional (moonlighting) capabilities (7, 14). A small proportion of TRP120 is translocated to the host cell nucleus, where it directly binds host cell DNA and targets genes associated with transcriptional regulation, apoptosis, and vesicle trafficking (4). In addition, TRP120 interacts with several host proteins involved in posttranslational modification, including enzymes required for ligation and conjugation of ubiquitin (Ub) and ubiquitin-like modifiers (7). Many TRP120-interacting proteins contain predicted small ubiquitin-like modifier (SUMO) interaction motifs (SIMs), which are protein motifs characterized as short hydrophobic stretches flanked by acidic residues involved in SUMO-mediated protein-protein interactions (15). The high frequency of interactions with predicted SIM-containing proteins suggests that SUMOylation may contribute to the underlying molecular basis of TRP120's host-pathogen molecular interactions.
SUMO shares structural homology and a similar three-step conjugation pathway with ubiquitin, but it engenders unique properties and functions for its targets (16). SUMOylation is a reversible regulatory modification that potentiates changes in protein conformation and stability, protein-protein interactions, and protein localization and translocation. More than 90% of the human proteome is thought to be targeted by the ubiquitin pathway, while significantly fewer proteins have been identified as substrates of the SUMO pathway (17–20). Identification of SUMO substrates has largely been hindered by the fact that only a small proportion of the available target protein is modified and the covalent modification is dynamic and quickly turned over, preventing capture of natively modified proteins (21–25). However, this small proportion of conjugated target protein renders significant structural and functional consequences, an observation coined the “SUMO enigma,” and suggests that this eukaryotic posttranslational modification (PTM) pathway is tightly regulated (16, 26).
The machinery required for SUMOylation and ubiquitination is unique to eukaryotes; however, pathogens, particularly those that survive in intracellular niches, exploit this pathway in various ways in order to modulate the host cell (27–29). Viruses and bacteria are known to modulate host cellular functions by mimicking, inhibiting, or serving as substrates of the SUMO pathway (30, 31). Some type 3 and type 4 secreted bacterial effectors modulate global host cell SUMOylation. Listeria monocytogenes-secreted LLO toxin mediates proteasome-independent degradation of the host SUMO E2 ligase (Ubc9) (32, 33), and Xanthomonas campestris secretes XopD, which mimics the activity of host SUMO-specific isopeptidases (SENPs) (34). These bacterial effectors disrupt and decrease global cellular SUMO conjugation, respectively, but have not been identified as substrates of the SUMO pathway. To date, descriptions of SUMO-conjugated pathogen proteins are limited to viruses, for which protein SUMOylation facilitates viral entry, nuclear translocation, changes in gene expression, and vesicular trafficking (35).
Here we report the first SUMOylated bacterial protein and demonstrate that the SUMO pathway plays an important role in intracellular survival of E. chaffeensis. The E. chaffeensis T1S TRP120 effector is conjugated with SUMO at a canonical consensus motif that enhances interactions with defined host protein targets and recruitment of host proteins to the ehrlichial vacuole. The importance of the host SUMO pathway in E. chaffeensis effector-host interactions and survival reveals a novel mechanism through which Ehrlichia, and potentially other intracellular bacteria, exploits host pathways to promote survival.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
Human cervical epithelial adenocarcinoma cells (HeLa; ATCC, Manassas, VA) were cultured in minimum essential medium (MEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT). Human monocytic leukemia cells (THP-1; ATCC) were grown in RPMI 1640 medium with l-glutamine, 1 mM sodium pyruvate (Sigma, St. Louis, MO), 25 mM HEPES (Invitrogen), 2.5 g/liter d-(+)-glucose (Sigma), and 10% FBS (HyClone). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (11).
Antibodies.
Polyclonal rabbit and mouse anti-TRP120 antibodies were generated against a peptide (SKVEQEETNPEVLIKDLQDVAS) and were previously described (36). Other antibodies used in this study were anti-pan-SUMO (Abgent, San Diego, CA), anti-SUMO1 (Enzo Life Sciences), anti-SUMO2/3 (Enzo Life Sciences), anti-Aequorea coerulescens green fluorescent protein (anti-GFP; Clontech, Mountain View, CA), anti-hemagglutinin (anti-HA; Thermo Scientific, West Palm Beach, FL), anti-Golgi-associated gamma adaptin ear-containing ARF binding protein 1 (anti-GGA1; Thermo Scientific, West Palm Beach, FL), anti-actin gamma 1 (anti-ACTG1; Santa Cruz Biotechnology, Santa Cruz, CA), anti-myosin-X (anti-MyoX; Millipore, Billerica, MA), anti-polycomb group ring finger 5 (anti-PCGF5; Abcam, Cambridge, MA), anti-ubiquitin-like modifier activating enzyme 2 (anti-Uba2; Santa Cruz Biotechnology), and anti-ubiquitin conjugating enzyme (anti-Ubc9; Santa Cruz Biotechnology).
Immunofluorescence microscopy.
Uninfected or E. chaffeensis-infected THP-1 cells were cytocentrifuged onto glass slides, fixed with ice-cold acetone for 10 min, incubated with E. chaffeensis-infected dog serum (1:1,000), mouse anti-TRP120 (1:1,000), rabbit anti-pan-SUMO (1:100), or rabbit anti-SUMO1 or anti-SUMO2/3 (1:100) for 1 h, washed, and stained with fluorescein isothiocyanate (FITC)-conjugated or Alexa Fluor 568-IgG(H+L) and Alexa Fluor 488-IgG(H+L) secondary antibodies (1:100; Molecular Probes) for 30 min. HeLa cells were collected 24 h following transfection (Lipofectamine 2000; Invitrogen), fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized, and blocked with 0.3% Triton X-100 and 2% bovine serum albumin in PBS for 2 h. Cells were then incubated with anti-TRP120 (1:20,000) and anti-GGA1, anti-ACTG1, or anti-Myo10 (1:100) primary antibodies for 1 h, washed, and treated with Alexa Fluor 568-IgG(H+L) and Alexa Fluor 488-IgG(H+L) secondary antibodies (1:100; Molecular Probes) for 30 min. All slides were washed and mounted with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen). Images were obtained using an Olympus BX61 epifluorescence microscope and were analyzed using Slidebook software (version 5.0; Intelligent Imaging Innovations, Denver, CO).
2DE.
Two-dimensional electrophoresis (2DE) was performed by Kendrick Labs, Inc. (Madison, WI), as previously reported (37, 38). Briefly, THP-1 cells were collected and lysed (uninfected or at 72 h postinfection [p.i.]) in 50 mM N-ethylmaleimide (NEM; Sigma). Using the carrier ampholyte method, isoelectric focusing was carried out using 2% Servalyt pH 4-8 mix (Serva, Heidelberg, Germany) for 20,000 V-h, followed by SDS slab gel electrophoresis (7% acrylamide gel) for 5 h at 25 mA/gel. After slab gel electrophoresis, gels were transblotted onto polyvinylidene difluoride (PVDF) membranes for immunoblotting with anti-TRP120 or anti-pan-SUMO antibody.
Expression constructs and site-directed mutagenesis.
As previously described (4), the full coding sequence of TRP120 (GenBank accession number U49426) was cloned into pAcGFP1-C (Clontech), which encodes an N-terminal Aequorea coerulescens green fluorescent protein fusion. Generation of the TRP120 bacterial expression construct (pBAD/Thio-TRP120) was described previously (4). TRP120 point mutants were generated with pAcGFP1 and pBAD/Thio constructs by using QuikChange II site-directed mutagenesis (Agilent, Santa Clara, CA) with sense and antisense primers (TRP120 K432R sense primer, 5′-AAATGTTTGCACCTTCATTTAATCCAATCGTTATACGGGAGGAAGATAAAGTTTG-3′; TRP120 K432R antisense primer, 5′-CAAACTTTATCTTCCTCCCGTATAACGATTGGATTAAATGAAGGTGCAAACATTT-3′; TRP120 E434A sense primer, 5′-TAATCCAATCGTTATAAAGGAGGCAGATAAAGTTTGTGAAACTTGCG-3′; and TRP120 E434A antisense primer, 5′-CGCAAGTTTCACAAACTTTATCTGCCTCCTTTATAACGATTGGATTA-3′). HA-tagged SUMO constructs were acquired from Addgene (pcDNA3-HA-SUMO1 [plasmid 21154] [39], SRa-HA-SUMO2 [plasmid 17360] [40], and pcDNA3-HA-SUMO3 [plasmid 17361] [40]). ProLabel-tagged expression constructs were generated for selected host proteins known to interact with TRP120, identified in previous yeast two-hybrid (Y2H) studies (4). Briefly, these host genes were amplified from pGADT7-prey constructs (Matchmaker Gold yeast two-hybrid system; Clontech) and cloned in frame downstream of the ProLabel tag (6-kDa fragment of β-galactosidase) in the pProLabel-C vector (Clontech).
Expression and purification of recombinant TRP120.
The pBAD/TOPO-Thio.TRP120 bacterial expression vector was transformed into Escherichia coli TOP10 (Invitrogen). Overnight cultures were diluted 1:20 in LB plus ampicillin (Amp) and grown for 2 h with agitation at 37°C, and protein expression was induced with 0.5% arabinose for 3 h at 37°C. For purification, cells were lysed by sonication in PBS (Sigma) with protease inhibitors (Complete mini, EDTA-free; Roche) and then centrifuged at 4°C and 12,000 × g (Eppendorf 5430R centrifuge with model FA 45-30-11 rotor) for 30 min, and the supernatant was transferred to washed Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). Following 2 h of incubation at 4°C, resin was washed twice in PBS with 10 mM imidazole and twice in PBS with 20 mM imidazole prior to 15 min of incubation in PBS with 250 mM imidazole elution buffer. Imidazole was removed by overnight dialysis in PBS. TRP120 expression and purification were confirmed with Coomassie-stained SDS-PAGE gels.
In vitro SUMOylation assay.
TRP120 SUMOylation was performed with recombinant TRP120 and an in vitro SUMOylation kit (Enzo Life Sciences, Farmingdale, NY). Briefly, TRP120 (200 nM) was added to SUMO buffer, SUMO protein (isoform 1, 2, or 3), E1 ligase, and E2 ligase, with or without Mg-ATP. Reaction mixtures were incubated at 30°C for 1 h and then boiled with 1× lithium dodecyl sulfate (LDS) sample buffer and beta-mercaptoethanol. Samples were analyzed by SDS-PAGE and Western blotting with rabbit anti-TRP120 (1:5,000) or anti-pan-SUMO (1:1,000) primary antibody and alkaline phosphatase-conjugated anti-rabbit secondary antibody (1:10,000; Kirkegaard & Perry Laboratories, Gaithersburg, MD). Bound antibodies were visualized after incubation with 5-bromo-4-chloro-3-indoylphosphate and nitroblue tetrazolium (BCIP/NBT) substrate (Kirkegaard & Perry Laboratories).
Cotransfection and HA immunoprecipitation.
HeLa cells were cotransfected with pAcGFP1-C or pAcGFP1-TRP120 and HA-SUMO constructs by use of Lipofectamine 2000 (Invitrogen). Cells were collected and lysed in 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 20 mM NEM (covalent isopeptidase inhibitor; Sigma), 20 mM iodoacetamide (isopeptidase inhibitor; Sigma), 1% Triton X-100, and protease inhibitors (Complete mini, EDTA-free; Roche) at 24 h posttransfection. Lysates were then centrifuged at 4°C and 16,000 × g (Eppendorf 5430R centrifuge with model FA 45-30-11 rotor) for 20 min, and the supernatants were transferred to washed anti-HA agarose (Thermo Scientific). Following overnight incubation at 4°C, resin was washed with lysis buffer, boiled in LDS buffer, and analyzed by SDS-PAGE and Western blotting.
Cotransfection of mammalian cells and coimmunoprecipitation.
HeLa cells were cotransfected with pAcGFP1-C or pAcGFP1-TRP120 and pProLabel constructs by use of Lipofectamine 2000 (Invitrogen). GFP expression was confirmed 24 h after transfection by using an inverted fluorescence microscope (IX71; Olympus). Interacting proteins were coimmunoprecipitated using a Matchmaker chemiluminescence coimmunoprecipitation assay kit (Clontech), and ProLabel activity (in relative light units) was measured at 15-min intervals following addition of substrate, using a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA).
Small-molecule dose-response assay and real-time qPCR.
THP-1 cells were pretreated with dimethyl sulfoxide (DMSO; 1% final concentration) or different concentrations of anacardic acid (AA; Enzo Life Sciences) for 4 h and then infected with E. chaffeensis. Samples were collected at 24-h intervals following infection, for up to 72 h. Diff-Quik-stained slides were prepared and morula counts per cell determined for 30 cells per treatment condition. Bright-field images of these slides were collected with an Olympus BX61 epifluorescence microscope using a color camera. The remainder of the time point samples were pelleted, lysed in SideStep lysis and stabilization buffer (Agilent) for 30 min at room temperature, and analyzed for bacterial load by real-time quantitative PCR (qPCR). Amplification of the integral ehrlichial gene dsb was performed using Brilliant II SybrGreen master mix (Agilent), 200 nM forward primer (5′-GCTGCTCCACCAATAAATGTATCCCT-3′), and 200 nM reverse primer (5′-GTTTCATTAGCCAAGAATTCCGACACT-3′). The dsb copy number was quantitated using a standard curve and normalized to qPCR-detected levels of the host genomic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. Statistical differences between control and experimental groups were assessed with two-tailed Student's t test. Significance is indicated for P values of <0.05. The qPCR thermal cycling protocol (denaturation at 95°C for 10 min and then 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min) was performed on a Mastercycler EP Realplex2 S machine (Eppendorf) and was optimized to ensure specific dsb amplicon formation.
RESULTS
Colocalization of host SUMO proteins with E. chaffeensis vacuole without changes in global SUMOylation levels.
By use of immunofluorescence microscopy, SUMOylated proteins were observed to colocalize with E. chaffeensis at 72 h p.i. in a cell line derived from acute human monocytic leukemia (THP-1) cells (Fig. 1). Using a pan-SUMO antibody, we observed colocalization of SUMOylated proteins with ehrlichial inclusions, in contrast to the largely punctate cytosolic and nuclear localization in uninfected THP-1 cells (Fig. 1A and D). In vertebrates, four SUMO isoforms have been identified (41). SUMO1 is typically associated with facilitating nuclear translocation and, in cells, is largely constitutively substrate conjugated (42). SUMO2 and -3 share 97% sequence identity (yet only 50% identity with SUMO1) and are often referred to as SUMO2/3 because they are indistinguishable by antibody identification. The SUMO2/3 proteins are often conjugated to proteins in response to a signaling or stress stimulus (17, 43). SUMO4 was recently identified, and polymorphisms of this isoform have been linked to several disease states (44, 45); however, it remains ambiguous whether SUMO4 is a direct protein modifier or a pseudogene (46, 47).
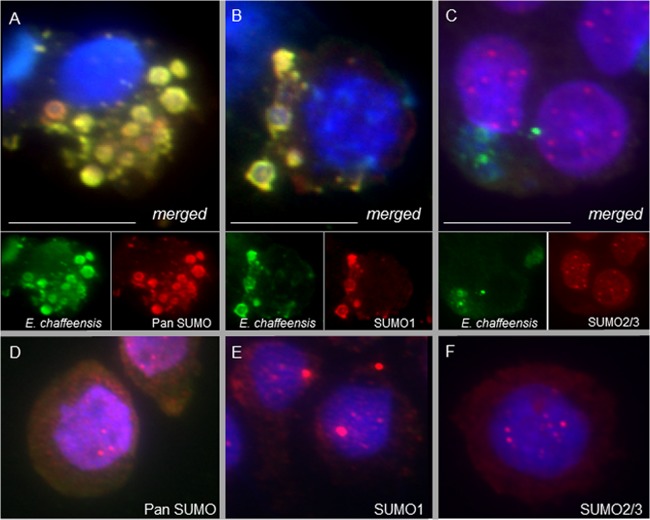
FIG 1.
SUMOylated proteins colocalize with E. chaffeensis inclusions. THP-1 cells were fixed (72 h p.i.), probed with polyclonal anti-E. chaffeensis antibody (green), anti-pan-SUMO (A; red), anti-SUMO1 (B; red), or anti-SUMO2/3 (C; red), and DAPI (DNA; blue), and then visualized by immunofluorescence microscopy (magnification, ×40). SUMO protein colocalization with ehrlichial inclusions (yellow), specifically the SUMO1 isotype, was observed compared to uninfected THP-1 cells probed with pan-SUMO (D), SUMO1 (E), or SUMO2/3 (F) antibody. Bars, 10 μm.
Isotype-specific antibodies demonstrated that the SUMO protein colocalization observed at ehrlichial vacuoles was dominated by SUMO1 proteins (Fig. 1B and E), while the most intense SUMO2/3 signals were observed at nuclear puncta (Fig. 1C and F). Interestingly, closer analysis of infected THP-1 cells, using an increased magnification (×100) and fluorescence exposure (increased 2.5× from Fig. 2A to Fig. 2B), showed low-abundance cytosolic SUMO2/3 proteins surrounding the ehrlichial vacuole, giving a ring-like appearance at the periphery and colocalizing with the ehrlichial effector TRP120 (Fig. 2A). Collectively, these immunofluorescence microscopic studies demonstrated that host SUMO proteins, regardless of isotype, colocalize with the vacuole during E. chaffeensis infection; however, while SUMO1 accounts for the greatest colocalization, SUMO2/3 proteins are also observed in low abundance at the periphery of the ehrlichial vacuole. These differences in colocalization may be mediated by isoform-selective recruitment of SUMOylated host proteins, host-mediated SUMO conjugation of ehrlichial effectors, or both.
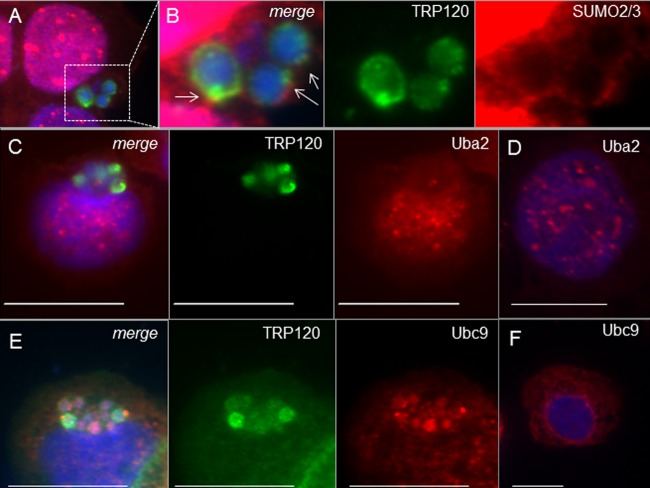
FIG 2.
SUMO2/3 and Ubc9 (E2), but not Uba2 (E1), colocalize with TRP120 at ehrlichial inclusions during E. chaffeensis infection. (A) THP-1 cells were probed (72 h p.i.) with anti-TRP120 (green), anti-SUMO2/3 (red), and DAPI (DNA; blue). (B) Increased fluorescence exposure (2.5×; magnification, ×100) reveals that SUMO2/3 (red) colocalizes with TRP120 at the periphery of ehrlichial inclusions (highlighted by white arrows). Uba2 (red) does not colocalize with TRP120 (green; C), while Ubc9 (red) redistributes to ehrlichial inclusions (E) compared to the case in uninfected THP-1 cells probed with Uba2 (red; D) or Ubc9 (red; F) antibody. Bars, 10 μm.
Surprisingly, SUMO protein colocalization with E. chaffeensis inclusions did not coincide with global changes in host cell SUMOylation patterns (see Fig. S1 in the supplemental material), as occurs with other intracellular bacterial infections known to modulate host SUMO pathways (32). SUMO isoform-specific immunoblot analysis of THP-1 whole-cell lysates at 72 h p.i. did not detect changes in SUMO1 (see Fig. S1A) or SUMO2/3 (see Fig. S1B) conjugation levels in E. chaffeensis-infected cell lysates.
Colocalization of SUMO with ehrlichial inclusions without changes in global SUMOylation levels suggested that SUMO-modified host proteins are recruited to the vacuole during infection or selectively modified at the vacuole at levels not detected in global SUMOylation immunoblot analysis. In the latter case, the host SUMOylation machinery would be expected to colocalize with ehrlichial vacuoles. Immunofluorescence microscopy of THP-1 cells at 72 h p.i. demonstrated that the E1 heterodimer (visualized with anti-Uba2) did not redistribute from nuclear puncta observed in uninfected cells (Fig. 2C and D) but that the E2 ligase (visualized with anti-Ubc9) did colocalize with TRP120 at ehrlichial inclusions (Fig. 2E and F). Overall protein levels of the SUMO machinery were unchanged in THP-1 whole-cell lysates with or without E. chaffeensis (see Fig. S1C and D in the supplemental material). Uba2 is itself a target of SUMOylation and other PTMs, resulting in Uba2 proteins of laddered molecular weights (48) (see Fig. S1C). Recently, Truong et al. reported that in normal cells, a proportion of Uba2 is auto-SUMOylated at lysine 236 (K236) (49). This modification inhibits interaction with E2 ligase (Ubc9) and results in cellular pools of inactive E1. During periods of cell stress, these pools are mobilized by de-SUMOylation at K236, resulting in Uba2-Ubc9 interaction and subsequent PTM and activation of Uba2 (49). Immunoblot analysis of E. chaffeensis-infected whole-cell lysate revealed an absence of the lowest-molecular-weight Uba2 protein band (see Fig. S1C), predicted to be mono-SUMOylated. This suggested that despite the absence of Uba2 redistribution during infection, inactive pools of this enzyme may be activated to dynamically modulate host SUMOylation. Protein levels of Ubc9 were unchanged with or without E. chaffeensis infection (see Fig. S1D), suggesting that E2 ligase is recruited to inclusions rather than upregulated.
Ubc9 colocalization with ehrlichial vacuoles suggested that select proteins may be SUMO modified at the vacuole. We were interested in determining whether ehrlichial effectors, including TRP120, are substrates of host SUMOylation and are modified at low levels not detected by one-dimensional SDS-PAGE. To assess this possibility, two-dimensional gel electrophoresis (2DE) and immunoblot analyses were performed using pan-SUMO and TRP120 antibodies (see Fig. S1E). The majority of SUMOylated proteins in both uninfected and infected lysates were observed to be large (>75 kDa), with a neutral to basic pI. However, immunoblot analysis revealed several unique SUMO proteins, including those with acidic or slightly acidic pIs, present only in E. chaffeensis-infected lysates compared to uninfected THP-1 cell lysates, suggesting the presence of additional novel SUMOylated proteins during infection.
Unmodified TRP120 is acidic (pI 4.1) due to the serine-rich tandem repeats and has an observed molecular mass of ∼95 kDa. SUMO modification of TRP120 was predicted to result in a 15-kDa increase in molecular mass, with only a minimal impact on pI (predicted modified pI = 4.2). 2DE immunoblot analysis performed with anti-TRP120 detected acidic proteins (pI 5 to 7) (see Fig. S1E in the supplemental material), suggesting that this effector is a target of PTMs that significantly shift pI. In addition, faint detection of TRP120 was also observed at pI 4.1 to 4.2 and a molecular mass of ∼110 kDa (see Fig. S1E), suggesting that TRP120 is a target of a PTM that increases molecular mass without greatly affecting pI. A series of distinct spots were also uniquely present in this region of the anti-SUMO E. chaffeensis immunoblot (see Fig. S1E). Differences in patterns observed between these immunoblots may be a function of antibody sensitivity. Cumulatively, the data support TRP120 as a target of SUMOylation during E. chaffeensis infection. However, levels of SUMOylated TRP120 were not sufficient for mass spectrometry identification or capture via immunoprecipitation. As this is a common issue reported by others, in vitro and ectopic overexpression methods, previously employed to characterize novel SUMOylation targets (21, 22), were utilized to demonstrate that TRP120 is a target of host SUMOylation.
The E. chaffeensis effector TRP120 is conjugated with SUMO2/3 at a carboxy-terminal canonical consensus SUMO motif.
Protein substrates of SUMO pathways often encompass the canonical consensus motif ψKxD/E (where ψ is a hydrophobic residue and “x” is any residue), through which the Ubc9 E2 ligase directly interacts with and covalently transfers SUMO to the lysine residue in the motif (50, 51). The E. chaffeensis TRP120 protein sequence was analyzed for consensus SUMO conjugation motifs by use of SUMOsp 2.0 (52, 53). A single consensus motif, IKEE (TRP120 amino acids 431 to 434), was identified downstream of the tandem repeats in the carboxy-terminal region of the protein, suggesting that lysine 432 is a target of SUMOylation (Fig. 3A).
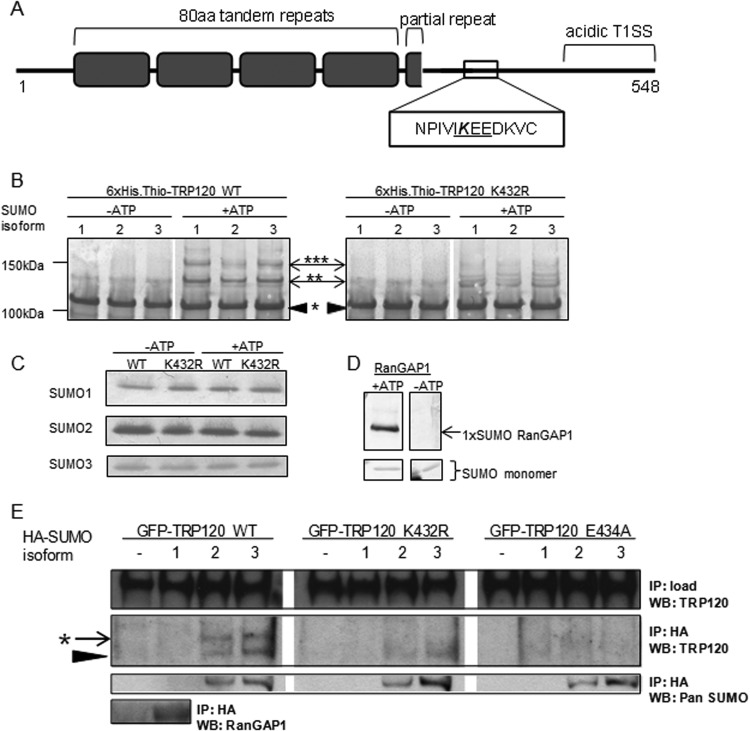
FIG 3.
E. chaffeensis TRP120 encompasses a carboxy-terminal canonical consensus SUMO motif that is modified in vitro and in human cells. (A) Illustration of TRP120 protein domains, including tandem repeats (gray boxes) and an acidic T1SS at the carboxy terminus, highlighting the canonical consensus SUMO motif (ψKxD/E, where ψ is a hydrophobic residue and “x” is any residue), in which lysine 432 is predicted to be SUMO conjugated. (B) In vitro SUMOylation assay (Enzo Life Sciences) of recombinant 6×His-thioredoxin-tagged wild-type TRP120 or the K432R mutant, with or without ATP. Western blot analysis performed with anti-TRP120 shows higher-molecular-weight laddering of wild-type TRP120 in the presence of ATP, indicative of multiple SUMO modification events. *, unmodified TRP120; **, mono-SUMOylation; ***, poly- or multi-SUMOylation. Representative data are shown (n = 3). TRP120 K432R yielded a diminished laddering pattern, demonstrating that the lysine residue is the primary site of SUMO conjugation in vitro. (C) Immunoblot performed with SUMO antibodies, demonstrating equal loading of each recombinant SUMO isoform for conditions with and without ATP. (D) Purified recombinant RanGAP1 was used as a positive control for in vitro SUMOylation. Immunoblot analysis with anti-pan-SUMO demonstrates a single SUMOylation of RanGAP1, as previously reported (87). (E) In cellular studies, GFP-TRP120 WT or the K432R or E434A mutant was ectopically expressed alone or with HA-SUMO isoforms in HeLa cells. Anti-HA immunoprecipitation (IP) and analysis by anti-TRP120 immunoblotting (WB) demonstrated that wild-type TRP120 was conjugated by a single SUMO2/3 protein in cells (*), while TRP120 K432R and TRP120 E434A were not modified. Representative data are shown (n = 6). Unmodified TRP120 (black arrowhead) coimmunoprecipitated with HA-SUMO2 and HA-SUMO3, suggesting a noncovalent interaction with SUMO or SUMO-modified proteins.
The potential for TRP120 to serve as a substrate of host cell SUMO pathways was first assessed in vitro. SUMOylation pathways are unique to eukaryotes; therefore, unmodified recombinant 6×His.thioredoxin-tagged TRP120 (observed molecular mass, 105 kDa) was expressed and purified from E. coli. The in vitro SUMOylation assay (Enzo Life Sciences) comprises the purified E1 heterodimer (Uba1-Uba2), E2 ligase (Ubc9), and individual SUMO isoforms 1, 2, and 3. In the presence of ATP, the E1 heterodimer covalently binds SUMO and transfers the modifier to Ubc9, facilitating SUMO conjugation of target proteins. Immunoblot analysis of assay products with anti-TRP120 demonstrated multiple higher-molecular-mass species of TRP120 in the presence of ATP and each of the SUMO isoforms, whereas these bands were not detected in the absence of ATP. Incremental higher-molecular-mass band shifts of 14 to 18 kDa are consistent with covalent SUMO conjugation of the target protein (Fig. 3B). These in vitro results demonstrated SUMOylation of recombinant TRP120 by all three SUMO isoforms in the presence of ATP, with equal loading of each individual recombinant SUMO isoform maintained with or without ATP (Fig. 3C). The presence of laddered, higher-molecular-mass bands suggested either multiple mono-SUMOylation events (multi-SUMOylation), in which TRP120 may be SUMOylated at more than one lysine residue, or poly-SUMOylation, in which SUMO proteins self-conjugate to form poly-SUMO chains emanating from a single target protein lysine residue. In vitro and in cells, SUMO2/3 proteins self-conjugate, allowing for poly-SUMO modifications with functionally distinct properties compared to those of mono-SUMOylation. Poly-SUMO1 conjugates, which are readily observed in vitro, are rarely observed in cells (54). RanGAP1, the first substrate identified for the SUMO pathway (55, 56), was used as a positive control in these experiments and was mono-SUMOylated as previously reported (Fig. 3D).
To assess TRP120 lysine 432 in the consensus SUMO motif as the target for modification, a conservative point mutation of lysine 432 to arginine (K432R) was performed. The in vitro SUMOylation assay was performed using purified recombinant 6×His.thioredoxin-tagged TRP120 K432R. Western immunoblot analysis (57) of in vitro samples with anti-TRP120 revealed a substantial decrease in higher-molecular-weight protein bands for TRP120 K432R in the presence of ATP compared to the case with wild-type (WT) TRP120, demonstrating that mutation of K432 diminishes SUMOylation and that this lysine is therefore the primary target for TRP120 modification in vitro (Fig. 3B). Lysine residues in the TR domain are predicted to be noncanonical sites for SUMOylation (52) and are ubiquitinated during ectopic expression (58). SUMO modification at these alternate residues may contribute to the low levels of protein laddering observed in vitro for TRP120 K432R.
Confirmation that TRP120 is a substrate of host SUMOylation pathways was performed in human cells. GFP-tagged WT TRP120 and TRP120 K432R were transiently coexpressed in HeLa cells with HA-tagged SUMO isoform 1, 2, or 3. At 24 h posttransfection, these cells were lysed in the presence of isopeptidase inhibitors (N-ethylmaleimide and iodoacetamide), and anti-HA immunoprecipitation was performed. Immunoprecipitated samples were analyzed by immunoblotting with anti-TRP120. GFP-tagged TRP120 coimmunoprecipitated with HA-SUMO2 and HA-SUMO3, but not HA-SUMO1, and exhibited a single protein band ∼15 kDa higher than that of unmodified WT GFP-TRP120, consistent with a single SUMOylation modification (Fig. 3E). Mutation of K432 entirely abolished TRP120 SUMOylation in cells, demonstrating that this lysine residue within the consensus motif is the sole target for TRP120 SUMO conjugation in cells. Unmodified GFP-TRP120, either the WT or the K432R mutant, coimmunoprecipitated with HA-SUMO isoforms, suggesting that this effector interacts noncovalently with SUMO or SUMO-modified host proteins. Pan-SUMO and RanGAP1 immunoblot analyses were also performed to confirm HA immunoprecipitation of SUMO2/3 monomers and SUMO1 (conjugated to RanGAP1), respectively (Fig. 3E).
In contrast to in vitro SUMOylation by the three isoforms, WT TRP120 in cells was mono-conjugated with HA-SUMO2 and HA-SUMO3 but not HA-SUMO1. The discrepancy in SUMO isoform selectivity between in vitro and cellular studies is consistent with results for other SUMOylated proteins, such as RanGAP1, which is SUMOylated by all three isoforms in vitro but selectively conjugated with SUMO1 in cells (59). This suggests that cellular regulatory mechanisms, such as E3 ligases or de-SUMOylating isopeptidases (sentrin/SUMO-specific cleavage [SENP]), influence TRP120 modification in cells. SUMO conjugation of TRP120 in vitro and in human cells demonstrates that this effector is a substrate of this host PTM pathway. TRP120 SUMOylation in the absence of the whole bacterium demonstrates that inherent properties of this effector, including the presence of a canonical consensus SUMO motif, facilitate this PTM.
In addition to SUMOylation, lysine residues are also the site of other PTMs, including ubiquitination and acetylation. To confirm that K432 is the target for SUMOylation and that mutation of K432 specifically disrupts this modification, the consensus SUMO motif was mutated to selectively prevent interaction with E2 ligase (Ubc9). Glutamate residue 434 was mutated to alanine (E434A), ablating the TRP120 consensus SUMO motif (IKEE to IKEA). TRP120 E434A was screened for SUMOylation in vitro (data not shown) and in HeLa cells by anti-HA immunoprecipitation (Fig. 3E). Consistent with results for TRP120 K432R, mutation of the consensus SUMO motif abolished SUMOylation of GFP-tagged TRP120 E434A ectopically expressed in human cells. The SUMO-null mutant TRP120 E434A was subsequently used in cellular studies to determine the functional implications of TRP120 SUMOylation.
TRP120 SUMOylation enhances interactions with host proteins.
In mammalian cells, target proteins are SUMO modified in an isoform-selective manner that elicits distinct functional consequences, including differences in protein interaction, often mediated through noncovalent interactions with SIMs (60–63). This has also been reported for a pathogen protein, cytomegalovirus IE1, for which host protein interactions were found to vary for SUMOylated versus unmodified forms of the effector (35, 64). SUMOylation can also render changes in target protein conformation that may indirectly influence function and subsequent protein interactions (65). Functionally, SUMO2/3 conjugation of TRP120 may facilitate recruitment of host proteins via direct noncovalent interactions with host protein SIMs or through indirect interactions mediated by conformational changes that occur in SUMOylated TRP120.
Recently, we used a yeast two-hybrid (Y2H) approach to demonstrate that ehrlichial TRP120 interacts with a diverse array of host proteins (7); however, the molecular basis for these diverse host-effector interactions was not determined. Protein sequence analysis using GPS-SBM 1.0 and GPS-SUMO (53) predicted a common motif, the SIM, in a subset of these host proteins, including components of the cytoskeleton—γ-actin and myosin-X (unconventional myosin; also known as Myo10)—as well as a protein involved in regulating recruitment and endosomal trafficking of select cargo proteins—GGA1 (Golgi-localizing, γ-adaptin ear domain homology, Arf-binding protein). In Myo10 and GGA1, the predicted SIM is encompassed in the carboxy-terminal host protein domain that interacts with TRP120, whereas in γ-actin, the predicted SIM is adjacent to the interacting domain (illustrated in Fig. S2 in the supplemental material).
The role of SUMOylation in mediating TRP120 interactions with Myo10, GGA1, and γ-actin was assessed in transfected HeLa cells by coimmunoprecipitation with WT TRP120 and TRP120 E434A constructs. Measurements of coimmunoprecipitated Myo10 and GGA1 were higher in samples with WT TRP120 than in those with the TRP120 E434A mutant (Fig. 4A and B, respectively). These differences in interaction were confirmed by immunofluorescence microscopy, by which we compared WT TRP120 and TRP120 E434A colocalization with endogenous Myo10 and GGA1 (see Fig. S3A and B in the supplemental material). In these microscopic studies, greater colocalization with endogenous host proteins was observed for WT TRP120 than for TRP120 E434A. In fact, host proteins appeared to redistribute and colocalize with WT TRP120 to a greater extent than that with the E434A construct.
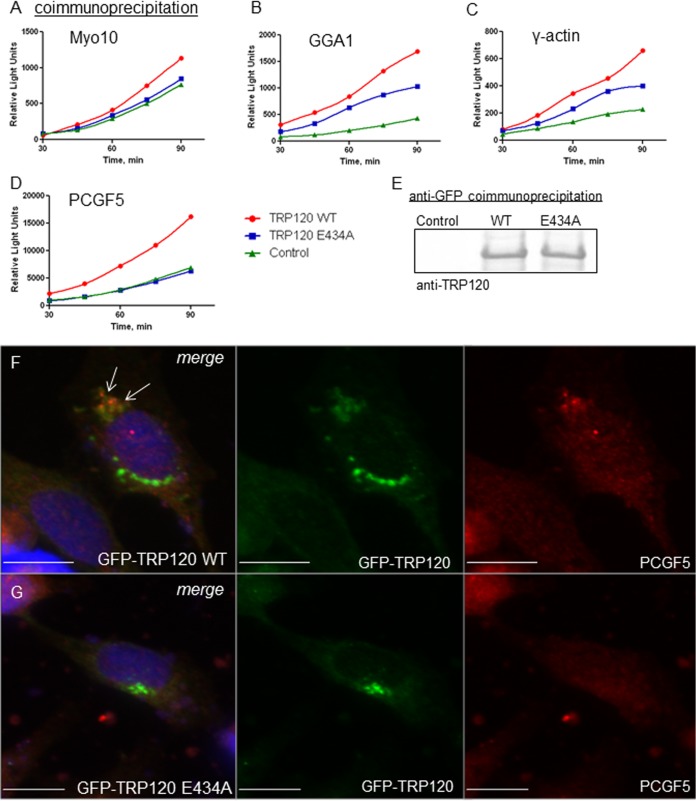
FIG 4.
Disruption of TRP120 SUMOylation perturbs interactions with host proteins. HeLa cells were transiently cotransfected with GFP-vector (control), GFP-TRP120 wild type (WT), or GFP-TRP120 E434A and ProLabel-tagged host protein domains (as previously reported [7]). Coimmunoprecipitation was performed with anti-GFP antibody at 24 h posttransfection, and ProLabel activity was measured (relative light units) over 90 min to determine the relative impact of SUMOylation on TRP120-host interactions. Representative data are shown (n = 3); differences in absolute relative light units varied between experiments. (A) γ-Actin; (B) GGA1; (C) Myo10; (D) PCGF5. (E) Anti-TRP120 immunoblot analysis of GFP coimmunoprecipitations demonstrates that wild-type TRP120 and the SUMO-null mutant (E434A) were pulled down with similar efficiencies and that pulldown efficiency did not contribute to observed differences in ProLabel activity. Colocalization of ectopically expressed GFP-TRP120 WT (green; F) and native PCGF5 (red) was observed in HeLa cells by use of immunofluorescence microscopy (white arrows), while colocalization was not observed with the TRP120 E434A SUMO-null mutant (green; G). Bars, 10 μm.
SUMOylation enhances TRP120 molecular interaction with Myo10 and GGA1, possibly through predicted SIMs encompassed in these host proteins. However, γ-actin also showed an enhanced interaction with WT TRP120 compared to TRP120 E434A (Fig. 4C). The γ-actin construct used in coimmunoprecipitation studies was composed of the carboxy terminus, the domain identified in Y2H studies to interact with TRP120. This construct did not contain a predicted SIM in the primary amino acid sequence, according to GPS-SBM 1.0 and GPS-SUMO (53). Therefore, differences in molecular interactions with WT TRP120 versus TRP120 E434A suggest that SUMOylation indirectly enhances interactions with host proteins, possibly through as yet uncharacterized conformational changes. SUMO-mediated conformational changes in the target protein are known to indirectly affect protein interactions (65). Immunofluorescence micrographs support the role of SUMOylation in enhancing TRP120 interaction with γ-actin, demonstrating an increased redistribution and colocalization of native γ-actin with WT TRP120 compared to those with TRP120 E434A (see Fig. S3C in the supplemental material).
Evidence that TRP120 SUMOylation indirectly enhances host protein interactions in the absence of a predicted SIM led us to determine the role of SUMOylation in mediating interactions with a protein lacking a predicted SIM in the primary amino acid sequence. PCGF5, a component of the polycomb repressive complex (PRC1) that mediates epigenetic modification of histones to regulate gene expression, robustly interacts with TRP120 (7). Prediction analysis of PCGF5 by GPS-SBM 1.0 and GPS-SUMO did not identify a SIM in the primary amino acid sequence (53). In HeLa cells transiently coexpressing truncated PCGF5 and TRP120 constructs, PCGF5 did not coimmunoprecipitate with TRP120 E434A compared to the WT (Fig. 4C). This ablation in interaction was not explained by differences in TRP120 construct immunoprecipitation efficiency (Fig. 4E). Immunofluorescence microscopy supported these data, demonstrating a redistribution of native PCGF5 to colocalize with a pool of WT TRP120, whereas colocalization was not observed with TRP120 E434A (Fig. 4F). These studies suggest that TRP120 SUMOylation mediates interaction with PCGF5 and that ablation of TRP120 SUMOylation impairs this interaction, despite the absence of a predicted SIM in the PCGF5 protein sequence.
In addition to enhancing or mediating protein interactions, SUMOylation is often associated with regulating target protein subcellular localization, specifically to the nucleus (57) or the plasma membrane (66). Immunofluorescence microscopy did not reveal differences in subcellular localization for ectopically expressed WT TRP120 and TRP120 E434A (Fig. 4F and G, respectively). However, as only a small proportion of WT TRP120 is modified relative to total TRP120, even when ectopically coexpressed with SUMO (Fig. 3E), changes in subcellular localization may not be detected via gross whole-cell microscopy.
These studies demonstrate that TRP120 SUMO conjugation significantly enhances, and in some cases entirely mediates, select host protein interactions via domains encompassing predicted SIMs (Myo10 and GGA1), as well as domains devoid of a predicted SIM in the primary protein sequence (γ-actin and PCGF5). In the latter situation, SUMOylation may indirectly affect interactions with host proteins through conformational effects on TRP120 that are as yet undefined.
SUMOylation mediates host protein recruitment to the ehrlichial vacuole.
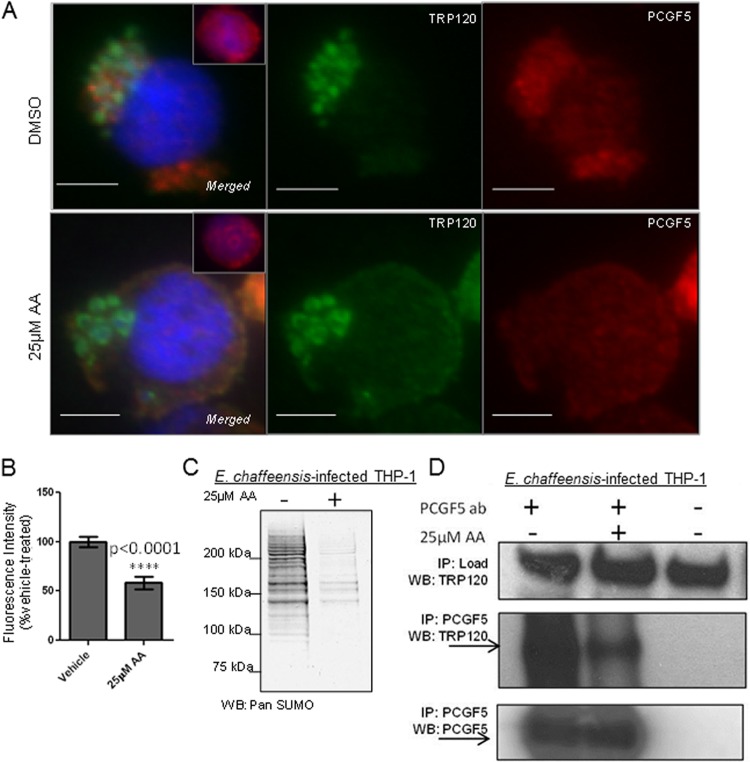
In vitro and ectopic analyses of TRP120 demonstrated that this effector is a direct target of the host SUMO pathway and that this modification facilitates interactions with select host proteins. It was important to demonstrate that this PTM is functionally important during E. chaffeensis infection. However, given technical limitations in the ability to genetically modify this bacterium, we utilized a small-molecule inhibitor to briefly block host SUMOylation and to assess changes in TRP120 protein interactions. Anacardic acid (AA; 15:0), a cell-permeating analog of salicylic acid, reversibly inhibits the E1 heterodimer (Uba1-Uba2) (67, 68). THP-1 cells at 72 h p.i. were treated with vehicle (DMSO) or 25 μM AA for 4 h, fixed, and observed using immunofluorescence microscopy with TRP120 and PCGF5 antibodies. Compared to vehicle-treated cells, in which PCGF5 robustly colocalized with TRP120 at ehrlichial inclusions, AA-treated cells showed significantly less PCGF5 colocalization (Fig. 5A and B). Immunoblot analysis demonstrated that 4 h of treatment with 25 μM AA decreased global SUMO levels in THP-1 whole-cell lysates at 72 h p.i. (Fig. 5C). Similarly, a brief (4 h) treatment with viomellein, a newly described small-molecule inhibitor of Ubc9 (E2 ligase) (69), also resulted in decreased colocalization of PCGF5 with TRP120 at the ehrlichial vacuole (see Fig. S4A and B in the supplemental material). Recruitment of PCGF5 to ehrlichial inclusions was diminished but not ablated following inhibitor treatment, likely due to continued interactions between PCGF5 and other ehrlichial effectors. We previously reported Y2H studies in which PCGF5 was identified as an interacting protein for TRP47 (5), a T1S ehrlichial effector present at ehrlichial inclusions that does not encompass a canonical consensus SUMO motif. To confirm that decreased colocalization was due to disruption of TRP120-PCGF5 interaction, we measured TRP120 that coimmunoprecipitated with PCGF5 following inhibitor treatment (Fig. 5D). Following AA treatment, there was decreased TRP120 coimmunoprecipitation with PCGF5 compared to the case with vehicle. These studies demonstrate that during E. chaffeensis infection, SUMOylation is important for PCGF5 interaction with TRP120 and for recruitment to ehrlichial inclusions.
FIG 5.
SUMOylation facilitates host protein recruitment to the ehrlichial vacuole. (A) Immunofluorescence microscopy visualization shows that PCGF5 recruitment to the ehrlichial vacuole decreased with a short (4 h) treatment with the SUMO pathway inhibitor anacardic acid (AA; 25 μM) compared with vehicle (DMSO) treatment. Bars, 10 μm. Decreased colocalization of PCGF5 (red) with TRP120 (green) was observed following AA treatment. (B) Decreased PCGF5 at the ehrlichial vacuole was graphed as a percentage of the vehicle control fluorescence intensity. Fluorescence at the ehrlichial inclusion was measured for 20 cells (Student's t test; P < 0.0001). (C) Immunoblot analysis with pan-SUMO antibody demonstrates decreased global SUMOylation levels following AA treatment (4 h; 25 μM). Representative data are shown (n = 3). (D) Coimmunoprecipitation of TRP120 with PCGF5 is significantly decreased following AA treatment.
Modulation of host SUMOylation decreases E. chaffeensis infection.
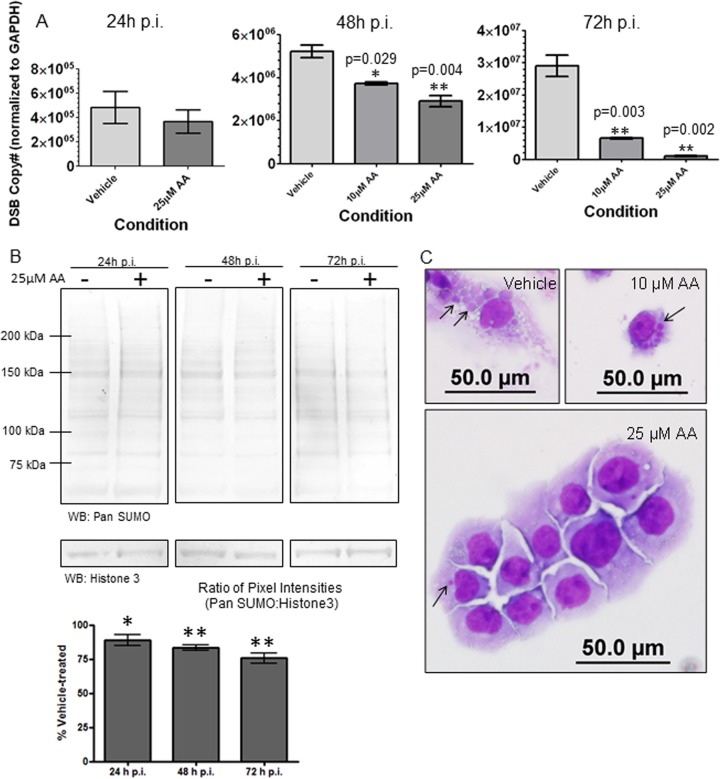
We have demonstrated that TRP120 is a target of host SUMOylation pathways and that this modification mediates interactions with and recruitment of select host proteins to the ehrlichial inclusion during E. chaffeensis infection. Therefore, disruption of global host SUMOylation resulting in diminished effector-host interactions likely affects overall E. chaffeensis infection and survival. To determine the impact of diminished host SUMOylation on E. chaffeensis infection, cells were exposed to prolonged treatment with AA. THP-1 cells were pretreated with AA for 4 h prior to infection with E. chaffeensis and cultured in the presence of AA for the duration of infection. Samples were collected at 24-h intervals for up to 72 h p.i. for analysis. AA did not affect bacterial entry, as there was no difference in bacterial loads at 24 h p.i. for vehicle (DMSO) and 25 μM AA treatment (Fig. 6A). However, at 48 h p.i. and 72 h p.i., significant concentration-dependent decreases in bacterial load, as measured by real-time qPCR analysis of the dsb copy number normalized to the host GAPDH gene, were observed in AA-treated cells relative to vehicle-treated cells (Fig. 6A). From these data, we observed >50% inhibition and >90% inhibition of bacterial loads with prolonged treatment with 10 μM AA and 25 μM AA, respectively. The 50% inhibitory concentration (IC50) for AA inhibition of ehrlichial loads was <10 μM at 72 h p.i., which is in the range demonstrated to inhibit the SUMO E1 ligase (IC50 = 2.2 μM) (67). Despite the low IC50 for E1 ligase inhibition published for AA, immunoblot analysis of global host SUMOylation demonstrated decreases with 25 μM AA but no ablation of SUMOylation levels compared to the case with vehicle-treated cells over the 72-h time course (Fig. 6B). This suggests that a portion of the host SUMOylated protein population is not quickly turned over.
FIG 6.
Small-molecule inhibition of host cell SUMOylation decreases E. chaffeensis survival. (A) THP-1 cells were treated with DMSO (vehicle) or AA, a SUMO E1 ligase inhibitor, for 4 h and then infected with E. chaffeensis. Bacterial loads were determined by qPCR analysis of the dsb gene normalized to the GAPDH gene and compared to those in vehicle-treated infected cells (24 h, 48 h, or 72 h p.i.) (Student's t test; P < 0.05; n = 3). (B) Immunoblot analysis was performed with pan-SUMO antibody (or histone 3 antibody for a loading control) and THP-1 whole-cell lysate, with or without 25 μM AA, at 24-h time intervals up to 72 h posttreatment (n = 4). Pixel intensities (determined using ImageJ software) were normalized to the loading control (histone 3) and graphed as a percentage of the vehicle-treated level. Wilcoxon signed-rank statistical analysis was performed to determine differences from vehicle-treated cells (100%). *, P < 0.05; **, P < 0.005. (C) Bright-field images (magnification, ×40) of Diff-Quik-stained samples collected at 72 h p.i. demonstrate a decreased number of ehrlichial inclusions per cell following treatment with increasing concentrations of AA.
At 72 h p.i., significantly fewer ehrlichial inclusions per cell were observed for AA-treated cells than for vehicle-treated cells (see Fig. S4C in the supplemental material). Diff-Quik-stained images of E. chaffeensis-infected THP-1 cells show this concentration-dependent decrease in ehrlichial inclusions (Fig. 6C, black arrows). Decreased ehrlichial inclusions and bacterial loads were not a function of AA toxicity to the host cell, as cell viability measured by trypan blue was not affected by prolonged AA treatment (see Fig. S4D). This is consistent with previous studies in which knockdown of the E1 heterodimer by RNA interference (RNAi) demonstrated sustained cell viability despite diminished SUMOylation (70). This was an important consideration, as prolonged treatment with the E2 inhibitor viomellein resulted in significant host cell death (data not shown). In total, these studies demonstrate that AA inhibits ehrlichial replication at a concentration consistent with a mechanism involving inhibition of the host SUMOylation pathway.
Another important consideration is that AA and similar plant-based derivatives are known antimicrobials with reported activity against Gram-positive bacteria (reviewed in reference 68). However, inhibitory activity specific for the parent compound (AA; 15:0) used in the present study was not observed for Gram-negative bacteria, including Helicobacter pylori and Escherichia coli (68). To confirm that AA inhibits ehrlichial replication through disruption of host SUMOylation rather than through direct antimicrobial mechanisms, targeted small interfering RNAs (siRNAs) were used to knock down expression levels of host SUMO pathway components. Prior to infection (24 h), THP-1 cells were transfected with siRNA targeting SUMO2/3 or Ubc9. Diff-Quik staining (at 72 h p.i.) demonstrated significantly decreased numbers of ehrlichial inclusions in THP-1 cells transfected with SUMO2/3- or Ubc9-targeting siRNA compared to those transfected with a nontargeting scrambled siRNA (see Fig. S5A in the supplemental material). Knockdown efficiency, assessed at 96 h posttransfection (72 h p.i.), showed decreased expression levels of SUMO2/3 and Ubc9 compared to the case with the scrambled siRNA control (see Fig. S5B). To confirm that host cell death did not account for decreased ehrlichial inclusions, differences in cell viability were assessed at 96 h posttransfection (72 h p.i.) (see Fig. S5C). Knockdown of SUMO2/3 significantly increased host cell viability compared to that with the nontargeting siRNA control, as would be expected in cells with a decreased bacterial load. Ubc9 knockdown did not yield a significant difference in host cell viability relative to that with the siRNA control, despite a significantly decreased bacterial load. This suggests that prolonged knockdown of Ubc9 is deleterious to the host cell, which is consistent with our observation that prolonged treatment with viomellein (a Ubc9 inhibitor) resulted in host cell cytotoxicity. Global modulation of the host SUMOylation pathway unquestionably affects host cell processes beyond TRP120-host interactions; however, collectively, the studies herein support the significance of this pathway in TRP120 effector function and E. chaffeensis infection.
DISCUSSION
There is an increasing appreciation for the role of host PTM pathways in bacterial effector function and turnover (27, 30, 71–73). While bacterial effectors have previously been reported to be substrates of the host Ub pathways, targeting by the SUMO pathway has not been demonstrated. More than 90% of mammalian proteins are targeted by Ub PTM (74), but a significantly smaller number of proteins have been identified as substrates of SUMOylation. This striking difference attests to the highly specific, regulated, and dynamic nature of this PTM. Herein we have demonstrated the first bacterial effector protein modified by the host SUMO pathway. TRP120 modification occurs at a consensus SUMO motif in the carboxy terminus and, in cells, is modified selectively by SUMO2/3. TRP120 is a multifunctional protein that is critical for ehrlichial entry (10) and which we previously reported to interact with a network of 98 functionally diverse host proteins and host DNA (4, 7). Herein we have demonstrated that SUMOylation enhances effector interactions with select host proteins and functions to recruit host proteins to the ehrlichial inclusion during E. chaffeensis infection.
During E. chaffeensis infection (72 h p.i.), intense SUMO1 fluorescence was observed at ehrlichial inclusions (Fig. 1B); however, global patterns of SUMO1 modification were unchanged (see Fig. S1A in the supplemental material), suggesting that redistribution of SUMO1-modified proteins from nuclear puncta (Fig. 1E) is the predominant mechanism for this observed localization. Low levels of SUMO2/3 at the periphery of the ehrlichial inclusions (Fig. 2B) may also involve a limited redistribution of SUMO2/3-modified proteins or may reflect low levels of protein modification not detected in global SUMOylation patterns (see Fig. S1B). TRP120 colocalizes with Ubc9 (E2 ligase) and with SUMO2/3 at the inclusion periphery. These data, in conjunction with pan-SUMO and TRP120 2DE immunoblots (see Fig. S1E), suggest that low levels of this effector are modified at the inclusion. Steady-state levels of modified effector are likely low during E. chaffeensis infection, as we were only able to indirectly demonstrate SUMOylation of native TRP120. Ectopically, the proportion of TRP120 that is SUMOylated is small relative to total TRP120 (Fig. 3E). This phenomenon is referred to as the SUMO enigma (26), where only a small proportion of the total target protein is modified at a given time, resulting in significant functional consequences. The small population of target protein that is modified suggests that this pathway is dynamic and is highly regulated by substrate-specific E3 SUMO ligases and quickly turned over by SENP isopeptidases. In the case of TRP120, E3 SUMO ligases may regulate the specific SUMO2/3 modification that is observed in cellular studies, in contrast to the case of the reconstituted assay, in which purified E3 ligase is not included. A limited number of SUMO E3 ligases have been identified. None of the 98 interacting host proteins identified in the TRP120 yeast two-hybrid screen are known to function as E3 SUMO ligases. However, because of the technical challenges in identifying transient substrates of the SUMO pathway and the frequent dual functioning of such enzymes, increasing numbers of E3 SUMO ligases are continually being identified.
Functionally, TRP120 SUMOylation serves to modulate host protein interactions. SUMO modification enhances or entirely facilitates TRP120 interaction with selected host proteins, chosen for evaluation herein based on robust interactions reported in Y2H studies (7). TRP120 interacts with GGA1 (7), which is involved in recognition of cargo protein sorting and clathrin-dependent vesicle formation for trans-Golgi network (TGN)-to-endosome trafficking (75). TRP120 interaction occurs with the GGA1 γ-adaptin ear domain, which encompasses a predicted SIM. WT TRP120 exhibited increased interaction with GGA1 at this domain relative to that with the SUMO-null mutant (Fig. 4B). In the cellular context, enhanced TRP120-GGA1 interaction may alter cargo protein sorting or clathrin vesicle formation to modulate TGN-endosome trafficking. TRP120 SUMOylation may serve as a means of regulating effector interaction with these protein trafficking processes to benefit E. chaffeensis.
TRP120 interacts with the cytoskeletal proteins Myo10 and γ-actin, which are host proteins involved in microtubule cargo trafficking and filopodium formation, respectively. TRP120 interacts with the carboxy terminus of Myo10, which encompasses a predicted SIM (76–78). WT TRP120 showed an increased interaction with this domain relative to that of the SUMO-null mutant (Fig. 4A). In the cellular context, enhanced interaction with Myo10 may affect cytoskeletal reorganization or facilitate TRP120 trafficking along microtubules. WT TRP120 also yielded an enhanced interaction with γ-actin compared to that of the SUMO-null mutant (Fig. 4C). SUMOylated actin binding proteins have been reported, including drebrin and α-actinin 4 (79), both of which mediate changes in actin remodeling and filament organization. SUMOylated actin binding proteins, including TRP120, may modulate actin rearrangement and filopodium filament formation, which have been reported late in E. chaffeensis infection and may serve as a bacterial escape mechanism (80).
We have demonstrated that E. chaffeensis T1S effectors TRP47 and TRP120 robustly interact with PCGF5 (5, 7), a component of the PRC1 E3 Ub ligase complex, which regulates histone modification for epigenetic control of gene transcription. TRP interactions occur at the PCGF5 RING domain, which does not encompass a predicted SIM. The striking difference observed in PCGF5 interaction with WT TRP120 versus the SUMO-null mutant suggests that PCGF5 may recognize only the SUMOylated TRP120 conformation. Previous studies demonstrated that TRP120-PCGF5 interaction occurs through the TRP120 TR domain (7). In fact, expression and coimmunoprecipitation of a truncated TRP120 construct composed entirely of the TR region demonstrated robust interaction with PCGF5, while amino- and carboxy-terminal constructs did not interact (7). Collectively, our studies suggest that TRP120 SUMOylation may result in conformational changes that expose the TR domain for robust interaction with PCGF5 and that, in the absence of this modification, the TR domain may not be accessible.
SUMOylation also regulates PCGF5 recruitment to the ehrlichial vacuole. Brief inhibition of the host SUMO pathway significantly decreases levels of PCGF5 colocalizing with ehrlichial inclusions. Coimmunoprecipitation data demonstrate that SUMO inhibition disrupts PCGF5 interaction with TRP120. Ectopic expression data obtained using SUMO-null mutants suggest that this is due to the necessity of TRP120 SUMO modification for PCGF5 interaction. The polycomb group protein paralog PCGF4 shares sequence homology with PCGF5 and was recently found to be a target of mammalian SUMOylation (81). However, PCGF5 does not encompass a lysine residue in the region that is modified in PCGF4; therefore, it is unlikely that PCGF5 is a direct target of the SUMO pathway. Instead, we posit that the decreased redistribution observed in the presence of AA is due to decreased interaction with TRP120 in the absence of effector modification.
Late in E. chaffeensis infection (72 h), significant colocalization of SUMO proteins at the ehrlichial vacuole was observed. SUMOylation, including TRP120 modification, facilitated recruitment of host proteins to ehrlichial inclusions. Therefore, it was not surprising that inhibition of host SUMOylation with the small molecule AA significantly decreased E. chaffeensis infection and survival. A natural derivative of salicylic acid, AA was previously described as an antimicrobial molecule at low micromolar concentrations (82, 83). At higher concentrations (>20 μM), AA is also a histone acetyltransferase inhibitor (84, 85), and it is cytotoxic to cancer cell lines at 40 to 100 μM (86). In the studies described here, the AA IC50 for bacterial load at 72 h p.i. was <10 μM, suggesting that the inhibitory effect on E. chaffeensis replication occurs through inhibition of host SUMOylation. siRNA knockdown of the host SUMOylation machinery confirmed that inhibition of this pathway decreases ehrlichial replication. Mechanistic characterization of the relevance of this PTM to effector function, combined with small-molecule and siRNA studies, suggests that this pathway is integral for E. chaffeensis infection and replication and may be a novel target for antiehrlichial therapeutics.
These studies represent the first report of a bacterial effector protein modified by the host SUMOylation pathway, which we demonstrate is the underlying molecular basis for interactions with select host proteins, and this PTM has a significant impact on ehrlichial survival. Acute changes in global protein SUMOylation were observed in 2DE and immunoblot analyses of infected whole-cell lysates. Preliminary proteomic examination of these proteins suggests that other bacterial effectors may also be targets of this host PTM (J. W. McBride and P. S. Dunphy, unpublished data). In addition, other ehrlichial TRPs encompass consensus SUMO motifs; hence, TRP120 is likely not the only example of a bacterial substrate of this pathway. The TRP120 ortholog in Ehrlichia canis (TRP140) contains consensus SUMO motifs in the TR region that may also serve as a substrate of this host PTM pathway. However, differences in motif position suggest that should these sites be targets of this PTM, the functional consequences may differ from those associated with SUMOylated TRP120. Thus, further studies will reveal the role of SUMOylation in various effector interactions with host proteins and its role in promoting replication and survival of Ehrlichia and other intracellular pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI105536 and AI106859. Paige Selvy Dunphy was supported by NIH biodefense training grant T32 AI007536.
We thank David Walker (University of Texas Medical Branch) for reviewing the manuscript and providing helpful suggestions.
Footnotes
Published ahead of print 21 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01984-14.
REFERENCES
- 1. Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8:328–339. 10.1038/nrmicro2318 [DOI] [PubMed] [Google Scholar]
- 2. McBride JW, Walker DH. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev. Mol. Med. 13:e3. 10.1017/S1462399410001730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunphy PS, Luo T, McBride JW. 2013. Ehrlichia moonlighting effectors and interkingdom interactions with the mononuclear phagocyte. Microbes Infect. 15:1005–1016. 10.1016/j.micinf.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. 2011. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect. Immun. 79:4370–4381. 10.1128/IAI.05422-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wakeel A, Kuriakose JA, McBride JW. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77:1734–1745. 10.1128/IAI.00027-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo T, McBride JW. 2012. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect. Immun. 80:2297–2306. 10.1128/IAI.00154-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect. Immun. 79:4382–4391. 10.1128/IAI.05608-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front. Cell. Infect. Microbiol. 1:22. 10.3389/fcimb.2011.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wakeel A, Zhu B, Yu XJ, McBride JW. 2010. New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 12:337–345. 10.1016/j.micinf.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Popov VL, Yu X, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28:71–80. 10.1006/mpat.1999.0327 [DOI] [PubMed] [Google Scholar]
- 11. Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. 2011. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One 6:e24136. 10.1371/journal.pone.0024136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuriakose JA, Zhang X, Luo T, McBride JW. 2012. Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins. Microbes Infect. 14:1054–1063. 10.1016/j.micinf.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jorgensen I, Bednar MM, Amin V, Davis BK, Ting JP, McCafferty DG, Valdivia RH. 2011. The Chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe 10:21–32. 10.1016/j.chom.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Copley SD. 2012. Moonlighting is mainstream: paradigm adjustment required. Bioessays 34:578–588. 10.1002/bies.201100191 [DOI] [PubMed] [Google Scholar]
- 15. Namanja AT, Li YJ, Su Y, Wong S, Lu J, Colson LT, Wu C, Li SS, Chen Y. 2012. Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J. Biol. Chem. 287:3231–3240. 10.1074/jbc.M111.293118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871. 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. 2009. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24. 10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- 18. Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F. 2013. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20:525–531. 10.1038/nsmb.2526 [DOI] [PubMed] [Google Scholar]
- 19. Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC. 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39:641–652. 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- 20. Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. 2011. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12:142–148. 10.1038/embor.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sarge KD, Park-Sarge OK. 2009. Detection of proteins sumoylated in vivo and in vitro. Methods Mol. Biol. 590:265–277. 10.1007/978-1-60327-378-7_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hilgarth RS, Sarge KD. 2005. Detection of sumoylated proteins. Methods Mol. Biol. 301:329–338. 10.1385/1-59259-895-1:329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Filosa G, Barabino SM, Bachi A. 2013. Proteomics strategies to identify SUMO targets and acceptor sites: a survey of RNA-binding proteins SUMOylation. Neuromol. Med. 15:661–676. 10.1007/s12017-013-8256-8 [DOI] [PubMed] [Google Scholar]
- 24. Da Silva-Ferrada E, Lopitz-Otsoa F, Lang V, Rodriguez MS, Matthiesen R. 2012. Strategies to identify recognition signals and targets of SUMOylation. Biochem. Res. Int. 2012:875148. 10.1155/2012/875148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barysch SV, Dittner C, Flotho A, Becker J, Melchior F. 2014. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 9:896–909. 10.1038/nprot.2014.053 [DOI] [PubMed] [Google Scholar]
- 26. Hay RT. 2005. SUMO: a history of modification. Mol. Cell 18:1–12. 10.1016/j.molcel.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 27. Ribet D, Cossart P. 2010. SUMOylation and bacterial pathogens. Virulence 1:532–534. 10.4161/viru.1.6.13449 [DOI] [PubMed] [Google Scholar]
- 28. Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J. Virol. 86:642–654. 10.1128/JVI.06227-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bekes M, Drag M. 2012. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J. Innate Immun. 4:159–167. 10.1159/000335027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ribet D, Cossart P. 2010. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143:694–702. 10.1016/j.cell.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang YE, Pernet O, Lee B. 2012. Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin-related modifiers. Biol. Cell 104:121–138. 10.1111/boc.201100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ribet D, Hamon M, Gouin E, Nahori MA, Impens F, Neyret-Kahn H, Gevaert K, Vandekerckhove J, Dejean A, Cossart P. 2010. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature 464:1192–1195. 10.1038/nature08963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 8:360–368. 10.1016/j.tim.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 34. Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. 2003. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50:377–389. 10.1046/j.1365-2958.2003.03730.x [DOI] [PubMed] [Google Scholar]
- 35. Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 11:400–411. 10.1038/nrmicro3015 [DOI] [PubMed] [Google Scholar]
- 36. Luo T, Zhang X, McBride JW. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16:982–990. 10.1128/CVI.00048-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Farrell PH. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 38. Burgess-Cassler A, Johansen JJ, Santek DA, Ide JR, Kendrick NC. 1989. Computerized quantitative analysis of Coomassie-blue-stained serum proteins separated by two-dimensional electrophoresis. Clin. Chem. 35:2297–2304 [PubMed] [Google Scholar]
- 39. Terui Y, Saad N, Jia S, McKeon F, Yuan J. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 279:28257–28265. 10.1074/jbc.M403153200 [DOI] [PubMed] [Google Scholar]
- 40. Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. 1998. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273:3117–3120. 10.1074/jbc.273.6.3117 [DOI] [PubMed] [Google Scholar]
- 41. Citro S, Chiocca S. 2013. Sumo paralogs: redundancy and divergencies. Front. Biosci. 5:544–553. 10.2741/S388 [DOI] [PubMed] [Google Scholar]
- 42. Pichler A, Melchior F. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381–387. 10.1034/j.1600-0854.2002.30601.x [DOI] [PubMed] [Google Scholar]
- 43. Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258. 10.1074/jbc.275.9.6252 [DOI] [PubMed] [Google Scholar]
- 44. Wang CY, She JX. 2008. SUMO4 and its role in type 1 diabetes pathogenesis. Diabetes Metab. Res. Rev. 24:93–102. 10.1002/dmrr.797 [DOI] [PubMed] [Google Scholar]
- 45. Hou S, Kijlstra A, Yang P. 2012. The genetics of Behcet's disease in a Chinese population. Front. Med. 6:354–359. 10.1007/s11684-012-0234-2 [DOI] [PubMed] [Google Scholar]
- 46. Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, Wang J, Dong Z, Brusko T, Atkinson M, Pozzilli P, Zeidler A, Raffel LJ, Jacob CO, Park Y, Serrano-Rios M, Larrad MT, Zhang Z, Garchon HJ, Bach JF, Rotter JI, She JX, Wang CY. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36:837–841. 10.1038/ng1391 [DOI] [PubMed] [Google Scholar]
- 47. Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. 2005. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337:517–520. 10.1016/j.bbrc.2005.09.090 [DOI] [PubMed] [Google Scholar]
- 48. Truong K, Lee TD, Li B, Chen Y. 2012. Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J. Biol. Chem. 287:42611–42619. 10.1074/jbc.M112.420877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Truong K, Lee TD, Chen Y. 2012. Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J. Biol. Chem. 287:15154–15163. 10.1074/jbc.M112.353789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sampson DA, Wang M, Matunis MJ. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669. 10.1074/jbc.M100006200 [DOI] [PubMed] [Google Scholar]
- 51. Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345–356. 10.1016/S0092-8674(02)00630-X [DOI] [PubMed] [Google Scholar]
- 52. Ren JGX, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. 2009. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9:3409–3412. 10.1002/pmic.200800646 [DOI] [PubMed] [Google Scholar]
- 53. Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42(Web Server issue):W325–W330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. 2008. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7:132–144. 10.1074/mcp.M700173-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matunis MJ, Coutavas E, Blobel G. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. 10.1083/jcb.135.6.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107. 10.1016/S0092-8674(00)81862-0 [DOI] [PubMed] [Google Scholar]
- 57. Rodriguez JA. 2014. Interplay between nuclear transport and ubiquitin/SUMO modifications in the regulation of cancer-related proteins. Semin. Cancer Biol. 27C:11–19. 10.1016/j.semcancer.2014.03.005 [DOI] [PubMed] [Google Scholar]
- 58. Dunphy PS, McBride JW. 2013. Host post translational modification pathways mediate nuclear translocation of Ehrlichia chaffeensis type 1 secreted effectors, abstr 15 Abstr. 26th Meet. Am. Soc. Rickettsiol. American Society for Rickettsiology, Portland, ME [Google Scholar]
- 59. Zhu S, Goeres J, Sixt KM, Bekes M, Zhang XD, Salvesen GS, Matunis MJ. 2009. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell 33:570–580. 10.1016/j.molcel.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kerscher O. 2007. SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8:550–555. 10.1038/sj.embor.7400980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U. S. A. 101:14373–14378. 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song J, Zhang Z, Hu W, Chen Y. 2005. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280:40122–40129. 10.1074/jbc.M507059200 [DOI] [PubMed] [Google Scholar]
- 63. Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428:133–145. 10.1042/BJ20100158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76:2990–2996. 10.1128/JVI.76.6.2990-2996.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ulrich HD. 2005. SUMO modification: wrestling with protein conformation. Curr. Biol. 15:R257–R259. 10.1016/j.cub.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 66. Tossidou I, Himmelseher E, Teng B, Haller H, Schiffer M. 18 June 2014. SUMOylation determines turnover and localization of nephrin at the plasma membrane. Kidney Int. 10.1038/ki.2014.198 [DOI] [PubMed] [Google Scholar]
- 67. Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M. 2009. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 16:133–140. 10.1016/j.chembiol.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 68. Hemshekhar M, Sebastin Santhosh M, Kemparaju K, Girish KS. 2011. Emerging roles of anacardic acid and its derivatives: a pharmacological overview. Basic Clin. Pharmacol. Toxicol. 110:122–132. 10.1111/j.1742-7843.2011.00833.x [DOI] [PubMed] [Google Scholar]
- 69. Hirohama M, Kumar A, Fukuda I, Matsuoka S, Igarashi Y, Saitoh H, Takagi M, Shin-ya K, Honda K, Kondoh Y, Saito T, Nakao Y, Osada H, Zhang KY, Yoshida M, Ito A. 2013. Spectomycin B1 as a novel SUMOylation inhibitor that directly binds to SUMO E2. ACS Chem. Biol. 8:2635–2642. 10.1021/cb400630z [DOI] [PubMed] [Google Scholar]
- 70. Kanakousaki K, Gibson MC. 2012. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139:2751–2762. 10.1242/dev.082974 [DOI] [PubMed] [Google Scholar]
- 71. Ribet D, Cossart P. 2010. Post-translational modifications in host cells during bacterial infection. FEBS Lett. 584:2748–2758. 10.1016/j.febslet.2010.05.012 [DOI] [PubMed] [Google Scholar]
- 72. Steele-Mortimer O. 2011. Exploitation of the ubiquitin system by invading bacteria. Traffic 12:162–169. 10.1111/j.1600-0854.2010.01137.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Angot A, Vergunst A, Genin S, Peeters N. 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3:e3. 10.1371/journal.ppat.0030003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ciechanover A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13–21. 10.1016/0092-8674(94)90396-4 [DOI] [PubMed] [Google Scholar]
- 75. Boman AL. 2001. GGA proteins: new players in the sorting game. J. Cell Sci. 114:3413–3418 [DOI] [PubMed] [Google Scholar]
- 76. Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. 2000. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 113:3439–3451 [DOI] [PubMed] [Google Scholar]
- 77. Berg JS, Cheney RE. 2002. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4:246–250. 10.1038/ncb762 [DOI] [PubMed] [Google Scholar]
- 78. Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. 2004. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 6:523–531. 10.1038/ncb1136 [DOI] [PubMed] [Google Scholar]
- 79. Liu X. 2012. Identification of SUMOylated proteins and investigation of protein ubiquitination in the NF-kappa B pathway. Ph.D. thesis University of Kentucky, Lexington, KY [Google Scholar]
- 80. Thomas S, Popov VL, Walker DH. 2010. Exit mechanisms of the intracellular bacterium Ehrlichia. PLoS One 5:e15775. 10.1371/journal.pone.0015775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ismail IH, Gagne JP, Caron MC, McDonald D, Xu Z, Masson JY, Poirier GG, Hendzel MJ. 2012. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 40:5497–5510. 10.1093/nar/gks222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Muroi H, Kubo I. 1996. Antibacterial activity of anacardic acid and totarol, alone and in combination with methicillin, against methicillin-resistant Staphylococcus aureus. J. Appl. Bacteriol. 80:387–394. 10.1111/j.1365-2672.1996.tb03233.x [DOI] [PubMed] [Google Scholar]
- 83. Kubo I, Nihei K, Tsujimoto K. 2003. Antibacterial action of anacardic acids against methicillin resistant Staphylococcus aureus (MRSA). J. Agric. Food Chem. 51:7624–7628. 10.1021/jf034674f [DOI] [PubMed] [Google Scholar]
- 84. Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. 2003. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 278:19134–19140. 10.1074/jbc.M301580200 [DOI] [PubMed] [Google Scholar]
- 85. Sun Y, Jiang X, Chen S, Price BD. 2006. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 580:4353–4356. 10.1016/j.febslet.2006.06.092 [DOI] [PubMed] [Google Scholar]
- 86. Park WJ, Ma E. 2012. Inhibition of PCAF histone acetyltransferase, cytotoxicity and cell permeability of 2-acylamino-1-(3- or 4-carboxy-phenyl)benzamides. Molecules 17:13116–13131. 10.3390/molecules171113116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Swaminathan S, Kiendl F, Korner R, Lupetti R, Hengst L, Melchior F. 2004. RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J. Cell Biol. 164:965–971. 10.1083/jcb.200309126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.