Abstract
Salmonella enterica serovar Typhimurium is a primary cause of enteric diseases and has acquired a variety of virulence factors during its evolution into a pathogen. Secreted virulence factors interact with commensal flora and host cells and enable Salmonella to survive and thrive in hostile environments. Outer membrane vesicles (OMVs) released from many Gram-negative bacteria function as a mechanism for the secretion of complex mixtures, including virulence factors. We performed a proteomic analysis of OMVs that were isolated under standard laboratory and acidic minimal medium conditions and identified 14 OMV-associated proteins that were observed in the OMV fraction isolated only under the acidic minimal medium conditions, which reproduced the nutrient-deficient intracellular milieu. The inferred roles of these 14 proteins were diverse, including transporter, enzyme, and transcriptional regulator. The absence of these proteins influenced Salmonella survival inside murine macrophages. Eleven of these proteins were predicted to possess secretion signal sequences at their N termini, and three (HupA, GlnH, and PhoN) of the proteins were found to be translocated into the cytoplasm of host cells. The comparative proteomic profiling of OMVs performed in this study revealed different protein compositions in the OMVs isolated under the two different conditions, which indicates that the OMV cargo depends on the growth conditions and provides a deeper insight into how Salmonella utilizes OMVs to adapt to environmental changes.
INTRODUCTION
Salmonella enterica serovar Typhimurium (here referred to as S. Typhimurium) is one of the most common causes of gastroenteritis and results in approximately 1.3 billion cases of nontyphoidal infections and approximately 3 million deaths annually worldwide (1). In the course of evolution, this organism has acquired a plethora of genes to combat commensal microorganisms for limited nutrient availability and to compromise the host immune responses in threatening environments (2, 3). The best-known genetic feature associated with Salmonella virulence is the Salmonella pathogenicity island (SPI), which produces virulence factors and the cognate type 3 secretion systems (T3SSs) for the secretion of virulence factors into the environment. The virulence determinants secreted via SPI-1 T3SS are required for Salmonella to invade intestinal epithelial cells and to provoke enterocolitis, whereas those delivered through SPI-2 T3SS mediate the intracellular survival and replication of Salmonella after it enters the host cells (4). To date, more than 40 virulence factors that are secreted through T3SSs have been identified, but the precise roles of these factors are less known (5, 6).
As a tool to facilitate their interaction with neighboring microbes, host cells, and even residence environments, a number of pathogenic bacteria release vesicles enclosing mixtures of toxins, degradative enzymes, and virulence factors and disseminate the contents into the environment. Since the advent of electron-microscopic observation of spherical blebs present in Gram-negative bacteria (7), membrane vesicle (MV) production has been regarded as an evolutionarily conserved process across Gram-positive and Gram-negative bacteria and even archaea (8–11). Outer membrane vesicles (OMVs) produced by Gram-negative bacteria are spherical structures 10 to 300 nm in diameter that pinch off from the cell surface and encompass a broad range of molecules present in the outer membrane and the periplasm and occasionally in the inner membrane and the cytoplasm (12, 13). The OMV constituents are varied and include proteins, phospholipids, lipopolysaccharides (LPSs), lipoproteins, and DNA (14–16). Accumulative proteomic analysis data demonstrate that OMVs function as specialized secretion vehicles that selectively sort out cargo molecules, but the selection mechanism remains undefined (17–19). The released OMVs benefit bacterial pathogenicity in offensive and defensive ways. OMVs deliver protease-protected virulence factors at a distance and are also able to integrate into the host plasma membrane, trafficking toxins directly into host cells (20, 21). The delivered virulence factors mediate host cell apoptosis and manipulate host immune responses (8, 22). As a defensive strategy, OMVs serve as bacterium-like decoys that absorb antimicrobial peptides and neutralize host immune responses (23, 24). In addition, OMV production also protects bacteria under stressful conditions by eliminating accumulated damaged DNAs and proteins (18, 25). Although the roles of OMVs have been extensively investigated, the mechanisms of OMV biogenesis have not yet been clearly addressed (15, 26).
In this study, Salmonella OMVs were isolated under standard laboratory and phagosome-mimicking conditions in vitro and were subjected to proteomic profiling analysis. Comparative analysis revealed a group of OMV-associated proteins that was specifically secreted under the phagosome-mimicking condition and a group of proteins that was conserved in both OMV protein profiles. Comparative proteomic profiling not only suggested new virulence factors associated with OMV in Salmonella but also provided fruitful information for understanding the roles and formation of OMVs in further studies.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. enterica serovar Typhimurium 14028S (ATCC 14028) was used as the parent strain in all of the experiments and strain constructions performed in this study. All of the deletion and tagged strains were produced using a phage λ Red recombination system (27). In brief, the kanamycin resistance (kan) cassette of pKD13 (27), pMini-Tn5-cycler (28), or pKD13-2HA (29) was amplified by PCR with 40-nucleotide flanking sequences that are homologous to the target genes at both termini, and the subsequent PCR products were introduced into recipient cells harboring pKD46 to replace the target genes with a kan cassette or to insert a CyaA′ (C-terminal peptide of adenylate cyclase)- or hemagglutinin (HA)-coding fragment and a kan cassette immediately prior to the stop codon sequence of the genes of interest. FLP recombinase produced from pCP20 removed the antibiotic resistance gene via site-specific recombination and resulted in in-frame deletions (27). For the construction of pHupA, pGlnH, and pPhoN, DNA regions containing the hupA-, glnH-, and phoN-coding sequences were amplified by PCR and cloned in pACYC184 (30), pUHE21-lacIq (31), and pZC320 (32) plasmids, respectively, through BamHI and HindIII restriction enzyme sites. IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a concentration of 50 μM to induce hupA, glnH, or phoN in trans during macrophage infection. All of the primers used to construct the bacterial strains and plasmids are listed in Table S1 in the supplemental material.
Growth conditions.
To partially reproduce the intracellular environment, an acidic low-magnesium minimal medium (acidic MgM), which was previously termed AMM1 (33, 34), was used, whereas Luria-Bertani (LB) medium was used to represent the standard laboratory conditions. For acidic MgM cultivation, Salmonella at the stationary phase in LB broth was washed and incubated overnight in magnesium minimal medium (MgM; pH 7.0) and then diluted into acidic MgM at a ratio of 1:100. The formula for MgM is the following: 100 mM Tris-Cl, 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.2% glycerol, 0.1% Casamino Acids, and 8 mM MgCl2. The pH was adjusted to 5.0 using H2SO4. For the general mutagenesis studies, LB medium was used to grow the bacteria, and antibiotics were added when required as follows: kanamycin, 50 μg/ml; chloramphenicol, 50 μg/ml; and ampicillin, 50 μg/ml.
OMV purification.
Salmonella cells were grown to the log phase under LB (3 h) or acidic MgM (4 h) conditions, and the culture was centrifuged at 10,000 × g for 5 min. The cell-free supernatant was filtered through a polyvinylidene difluoride (PVDF) filter (Millipore) (0.45-μm pore size) to remove the remaining bacteria, and the filtrate was concentrated 50-fold by ultrafiltration using a stirred-diffusion cell (model 8200; Millipore) equipped with ultrafiltration disks (PLHK09005; Millipore) of a nominal molecular weight limit (NMWL) of 100. The retentate was filtered again through a 0.45-μm-pore-size membrane to remove any remaining bacteria and then subjected to ultracentrifugation at 150,000 × g and 4°C for 4 h in a P40ST rotor to pellet the vesicles (Himac CP100β; Hitachi). The supernatant was carefully removed, leaving 1 ml at the bottom, which was then diluted with 35 ml of 4× HEPES solution (40 mM HEPES, 3.4% NaCl, pH 7.4) and reultracentrifuged as described above. The OMV fraction (0.5 ml) at the bottom was mixed with 1.5 ml of OptiPrep (60% iodixanol; Sigma) to resuspend the vesicles in 45% OptiPrep–1× HEPES solution (10 mM HEPES and 0.85% NaCl, pH 7.4) and then layered at the bottom of a centrifuge tube. An OptiPrep gradient was prepared by slowly layering 40%, 35%, 30%, 25%, and 20% OptiPrep–1× HEPES solution over the vesicle fraction. The mixture was then subjected to ultracentrifugation at 150,000 × g, and equal fractions were removed sequentially from the top. A portion of each fraction was precipitated with 20% trichloroacetic acid (TCA), resuspended in 1× Laemmli loading buffer, and visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12%) followed by Coomassie brilliant blue R-250 (Sigma) staining or silver staining with a PowerStain silver stain kit (ELPIS Biotech). The desirable gradient fractions of middle densities were pooled and diluted in 1× HEPES solution, and the diluted fractions were then ultracentrifuged (150,000 × g, 2 h) to recover the OMVs. The purified OMVs were quantified using a bicinchoninic acid (BCA) protein assay kit (Pierce). The sterility of the OMV-containing solutions was checked occasionally throughout the procedure by spreading a portion on an LB plate to test the bacterial growth.
TEM.
For the microscopic analysis of the isolated OMVs, the vesicle fractions were mounted onto carbon-coated copper grids and incubated for 10 min for attachment to the grid. After the grid was washed twice with ultrapure water, 2% uranyl acetate (pH 4.0) was applied to the grid for negative staining and subsequently removed. The stained grid was subjected to transmission electron microscopy (TEM) analysis with LIBRA 120 (Carl Zeiss, Switzerland) at 80 kV.
Sample preparation for liquid chromatography-tandem mass spectrometry (LC-MS/MS).
The proteins in the purified OMVs were separated via SDS-PAGE (12%) and stained with Coomassie brilliant blue R-250. The excised gel slices were destained in destaining buffer containing 50% acetonitrile (ACN) and 25 mM NH4HCO3 and dehydrated in ACN. The proteins in the gel were reduced with 10 mM dithiothreitol (DTT) at 56°C for 60 min, alkylated with 55 mM iodoacetamide at room temperature for 45 min, washed twice with 100 mM NH4HCO3 and ACN, and then dried using a Speedvac. For in-gel digestion, the gels were reswelled in trypsin solution (sequencing-grade modified trypsin from Promega, Madison, WI) (12.5 ng/ml) at 37°C overnight. The digested peptides were extracted from the gel by incubating the gel in 5% formic acid solution three times at room temperature. The pooled peptides were dried using a Speedvac and reconstituted with 0.1% formic acid.
Proteomic profiling using LC-MS/MS.
Nano-LC-MS/MS analysis was performed on an Agilent 1100 Series nano-LC and linear trap quadropole (LTQ)-mass spectrometer (Thermo Electron, San Jose, CA). The capillary column used for the LC-MS/MS analysis (150 mm by 0.075 mm) was obtained from Proxeon (Odense M, Denmark), and the slurry was packed in-house using a 5-μm-, 100-Å-pore-size Magic C18 stationary-phase column (Michrom Bioresources, Auburn, CA). The mobile A phase for the LC separation was 0.1% formic acid–deionized water, and the mobile B phase was 0.1% formic acid–acetonitrile. The chromatography gradient was set up to obtain a linear increase from 5% B to 35% B in 100 min, from 40% B to 60% B in 10 min, and from 60% B to 80% B in 20 min. The flow rate was maintained at 300 nl/min after splitting. The mass spectra were acquired using data-dependent acquisition with a full-mass scan (400 to 1,800 m/z) followed by MS/MS scans. Each MS/MS scan acquired corresponded to an average of one microscan on the LTQ device. The temperature of the ion transfer tube was maintained at 200°C, and the spray was 1.5.0 to 2.0 kV. The normalized collision energy was set to 35% for MS/MS.
Data analysis for protein identification.
The collected mass spectrometry (MS/MS) spectra of the peptides were analyzed using the MASCOT algorithm (Matrixscience) against databases of S. enterica serovar Typhimurium LT2 and 14028S (NCBInr database downloaded on 1 November 2012). The search parameters were the following: (i) enzyme specificity, trypsin; (ii) maximum allowed missed cleavages of 2; (iii) carbamidomethylation at cysteine residues (C) as a fixed modification; (iv) oxidation at methionine residues as variable modification; (v) MS tolerance of 1.2 Da; and (vi) MS/MS tolerance of 0.6 Da. The MASCOT result files (.dat) were exported and filtered for a false-discovery rate (FDR) of less than 5% at the peptide level using “Rockerbox” (Version 2.0.6) (35). All of the peptide spectrum matches (PSMs) were filtered using the FDR-based algorithm. At least two unique peptides were required for the protein identification. The subcellular location of a protein was predicted by PSORTb (version 3.0.2; http://www.psort.org/psortb/) (36). The presence of secretion signals in a protein was inferred using PSORTb, LipoP 1.0 (www.cbs.dtu.dk/services/LipoP) (37), TatP 1.0 (www.cbs.dtu.dk/services/TatP/) (38), and SecretomeP 2.0 (www.cbs.dtu.dk/services/SecretomeP) (39).
Macrophage survival assay.
The RAW264.7 murine macrophage-like cell line (ATCC TIB-71) was seeded at 2 × 105 cells per well in 24-well tissue culture plates and incubated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) overnight at 37°C with 5% CO2. The S. Typhimurium strains were grown overnight in LB prior to infection, and the bacterial cells were washed with phosphate-buffered saline (PBS) and resuspended in DMEM. The macrophage cells in the 24-well plates were infected with bacteria at an input multiplicity of infection (MOI) of 100. The infections were initiated by centrifuging the bacteria onto the macrophage monolayers at 1,000 × g for 5 min, and the plates were then incubated at 37°C for 30 min. To remove any extracellular bacteria after 30 min of infection, the cells were washed with PBS and subsequently treated with DMEM containing 100 μg/ml gentamicin (Gibco) for 1 h. The infected macrophage cells were then washed three times with PBS and overlaid with DMEM containing 20 μg/ml gentamicin for the remainder of the experiment. To compare the rates of bacterial survival inside the macrophages, the cells were lysed with 1% Triton X-100–PBS, and the lysates were serially diluted and plated on LB agar.
cAMP assay.
To test whether the CyaA′-tagged proteins were translocated into the host cytoplasm, the macrophages were infected with S. Typhimurium strains expressing CyaA′ fusion proteins as described above, washed three times with PBS, and then lysed with 0.1 M HCl at 12 h postinfection. The intracellular cyclic AMP (cAMP) levels were measured as directed by the manufacturer's instructions for a Direct cAMP enzyme-linked immunosorbent assay (ELISA) kit (Enzo Life Science). The cell lysate was centrifuged at 5,000 × g for 2 min to remove any unbroken macrophages and bacterial debris, and the cytosolic fraction was used to determine the levels of cAMP. All of the cAMP assays were repeated at least three times using independently infected macrophages, and the cAMP levels were averaged.
Immunoblot analysis.
The isolated OMV fractions or whole-cell pellets were dissolved in Laemmli sample buffer and boiled for 5 min. The protein samples were loaded on a 12% SDS-polyacrylamide gel. The separated proteins on the gel were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked with 5% nonfat dry milk–1× Tris-buffered saline-Tween 20 (TBST) buffer. The membrane was probed with anti-HA antibody (Sigma) and anti-DnaK antibody (Enzo Life Science) as primary antibodies and then treated with anti-mouse IgG conjugated with peroxidase (Santa Cruz Biotechnology) as the secondary antibody in all of the immunoblot experiments. The chemiluminescent signals were developed with a West-Zol plus Western blot detection system (Intron Biotechnology, South Korea).
RESULTS
Differences in the protein compositions of OMVs under two growth conditions.
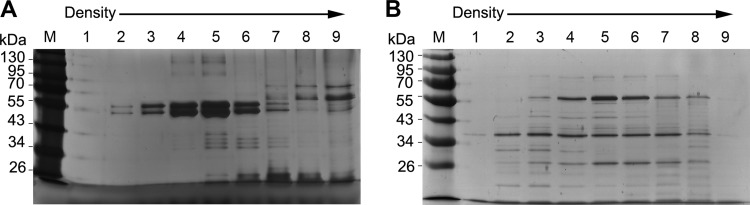
The nature of Salmonella pathogenic properties is attributable to the ability of Salmonella to resist hostile intracellular conditions and replicate inside phagocytic cells. To understand how Salmonella manipulates the OMV contents for its proliferation inside host cells, the bacteria were cultivated in an acidic minimal medium (MgM, pH 5.0) that partially reproduces the intracellular environment (33, 34), and the OMVs were isolated from the cell-free culture supernatant by ultracentrifugation as described in Materials and Methods. The OMVs were further purified through density gradient ultracentrifugation, and the subsequent gradient fractions were resolved by SDS-PAGE to visualize the OMV-associated proteins. OMVs were also harvested under a standard laboratory culture condition (LB) in parallel, and the repertoire of OMV proteins was compared with that obtained from the acidic minimal medium culture (Fig. 1). Gradient fractions 4, 5, and 6 exhibited comparable protein distribution patterns under both culture conditions, indicating that the OMV protein compositions were similar in these three fractions, although the amounts of the component proteins differed between the fractions. The comparison of the middle-density fractions (fractions 4, 5, and 6) under the two culture conditions revealed OMV protein compositions the differed under the LB and acidic MgM conditions. For example, the OMVs under the LB condition were enriched in proteins of approximately 50 kDa, whereas proteins of multiple sizes were abundant in the acidic MgM-derived OMVs. However, there was no significant morphological difference in the OMVs under the two conditions, as determined through transmission electron microscopy (see Fig. S1 in the supplemental material).
FIG 1.
SDS-PAGE protein profiles of OMVs isolated under two different conditions. S. Typhimurium OMV fractions isolated under the LB (A) and acidic MgM (B) growth conditions were further purified through density gradient ultrafiltration, and the resultant gradient fractions were loaded onto SDS-PAGE gels for electrophoresis. The whole gradient was divided into nine fractions from top to bottom, and a portion of each fraction was loaded from lanes 1 to 9. The proteins were visualized through silver staining and compared.
Determination of the OMV proteome.
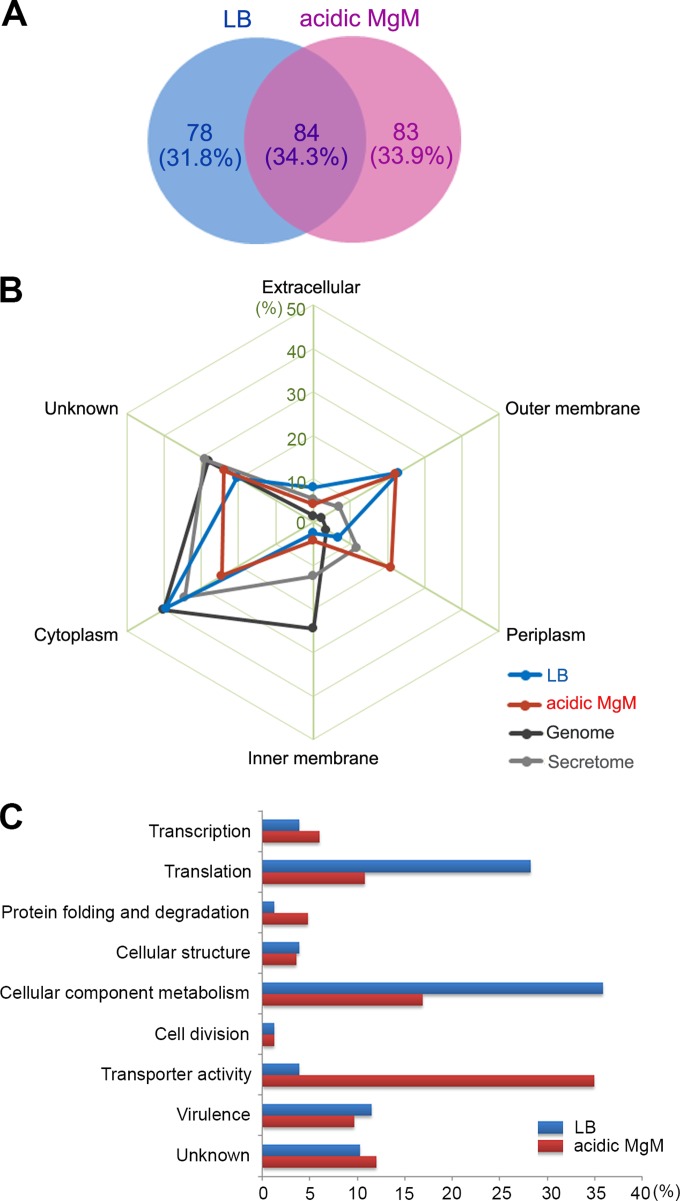
The OMVs purified under two different conditions were subjected to LC-MS/MS analysis to identify their associated proteins. To avoid the identification of false-positive proteins, those proteins with unique matches of two or more peptides after filtration of the raw data with a 5% FDR at the peptide level were selected, and the proteomic profiles are shown in Dataset S1 in the supplemental material. Totals of 162 and 167 proteins were identified in the LB-derived and acidic MgM-derived OMVs, respectively, and there were 84 proteins found in both OMV proteomic profiles (Fig. 2A). The subcellular locations of the identified proteins were predicted using PSORTb (36), and the possibility that these proteins possessed secretion signal peptides was assessed using PSORTb (36), LipoP (37), TatP (38), and SecretomeP (39) (see Dataset S1). The comparison of the subcellular locations of the OMV components with those of Salmonella genome-inferred proteins revealed that the OMVs derived under both the LB and acidic MgM conditions were enriched in outer membrane proteins, periplasmic proteins, and extracellular proteins (38% in the LB OMVs and 47% in the acidic MgM OMVs), whereas only 8% of the total Salmonella proteins were classified as extracellular, periplasmic, or outer membrane proteins (Fig. 2B). These subcellular prediction results demonstrate that the OMVs were successfully purified in our study. The comparison of the OMVs obtained under the two growth conditions revealed that more envelope proteins were identified in the OMVs isolated under the acidic minimal medium condition. Interestingly, the Salmonella OMVs also contained a large number of cytoplasmic proteins as cargo proteins (40% in the LB OMVs and 25% in the acidic MgM OMVs), similarly to the OMVs of other bacteria (12, 13), although some of the cytoplasmic proteins were experimentally relocalized to be in the outer membrane or periplasm (40), which inspires the notion that these proteins are more apt to be entrapped in OMVs. Some proteins with versatile roles may be localized in multiple sites on purpose as discussed below.
FIG 2.
Proteomic analysis of OMVs isolated under two different growth conditions. (A) Venn diagram of OMV-associated proteins from two proteomic profiles. A total of 245 proteins were identified from the LB-derived and acidic MgM-derived OMVs through LC-MS/MS analysis using the stringent criteria described in Materials and Methods. The proteins found under both conditions or under only one of the conditions were counted, and their numbers (and percentages) are shown. (B) Subcellular location of OMV-associated proteins. The proteins identified through LC-MS/MS profiling were processed using PSORTb (http://www.psort.org/psortb/) to predict their main location in the cell in addition to the OMV. The proteins in the LB-derived and acidic MgM-derived OMV profiles that were predicted to be in the extracellular, outer membrane, periplasm, inner membrane, or cytoplasm fractions were counted, and their percentages are depicted. The proteins identified through S. Typhimurium genome annotation and secretome analysis (see details in Discussion) were subjected to subcellular location prediction and compared with those found in the OMV profiles. (C) Classification of OMV-associated proteins using Gene Ontology (GO)-based functions. The OMV-associated proteins unique to the LB or acidic MgM conditions were grouped based on their biological processes and molecular functions, and their proportions under each condition are plotted.
Comparative proteomic analysis of OMVs under two different conditions.
A comparative analysis of two OMV profiles can provide more information about how Salmonella controls the OMV cargo in order to adapt to environmental stresses in host cells. Furthermore, the proteins conserved in both OMV fractions independently of the growth conditions may be associated with OMV biogenesis and may play important roles in the blebbing and release of OMVs. The majority (56%) of the 84 proteins conserved in both conditioned OMVs were predicted to constitute the bacterial cell envelope: 5 proteins that participate in the cell wall metabolic process, 6 proteins that participate in the flagella structure, and 14 proteins that participate in transport systems (Fig. 2A; see also Fig. S2 and Dataset S1 in the supplemental material). The number of proteins that were associated with OMVs only under the LB condition was 78, and the majority of these (45 of 78 proteins) were predicted to be located in the cytoplasm (Fig. 2A; see also Fig. S2 in the supplemental material). In contrast, 83 OMV proteins were found to be secreted only under the acidic MgM condition, and these included many envelope proteins (7 outer membrane proteins, 27 periplasmic proteins, and 5 inner membrane proteins) (Fig. 2A; see also Fig. S2).
To understand the roles of the OMV protein constituents under specific environmental conditions, the proteins were classified using their Gene Ontology (GO)-based biological processes and molecular functions (Fig. 2C; also see Dataset S1 in detail in the supplemental material). Although the numbers of OMV proteins obtained under the two conditions belonging to the functional groups of transcription, cellular structure, cell division, virulence, and unknown were comparable, proteins with transporter activity were highly sorted into the OMVs obtained under the acidic minimal condition (29 of 83 proteins), whereas the LB-derived OMVs were enriched with proteins involved in cellular component metabolism and translation (28 and 22, respectively, of 78 proteins). The dominant distribution of transporter-relevant proteins in the acidic MgM-derived OMVs may be a bacterial countermeasure to ensure their survival under nutrient-deficient conditions. Similarly, the large numbers of metabolism- and translation-related proteins in the LB-derived OMVs were likely caused by vigorous bacterial replication under the nutrient-rich condition. The genes responsible for ribosome synthesis and translation are fully upregulated through the mid-log phage in LB medium (41), and their secretion into OMVs and the extracellular culture supernatant has been previously reported (12, 42).
To our surprise, SPI effector and translocon proteins that are secreted through T3SS were also identified in the OMV fraction. Interestingly, SPI-1 proteins, including SipA, SipB, SipC, SopA, SopB, and SopE2, were selectively detected in the OMVs isolated under the LB condition, which partially mimics the nutrient-rich intestinal lumen, whereas SPI-2 proteins, such as SseB, SseC, and SseD, were trapped into OMVs only under the phagosome-mimicking acidic MgM condition (see Table S2 in the supplemental material). SPI effectors secreted via T3SS may be incorporated into extracellular OMVs or may traverse the inner membrane via an unknown pathway and be enclosed into OMVs. It would be worthwhile to examine the possibility of the OMV-mediated translocation of SPI effectors in a further study to obtain a better understanding of Salmonella virulence strategies.
The proteins that were secreted via OMVs under the phagosome-mimicking condition may participate in Salmonella virulence during its intracellular replication inside host cells. Therefore, we selected the OMV-associated proteins in the acidic MgM medium condition that exhibited a high confidence (unique peptides ≥ 4) and tested whether these are involved in Salmonella intracellular survival and selectively sorted into OMVs.
Surveying OMV-associated transport proteins important for Salmonella intracellular survival.
The majority of proteins associated with acidic MgM-derived OMVs were predicted to play roles in the transport of diverse molecules, such as amino acids, sugars, polyamines, metals, and ion compounds (see Dataset S1 in the supplemental material). The biased location of the transport machinery toward OMVs may be required for Salmonella to survive and replicate inside the Salmonella-containing vacuole, the phagosome-like nutrient-deficient intracellular reservoir (43). To test this possibility, 26 (22 with transporter activity and 4 with uncharacterized function) of the 47 top-matched proteins (unique peptides ≥ 4) were selected (see Table S3 in the supplemental material), and their effects on Salmonella survival inside macrophages were examined. Twenty-six strains lacking these candidate proteins were constructed and used to infect murine RAW264.7 macrophages as described in Materials and Methods. The intracellular bacteria were enumerated at 16 h postinfection, and their survival ability was compared with that of wild-type Salmonella. Five strains devoid of PstS, DppA, CysP, STM14_1918, or YcfM exhibited intracellular survival attenuated by more than 2-fold, whereas the rate of survival of six strains lacking RbsB, PotD, STM14_2773, YggE, MetQ, or YehZ was more than 2.5-fold higher than that of wild-type Salmonella (Fig. 3A). These 11 mutant strains and the wild-type stain showed comparable growth rates at 1.5 h postinfection (data not shown), and their growth rates were also similar under the acidic MgM condition (see Fig. S3 in the supplemental material). In terms of the host-pathogen equilibrium, bacterial outgrowth is also deleterious to long-term bacterial persistence in hosts, as demonstrated in other studies (44–47).
FIG 3.
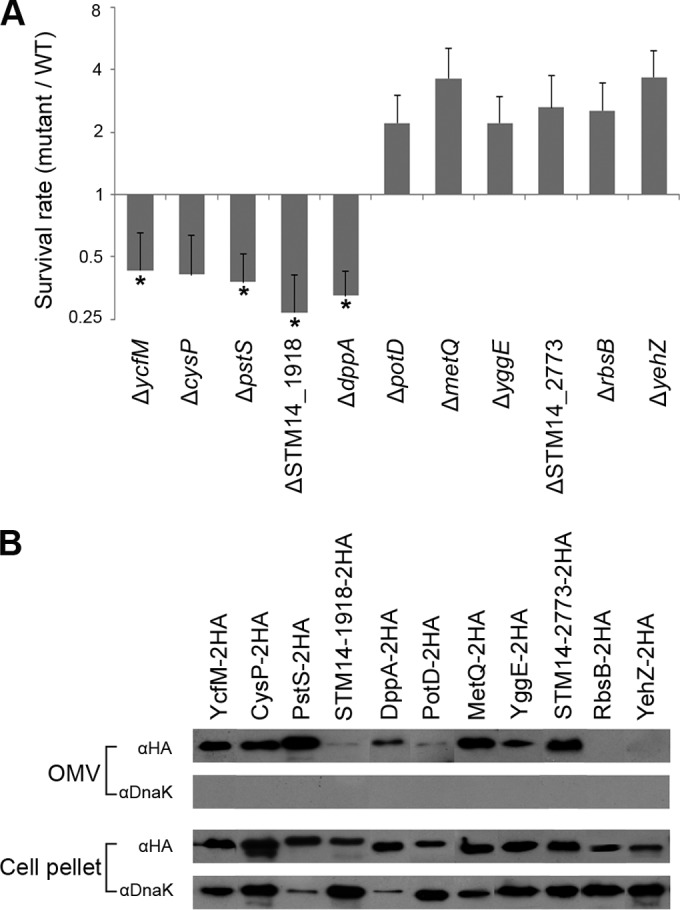
OMV-associated transport proteins and uncharacterized proteins important for Salmonella survival inside macrophages. (A) Intracellular survival assay of 11 mutants. The S. Typhimurium wild-type (WT) strain and 11 mutant strains lacking proteins with transporter activity or unknown function were used to infect RAW264.7 macrophages, and their rates of intracellular survival at 16 h postinfection were compared. The survival rates of a mutant strain relative to the wild-type bacteria from at least three independent infections were averaged, and the increase or decrease in survival was evaluated statistically through one-way analysis of variance (ANOVA) and Tukey's post hoc test. All of the mutant strains showed significant differences, with P values of less than 0.05. The cases denoted by asterisks (*) indicate that the P values were less than 0.005. (B) Immunoblot verification of OMV localization of proteome-inferred proteins. Eleven proteins were genetically tagged with a hemagglutinin (HA), and Salmonella strains producing the tagged proteins were grown under acidic MgM conditions. The OMV fractions from each Salmonella culture were purified and analyzed using an immunoblot assay with an anti-HA antibody to investigate the presence of the HA-tagged proteins in the OMVs. Bacterial whole-cell lysates were analyzed in parallel with the OMV fractions. An anti-DnaK antibody was also used to check the possibility of OMV contamination with whole cells.
The 11 proteins affecting bacterial intracellular survival include 7 proteins associated with transport systems and 4 with an uncharacterized function. The phosphate transport system (Pst), in which PstS acts as a periplasmic Pi-binding protein, is linked to the Pho regulon (48), and its absence has been reported to reduce virulence in a variety of pathogenic bacteria (48, 49). DppA is a substrate-binding protein in the dipeptide uptake transporter system that is responsible for the uptake of essential amino acids, heme production, chemotaxis, and sporulation (50–52), and the lack of DppA was found to reduce virulence in group B streptococci (53). CysP and Sbp are periplasmic sulfate/thiosulfate-binding components of ABC-type sulfate transporters and exhibit partially overlapping substrate-binding activities (54). However, the deletion of the sbp gene did not impair Salmonella survival in our macrophage infection assay (data not shown), which suggests a unique role of CysP in the intracellular growth of Salmonella. RbsB is known as the substrate-binding protein of the ribose ABC transporter system, RbsACB, with a high affinity for ribose but has also been found to sequester autoinducer 2 (AI-2) and modulate cellular responses mediated by quorum-sensing signals, including growth in Aggregatibacter actinomycetemcomitans (55, 56). PotD is the substrate-binding protein of the PotABCD ATP-dependent polyamine transporter, which transports spermidine and putrescine. Recently, polyamine was revealed to function as a signal molecule that regulates the virulence of intracellular pathogens, including S. Typhimurium (57). Spermidine is synthesized from putrescine or methionine in bacteria, which indicates a metabolic convergence between methionine and polyamine (57, 58). The hypervirulence phenotype of the ΔmetQ strain, which is defective in the transport of methionine, may be metabolically connected with the intracellular polyamine imbalance caused by ΔpotD. YehZ shows high sequence homology with OsmX, which is the osmoprotectant-binding protein of the OsmU transport system (45, 59). It was recently found that Salmonella strains lacking OsmU stimulate trehalose accumulation and thereby become resistant to intracellular stressors, such as oxidative stress and acidic pH (45).
To verify that these 11 proteins are released via OMVs, each protein was genetically tagged with a hemagglutinin (HA) peptide, and their presence in the OMV fraction was examined through Western blot analysis with an anti-HA antibody (Fig. 3B). When the tagged strains were cultivated under the acidic MgM condition, 9 of the 11 proteins were localized in the OMVs, and 2 proteins (RbsB and YehZ) were barely detected in the OMV fraction. The failure of the immunoblot verification of RbsB and YehZ may be due to their low production in the cells. To rule out the possibility of OMV contamination with whole cells, the presence of DnaK, an abundant cytoplasmic protein in S. Typhimurium (40), in the OMV fractions was checked in parallel.
Exploring the translocation of OMV-associated virulence proteins into the host cytoplasm.
In addition to importing materials that are essential for intracellular survival and resistance, pathogenic bacteria also translocate a variety of virulence factors, including toxins and enzymes, into host cells via OMVs. To identify the OMV-associated virulence factors that are translocated into the host cytoplasm, 28 of the 47 proteins that were confidently associated with acidic MgM-derived OMVs were selected as secreted candidates (see Table S3 in the supplemental material). The selection criteria for candidate screening were the following: the candidate was (i) predicted to be a non-outer membrane protein and (ii) not known as an SPI effector protein.
The candidate proteins were genetically tagged with the N-terminal catalytic domain of CyaA (adenylate cyclase) at their C termini, and Salmonella strains producing the CyaA′-tagged candidates were used to infect murine macrophages. The delivery of CyaA′ into the eukaryotic cell cytoplasm accelerates cAMP production and has been widely used for screening secreted proteins (28, 60). Of the 28 proteins tested, three proteins (HupA, GlnH, and PhoN) were found to be translocated into the host cytoplasm (Fig. 4A). HU is a heterodimeric DNA-binding protein composed of HupA and HupB and not only compacts the DNA of the bacterial nucleoid but also regulates the transcription of a variety of genes, including the virulence genes of S. Typhimurium (61) and other pathogens (62–64). Interestingly, HupA was preferentially associated with acidic MgM-derived OMVs, whereas HupB was detected in OMVs isolated under either condition, which suggests differential roles of HupA and HupB. Glutamine uptake and synthesis are important for full bacterial fitness and virulence. The virulence of Salmonella was severely attenuated when both glnA and glnH were inactivated (65). GlnA is glutamine synthetase, responsible for glutamine synthesis, and GlnH is the periplasmic glutamine-binding protein belonging to the GlnHPQ transport system. phoN, which encodes an acid phosphatase (PhoN), is regulated by PhoPQ, a representative of the virulence regulatory system in Salmonella, but the absence of phoN did not influence Salmonella virulence in mice (66, 67). However, the importance of acid phosphatase in bacterial virulence and intracellular survival has been observed in several pathogenic bacteria (68–70).
FIG 4.
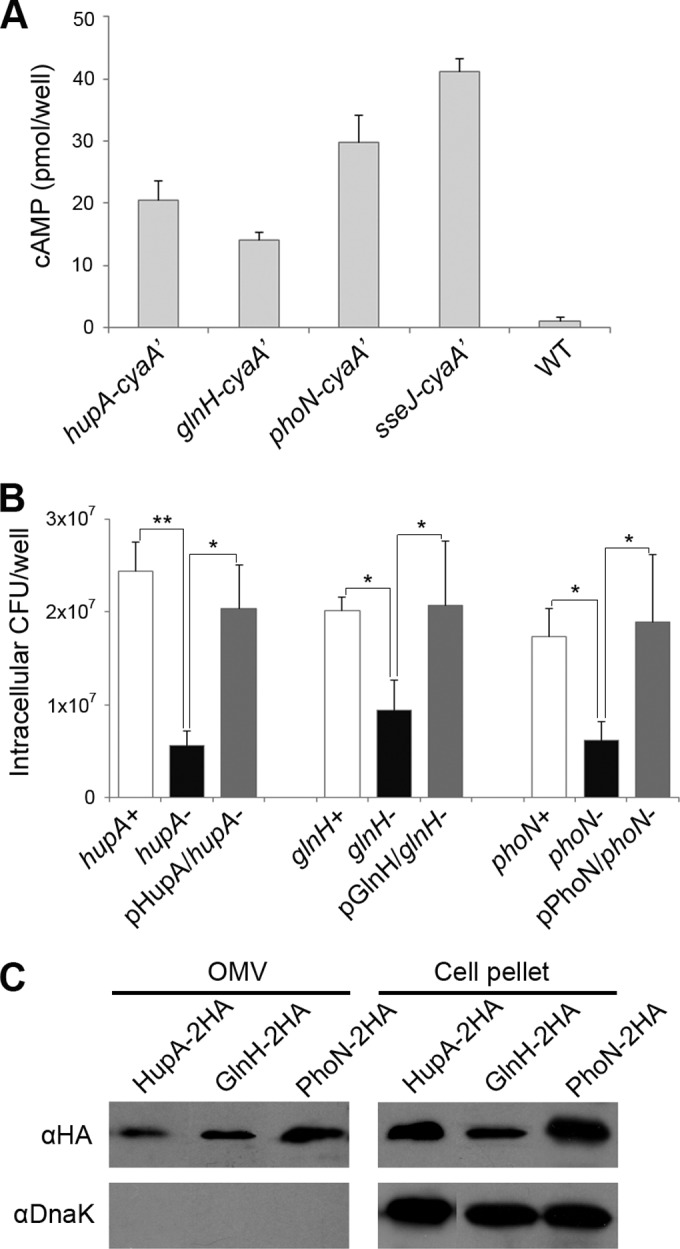
OMV-associated virulence proteins translocated into the macrophage cytoplasm. (A) Translocation of HupA, GlnH, and PhoN into the host cytosol. HupA, GlnH, and PhoN were tagged with CyaA′ at their C termini using genetic modification as described in Materials and Methods. Macrophages were infected with Salmonella strains producing CyaA′-tagged proteins for 12 h, and the intracellular cAMP levels were assayed to test the translocation of the CyaA′-tagged proteins into the host cytoplasm. SseJ, a well-known SPI-2 T3SS-secreted effector, was also tagged with CyaA′ and examined with three tested proteins. The intracellular cAMP levels averaged from three separate experiments are shown with their standard deviations. One-way ANOVA and Tukey's post hoc analysis were used for the statistics (P < 0.05). (B) Intracellular survival assay of ΔhupA, ΔglnH, and ΔphoN mutants. Macrophages were infected with Salmonella strains lacking HupA, GlnH, or PhoN, and the intracellular CFU were enumerated at 12 h postinfection. To complement the attenuated survival of the deletion strains, plasmids expressing HupA, GlnH, or PhoN were introduced into the mutant strains lacking the corresponding functional alleles. The averages and standard deviations of the results from three independent infections are shown. The asterisks indicate a significant difference, as determined by ANOVA and Tukey's post hoc test. *, P < 0.05; **, P < 0.005. (C) Immunoblot verification of OMV localization of HupA, GlnH, and PhoN. Salmonella strains producing HupA, GlnH, and PhoN tagged with HA were cultivated under acidic MgM conditions, and the OMVs were isolated. The OMV fractions and whole-cell lysates were analyzed through an immunoblot assay with an anti-HA antibody to verify the presence of HupA, GlnH, and PhoN in the OMVs. The presence of DnaK was also examined to evaluate the purity of the OMV fractions.
When macrophages were infected with Salmonella lacking HupA, GlnH, or PhoN and the number of intracellular bacteria was counted at 12 h postinfection, all three mutants showed a decrease in survival of at least 50% compared with wild-type Salmonella, which suggests the importance of the proteins for the survival of Salmonella inside macrophages. In addition, the attenuated survival was completely complemented by the introduction of a plasmid that produces HupA, GlnH, or PhoN (Fig. 4B). The three mutant strains did not exhibit impaired growth under the acidic minimal medium condition (see Fig. S3 in the supplemental material), which suggests the incomplete reproduction of the intracellular environment under the acidic MgM condition in vitro. The attenuation of the intracellular survival of the ΔphoN mutant is inconsistent with the previously reported phenotypes of phoN mutants in host animals (66, 67). However, a discrepancy between the virulence phenotypes in host cells and host animals is occasionally reported and is likely due to the complicated coordination of a variety of immune systems in host animals (44, 71–74).
These three proteins were determined to be located in the periplasm or cytoplasm (see Dataset S1 in the supplemental material), based on a computational prediction using PSORTb (36). Although cytoplasmic proteins located in OMVs have been observed in other OMV profiling studies, the presence of transcriptional regulators, such as HupA, in OMVs is curious because regulator-mediated transcriptional activity occurs in the cytoplasm harboring nucleic acids and other transcription machineries. Nie-mann et al. also observed three major nucleoid-associated proteins (HupA, HupB, and H-NS) in the culture supernatant (42). This unheralded location of cytoplasmic proteins in the extracellular milieu appears to be caused by a deliberate sorting mechanism and not by an accidental outflow from dead cells being lysed. We tagged three proteins (HupA, GlnH, and PhoN) with an HA-coding fragment and experimentally verified their presence in the OMV fractions using a Western blot assay (Fig. 4C). All three proteins were located in the OMVs isolated from the acidic MgM culture. Taken together, these results indicate that these three proteins, which are important for Salmonella virulence, can be secreted into the extracellular milieu via OMVs. Translocation of these proteins into the host cytoplasm might be caused by OMVs as observed under in vitro conditions, although reliable microscopic observations are required to verify the possibility. The unexpected identification of cytoplasmic proteins in OMVs suggests several possibilities regarding their location-dependent distinct roles during Salmonella infection and the existence of an unidentified secretion pathway for proteins lacking signal sequences, as discussed in detail in Discussion.
DISCUSSION
A comprehensive proteomic profiling analysis identified a total of 245 proteins associated with OMVs in S. Typhimurium and also revealed a group of 47 proteins that were confidently observed in acidic MgM-derived OMVs and are therefore potent candidates for virulence determinants in Salmonella pathogenicity (see Table S3 in the supplemental material). Of the 47 proteins, 22 proteins were assigned as transport proteins for a variety of molecules such as amino acids, sugars, and polyamines, and this finding seemed indicative of an active response of Salmonella to the possible lack of these molecules under the phagosome-mimicking conditions; hence, an insufficient supply of these molecules by malfunctioning transport systems may threaten Salmonella survival in host cells. However, Salmonella lacking some of the transport proteins instead outcompeted wild-type bacteria inside host cells. Both excessive bacterial growth and attenuated bacterial growth are unfavorable because these compromise the host-pathogen equilibrium required for long-term bacterial persistence and lead to their early elimination from the site of infection due to aggressive host immune responses. Therefore, pathogenic bacteria need to fine-tune their growth rate to ensure their fitness in a host. Increasing numbers of studies are analyzing the hypervirulence of Salmonella mutant strains lacking genes with diverse roles and, often, unknown function (44–47).
The comprehensive proteomic profiling of OMVs performed in this study provides a foundation for understanding the Salmonella OMV architecture and OMV-mediated virulence regulation. In addition, extensive proteomics efforts over the last few decades have been made to attempt to unravel the virulence mechanisms that are unique to Salmonella. Recently, a proteomic survey of the Salmonella extracellular proteins secreted into the supernatant of another acidic minimal medium {mLPM; 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 0.3% (vol/vol) glycerol, 0.00001% thiamine, 0.5 μM ferric citrate, 8 μM MgCl2, 337 μM PO43−, 80 mM MES [2-(N-morpholino)ethanesulfonic acid]-free acid adjusted to pH 5.8 with NaOH} culture was conducted (42). When the Salmonella OMV proteome (a total of 245 proteins) was compared with the secretome (a total of 300 proteins), only 94 proteins were found to be overlapping. This discordance may be due to the different methods used to prepare the protein samples. We concentrated the OMV fractions gradually by removing undesirable small materials through ultrafiltration and multiple centrifugations, whereas the researchers who conducted the above-mentioned study collected the secreted proteins from the filtered culture supernatants through serial solid-phase extractions with C4, C8, and C18 resins (42). OMVs with the hydrophilic polysaccharide moiety of LPS exposed on the spherical surface are less likely to bind to hydrophobic sorbents, such as C8 and C18.
The concomitant presence of cytoplasmic proteins and cell envelope proteins in OMVs has been one of the most intriguing puzzles that have challenged our understanding of OMV biogenesis because the bulging outward of the outer membrane is most likely to trap adjacent proteins, such as periplasmic proteins, into the vesicle, and cytoplasmic proteins without signal sequences are hardly accessible to the vesiculation site due to the rigid barrier of the inner membrane. Nevertheless, increasing numbers of cytoplasmic proteins have been observed in unexpected subcellular compartments, including the outer membrane, periplasm, and OMVs, in a variety of bacteria (12, 40, 75). We also identified a large number of cytoplasmic proteins in the OMV fraction. However, this unexpected location of cytoplasmic proteins did not appear to occur by the passive binding of the released cytoplasmic proteins to extracellular vesicle compartments as the result of cell death because the OMVs were isolated during the phase of exponential growth. Another explanation for this phenomenon may be related to a physiological role of OMVs, which have been found to help bacteria expel useless and harmful waste that has accumulated inside cells. As an envelope response to environmental stresses that lead to protease impairment and protein misfolding or denaturation, Gram-negative bacteria shed proteinaceous waste via OMVs (18). However, this speculation is not feasible, either, because more cytoplasmic proteins were found to be associated with the OMVs isolated under the nutrient-rich (LB) condition than with those isolated under the stressful nutrient-deficient (acidic MgM) condition (40% in the LB-derived OMVs versus 25% in the acidic MgM-derived OMVs; Fig. 2B).
Recently, many proteins associated with conserved cellular functions, such as metabolic processes and stress responses, were proven to perform additional biological activities in locations distinct from those they normally occupy (76, 77). These multitasking bacterial proteins, which are termed moonlighting proteins, include metabolic proteins/enzymes and molecular chaperones. A wide range of housekeeping proteins belonging to the glycolytic pathway, glyoxylate cycle, and protein-folding process were found to be located on the surface of pathogenic bacteria and to play a role in bacterial interaction with host cells by serving as adhesins and invasins even though they do not have the typical signal peptide sequences or membrane-anchoring domains (76, 78, 79). The moonlighting functions may not be conserved between homologous proteins (80), but we also found that S. Typhimurium OMVs contain 10 cytoplasmic proteins with homologues that behave as moonlighting proteins in other pathogenic bacteria. These proteins include six glycolysis proteins (GapA, GpmA, FbaB, TpiA, Pgk, and PykF), three superoxide dismutase proteins (SodA, SodB, and SodC), and one chaperone (FkpA). Their detailed functions have been reviewed by others (76, 79). The unexpected subcellular locations of DNA-binding proteins, such as HU and H-NS, in OMVs may also reflect their secondary roles in Salmonella virulence. Because most bacterial moonlighting proteins have been observed in Gram-positive pathogens, Salmonella cytoplasmic proteins that are unexpectedly found in OMVs may serve as good candidates for exploring moonlighting-protein-mediated virulence mechanisms in Gram-negative pathogens.
The comprehensive proteomic profiling of OMVs isolated under two different growth conditions demonstrated that Salmonella can manipulate the OMV cargo to cope with environmental stresses and also provided a range of OMV-associated proteins whose secretion was restricted to the phagosome-mimicking condition and are therefore likely required for S. Typhimurium virulence. This report may lay the groundwork for understanding the general physiological attributes of OMVs and the OMV-mediated virulence mechanisms in Salmonella.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Science, ICT and Future Planning (grant no. NRF-2012R1A1A3005712).
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01416-13.
REFERENCES
- 1. Chimalizeni Y, Kawaza K, Molyneux E. 2010. The epidemiology and management of non typhoidal salmonella infections. Adv. Exp. Med. Biol. 659:33–46. 10.1007/978-1-4419-0981-7_3 [DOI] [PubMed] [Google Scholar]
- 2. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thiennimitr P, Winter SE, Baumler AJ. 2012. Salmonella, the host and its microbiota. Curr. Opin. Microbiol. 15:108–114. 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galán JE. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86. 10.1146/annurev.cellbio.17.1.53 [DOI] [PubMed] [Google Scholar]
- 5. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66. 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- 6. McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124. 10.1016/j.mib.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knox KW, Vesk M, Work E. 1966. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 92:1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74:81–94. 10.1128/MMBR.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107:19002–19007. 10.1073/pnas.1008843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ellen AF, Zolghadr B, Driessen AM, Albers SV. 2010. Shaping the archaeal cell envelope. Archaea 2010:608243. 10.1155/2010/608243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deatherage BL, Cookson BT. 2012. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80:1948–1957. 10.1128/IAI.06014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, Kim KP, Gho YS. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153. 10.1002/pmic.200700196 [DOI] [PubMed] [Google Scholar]
- 13. Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11:3424–3429. 10.1002/pmic.201000212 [DOI] [PubMed] [Google Scholar]
- 14. Lindmark B, Rompikuntal PK, Vaitkevicius K, Song T, Mizunoe Y, Uhlin BE, Guerry P, Wai SN. 2009. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 9:220. 10.1186/1471-2180-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72:1395–1407. 10.1111/j.1365-2958.2009.06731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414–4420. 10.1128/AEM.66.10.4414-4420.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178:2767–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286:1269–1276. 10.1074/jbc.M110.185744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549. 10.1038/sj.emboj.7600471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharpe SW, Kuehn MJ, Mason KM. 2011. Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infect. Immun. 79:4361–4369. 10.1128/IAI.05332-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. 10.1371/journal.pone.0017027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. 10.1371/journal.ppat.1000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11:258. 10.1186/1471-2180-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, Chambers J, Arulanandam B, Haskins WE, Weitao T. 2012. Vesiculation from Pseudomonas aeruginosa under SOS. ScientificWorldJournal 2012:402919. 10.1100/2012/402919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geddes K, Worley M, Niemann G, Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 73:6260–6271. 10.1128/IAI.73.10.6260-6271.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. 2009. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One 4:e4809. 10.1371/journal.pone.0004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi J, Biek DP. 1995. A versatile low-copy-number cloning vector derived from plasmid F. Gene 164:55–58. 10.1016/0378-1119(95)00419-7 [DOI] [PubMed] [Google Scholar]
- 33. Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306. 10.1371/journal.ppat.1000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773. 10.1046/j.1365-2958.1999.01312.x [DOI] [PubMed] [Google Scholar]
- 35. van den Toorn HW, Munoz J, Mohammed S, Raijmakers R, Heck AJ, van Breukelen B. 2011. RockerBox: analysis and filtering of massive proteomics search results. J. Proteome Res. 10:1420–1424. 10.1021/pr1010185 [DOI] [PubMed] [Google Scholar]
- 36. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. 10.1186/1471-2105-6-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bendtsen JD, Kiemer L, Fausboll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. 10.1186/1471-2180-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown RN, Sanford JA, Park JH, Deatherage BL, Champion BL, Smith RD, Heffron F, Adkins JN. 2012. A comprehensive subcellular proteomic survey of Salmonella grown under phagosome-mimicking versus standard laboratory conditions. Int. J. Proteomics 2012:123076. 10.1155/2012/123076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron AD, Alston M, Stringer MF, Betts RP, Baranyi J, Peck MW, Hinton JC. 2012. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 194:686–701. 10.1128/JB.06112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Niemann GS, Brown RN, Gustin JK, Stufkens A, Shaikh-Kidwai AS, Li J, McDermott JE, Brewer HM, Schepmoes A, Smith RD, Adkins JN, Heffron F. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79:33–43. 10.1128/IAI.00771-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45. 10.1016/j.mib.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bowden SD, Ramachandran VK, Knudsen GM, Hinton JC, Thompson A. 2010. An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS One 5:e13871. 10.1371/journal.pone.0013871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pilonieta MC, Nagy TA, Jorgensen DR, Detweiler CS. 2012. A glycine betaine importer limits Salmonella stress resistance and tissue colonization by reducing trehalose production. Mol. Microbiol. 84:296–309. 10.1111/j.1365-2958.2012.08022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U. S. A. 102:17460–17465. 10.1073/pnas.0505401102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parsons DA, Heffron F. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73:4338–4345. 10.1128/IAI.73.7.4338-4345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lamarche MG, Dozois CM, Daigle F, Caza M, Curtiss R, III, Dubreuil JD, Harel J. 2005. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 73:4138–4145. 10.1128/IAI.73.7.4138-4145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Daigle F, Fairbrother JM, Harel J. 1995. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect. Immun. 63:4924–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elliott T. 1993. Transport of 5-aminolevulinic acid by the dipeptide permease in Salmonella typhimurium. J. Bacteriol. 175:325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manson MD, Blank V, Brade G, Higgins CF. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253–256. 10.1038/321253a0 [DOI] [PubMed] [Google Scholar]
- 52. Slack FJ, Mueller JP, Sonenshein AL. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J. Bacteriol. 175:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jones AL, Knoll KM, Rubens CE. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444–1455. 10.1046/j.1365-2958.2000.02099.x [DOI] [PubMed] [Google Scholar]
- 54. Sirko A, Zatyka M, Sadowy E, Hulanicka D. 1995. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 177:4134–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shao H, James D, Lamont RJ, Demuth DR. 2007. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J. Bacteriol. 189:5559–5565. 10.1128/JB.00387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. James D, Shao H, Lamont RJ, Demuth DR. 2006. The Actinobacillus actinomycetemcomitans ribose binding protein RbsB interacts with cognate and heterologous autoinducer 2 signals. Infect. Immun. 74:4021–4029. 10.1128/IAI.01741-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jelsbak L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE. 2012. Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One 7:e36149. 10.1371/journal.pone.0036149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Durand JM, Bjork GR. 2003. Putrescine or a combination of methionine and arginine restores virulence gene expression in a tRNA modification-deficient mutant of Shigella flexneri: a possible role in adaptation of virulence. Mol. Microbiol. 47:519–527. 10.1046/j.1365-2958.2003.03314.x [DOI] [PubMed] [Google Scholar]
- 59. Frossard SM, Khan AA, Warrick EC, Gately JM, Hanson AD, Oldham ML, Sanders DA, Csonka LN. 2012. Identification of a third osmoprotectant transport system, the osmU system, in Salmonella enterica. J. Bacteriol. 194:3861–3871. 10.1128/JB.00495-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ladant D, Ullmann A. 1999. Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 7:172–176. 10.1016/S0966-842X(99)01468-7 [DOI] [PubMed] [Google Scholar]
- 61. Mangan MW, Lucchini S, Ó Cróinín T, Fitzgerald S, Hinton JC, Dorman CJ. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology 157:1075–1087. 10.1099/mic.0.046359-0 [DOI] [PubMed] [Google Scholar]
- 62. Berger M, Farcas A, Geertz M, Zhelyazkova P, Brix K, Travers A, Muskhelishvili G. 2010. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 11:59–64. 10.1038/embor.2009.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bi H, Sun L, Fukamachi T, Saito H, Kobayashi H. 2009. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Curr. Microbiol. 58:443–448. 10.1007/s00284-008-9340-4 [DOI] [PubMed] [Google Scholar]
- 64. Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20–36. 10.1046/j.1365-2958.2001.02358.x [DOI] [PubMed] [Google Scholar]
- 65. Klose KE, Mekalanos JJ. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fields PI, Groisman EA, Heffron F. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243(Pt 1):1059–1062. 10.1126/science.2646710 [DOI] [PubMed] [Google Scholar]
- 67. Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054–5058. 10.1073/pnas.86.13.5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baca OG, Roman MJ, Glew RH, Christner RF, Buhler JE, Aragon AS. 1993. Acid phosphatase activity in Coxiella burnetii: a possible virulence factor. Infect. Immun. 61:4232–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mohapatra NP, Soni S, Reilly TJ, Liu J, Klose KE, Gunn JS. 2008. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect. Immun. 76:3690–3699. 10.1128/IAI.00262-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Remaley AT, Glew RH, Kuhns DB, Basford RE, Waggoner AS, Ernst LA, Pope M. 1985. Leishmania donovani: surface membrane acid phosphatase blocks neutrophil oxidative metabolite production. Exp. Parasitol. 60:331–341. 10.1016/0014-4894(85)90039-6 [DOI] [PubMed] [Google Scholar]
- 71. Buchmeier NA, Heffron F. 1989. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect. Immun. 57:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Conlan JW, North RJ. 1994. Neutrophils are essential for early anti-Listeria defense in the liver, but not in the spleen or peritoneal cavity, as revealed by a granulocyte-depleting monoclonal antibody. J. Exp. Med. 179:259–268. 10.1084/jem.179.1.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu G, Xia XP, Gong SL, Zhao Y. 2006. The macrophage heterogeneity: difference between mouse peritoneal exudate and splenic F4/80+ macrophages. J. Cell. Physiol. 209:341–352. 10.1002/jcp.20732 [DOI] [PubMed] [Google Scholar]
- 74. Yoon H, Gros P, Heffron F. 2011. Quantitative PCR-based competitive index for high-throughput screening of Salmonella virulence factors. Infect. Immun. 79:360–368. 10.1128/IAI.00873-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goulter-Thorsen RM, Gentle IR, Gobius KS, Dykes GA. 2011. The DNA protection during starvation protein (Dps) influences attachment of Escherichia coli to abiotic surfaces. Foodborne Pathog. Dis. 8:939–941. 10.1089/fpd.2011.0837 [DOI] [PubMed] [Google Scholar]
- 76. Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79:3476–3491. 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jeffery CJ. 2009. Moonlighting proteins—an update. Mol. Biosyst. 5:345–350. 10.1039/b900658n [DOI] [PubMed] [Google Scholar]
- 78. Pancholi V, Chhatwal GS. 2003. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 293:391–401. 10.1078/1438-4221-00283 [DOI] [PubMed] [Google Scholar]
- 79. Wang G, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, Chen H, Zhang H, Chen W. 2013. The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 16:15–22 [PubMed] [Google Scholar]
- 80. Gancedo C, Flores CL. 2008. Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72:197–210, table of contents. 10.1128/MMBR.00036-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.