Abstract
High-throughput RNA sequencing technology has found the 5′ untranslated region of sarA to contain two putative small RNAs (sRNAs), designated teg49 and teg48. Northern blot analysis disclosed that teg49 and teg48 were detectable within the P3-P1 and P1 sarA promoter regions, respectively. Focusing on teg49, we found that this sRNA, consisting of 196 nucleotides, is transcribed in the same direction as the sarA P3 transcript. The expression of both P3 and teg49 transcripts is dependent on sigB and cshA, which encodes a DEAD box RNA helicase. Within the sRNA teg49, there are two putative hairpin-loop structures, HP1 and HP2. Transversion mutation of the HP1 loop produced a smaller amount of sarA P3 and P2 transcripts and SarA protein than the corresponding HP1 stem and the HP2 stem and loop mutations, leading to lower RNAII transcription and derepression of aur transcription. The HP1 loop mutant also exhibited less biofilm formation than the parental and complemented strains. Complementation with shuttle plasmid pEPSA5 carrying teg49 was able to reestablish sarA P3 and P2 transcription and augment RNAII expression in the HP1 loop mutant. We thus conclude that teg49, embedded within the extended promoter regions of sarA, is modulated by sigB and cshA and plays an important trans-acting role in modulating the transcription and ensuing expression of sarA.
INTRODUCTION
Staphylococcus aureus, an opportunistic pathogen, is a major cause of life-threatening infections in humans. The pathogenicity of S. aureus is shaped by a variety of extracellular and cell wall-associated proteins that are expressed in a growth phase-dependent fashion (1, 2). The expression of these virulence factors is controlled by a complex network of global regulatory elements, including agr and the staphylococcal accessory regulator (SarA) of S. aureus. The sarA locus encodes a DNA binding protein, SarA, that functions as a repressor or an activator by binding to conserved AT-rich DNA motifs (ATTTTAT) in the promoter regions of target genes in agr-dependent and agr-independent manners (3, 4).
The sarA locus in S. aureus has an open reading frame (ORF) of 372 bp with a predicted molecular mass of 14,718 Da and a deduced pI of 8.52 (5). Notably, the 5′ untranslated region (UTR) of the sarA locus spans 860 bp with three promoters (6), encompassing three distinct but overlapping transcripts of 0.58 (P1), 0.84 (P3), and 1.15 (P2) kb, all encoding the SarA protein. The expression of these three transcripts is known to be growth phase dependent, with P1 and P2 expressed during the exponential phase and P3 expressed postexponentially (6), and is thought to be required for the complete restoration of SarA function (6). Because of the unusual length of the sarA promoter region, it was hypothesized that the 5′ UTR may possess elements (e.g., small RNAs [sRNAs]) that are involved in optimal expression of the sarA gene.
S. aureus expresses a large number of sRNAs, many of which do not possess well-described functions (7, 8, 9, 10, 11). Recent studies have demonstrated that sRNAs of S. aureus are known to play a pivotal role in a variety of regulatory processes by base pairing with target mRNAs and by modulating protein activity (12, 13, 14, 15). For instance, sRNA RsaE pairs with the Shine-Dalgarno sequence of mRNAs of opp-3B/opp-3A (amino acid and peptide transporter) and sucC (succinyl-coenzyme A synthase) and prevents the formation of a ribosomal initiation complex. It has been suggested that RsaE may coordinate the downregulation of central metabolism when carbon sources become limited (12). ArtR, a toxin-regulating sRNA, is involved in virulence regulation by activating alpha-toxin expression. ArtR also promotes degradation of sarT mRNA by RNase III and arrests the translation of SarT by direct binding to the 5′ UTR of the sarT mRNA (13). SprD, a sRNA expressed from S. aureus pathogenicity islands (PIs), represses translation initiation by base pairing with sbi mRNA, leading to impaired adaptive and innate host immune responses (14). Morrison et al. (15) showed that SSR42, which is expressed predominantly during stationary phase, regulates the expression of several virulence factors, including protein A, capsule, α-hemolysin, and Panton-Valentine leukocidin toxin. SSR42 is also involved in resistance to human polymorphonuclear leukocyte killing and pathogenesis in a murine model of S. aureus infection (15).
Here we report the presence of an sRNA designated teg49 in the sarA promoter region located within the P3-P1 promoter region. Analyses by primer extension and 3′ rapid amplification of cDNA ends (RACE) revealed that teg49 is 196 nucleotides (nt) in length and located at nt 667081 to 666886 of the reference S. aureus N315 genome. The teg49 transcript and the sarA P3 transcript were diminished in cells lacking sigB or the DEAD box RNA helicase gene cshA but restored in the complemented mutants. There are two putative hairpin structures, designated HP1 and HP2, within the teg49 sRNA. Replacement of the 7 nt in the HP1 loop of teg49 in the chromosome led to truncated transcripts from the P2 and/or P3 promoters, resulting in reduced sarA expression, agr expression, and biofilm formation, analogous to what has been found in a sarA mutant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The bacterial strains used in this study are listed in Table 1. S. aureus strain Newman and its derivatives ALC6094, SH1000, and RN4220 were grown at 37°C in tryptic soy broth (TSB) or tryptic soy agar. Escherichia coli strains were grown in LB broth or LB agar. The following antibiotics were used when appropriate: erythromycin (5 μg/ml), chloramphenicol (10 μg/ml), tetracycline (2.5 μg/ml), and kanamycin (50 μg/ml) for S. aureus strains and ampicillin (100 μg/ml) and kanamycin (100 μg/ml) for E. coli strains.
TABLE 1.
Strains used in this study
| S. aureus strain | Genotype and/or characteristic | Source or reference |
|---|---|---|
| RN4220 | Mutagenized strain 8325-4 that accepts foreign DNA | 17 |
| SH1000 | Functional rsbU derivative of 8325-4 rsbU+ | 19 |
| Newman | Isolated from a human infection in 1952 | 41 |
| ALC6094 | Strain Newman transduced with IPTGb-inducible T7 polymerase; Tetr | 42 |
| ALC7201 | ALC6094 ΔcshA::Kanr | This study |
| ALC7252 | ALC7201 ΔcshA::cshA | This study |
| ALC7286 | SH1000 sarA deletion | This study |
| ALC7288 | SH1000 HP1 stem mutation of teg49 | This study |
| ALC7289 | SH1000 HP1 loop mutation of teg49 (ATTGCGC→CGCTATA)a | This study |
| ALC7290 | SH1000 HP2 loop mutation of teg49 (GTCGATT→TATCGGC)a | This study |
| ALC7291 | SH1000 HP2 stem mutation of teg49 | This study |
| ALC7869 | Tn551::sigB in ALC6094 | This study |
| ALC7870 | ALC7869 containing pSK236::sigB | This study |
| ALC7909 | ALC7289 containing empty vector pEPSA5 | This study |
| ALC7911 | ALC7289 containing pEPSA5::teg49 | This study |
Nucleotides replaced in chromosomal DNA.
IPTG, isopropyl-β-d-thiogalactopyranoside.
DNA manipulations and transformation.
Standard procedures for DNA manipulations were used for cloning (16). Restriction endonucleases and DNA-modifying enzymes were purchased from New England BioLabs. E. coli and S. aureus plasmids were isolated with the QIAprep Spin Miniprep kit (Qiagen). Transformations of E. coli and S. aureus cells were carried out with a MicroPulser (Bio-Rad). Recombinant plasmids obtained from E. coli were first transformed into S. aureus RN4220 for proper DNA methylation to reduce restriction barriers (17). The plasmids purified from RN4220 were then electroporated into S. aureus Newman derivative ALC6094 (18) or SH1000 (19).
Construction of mutants and complementation in S. aureus.
Allelic exchange was carried out with the temperature-sensitive and Ermr pMAD shuttle vector (20). The cshA mutant of strain ALC6094, a strain Newman derivative that contains a T7 polymerase gene at the geh site (18), was created with a kanamycin resistance cassette cloned into pMAD. Briefly, PCR was used to amplify a 1.3-kb fragment comprising a 0.74-kb fragment upstream and a 0.56-kb fragment downstream of cshA with genomic DNA as the template. The PCR fragment was cloned into pMAD, resulting in pMAD-ΔcshA. A SmaI-digested 0.9-kb fragment containing the kanamycin resistance cassette was inserted into pMAD-ΔcshA to create pMAD-ΔcshA::Kanr. The recombinant shuttle vector was transformed into S. aureus ALC6094 (18). Transversion mutations of HP1 and HP2 loops and HP1 and HP2 stems in teg49 were introduced by PCR with primers with altered nucleotides, cloned into pMAD, and transformed into S. aureus SH1000. Allelic exchanges were performed as described previously (20). Selected mutants were subsequently complemented by reintroducing pMAD-cshA and pMAD-teg49 into the chromosome by homologous recombination as described previously (20). All of the mutant strains created were confirmed by PCR and sequencing.
Complementation of the HP1 loop mutant was also performed in trans. In this instance, we cloned teg49 immediately downstream of the xylose-inducible promoter in shuttle plasmid pEPSA5; this was followed by transformation into RN4220 and then into the HP1 loop mutant. Pilot Northern analysis indicated that teg49 was expressed in the HP1 loop mutant with recombinant plasmid pEPSA5::teg49 in the presence of 1% xylose. The pEPSA5 vector in the HP1 loop mutant served as the negative control.
RNA isolation, Northern blot analysis, and primer extension.
Isolation of RNA and Northern blot hybridization were performed as previously described (18). Briefly, cells grown in TSB at 37°C were harvested at optical densities at 650 nm (OD650s) of ∼0.2, ∼0.7, ∼1.1, and ∼1.7 with 18-mm borosilicate glass tubes in a Spectronic 20 (Spectronic Instrument), representing the early, mid-log, late log, and stationary phases, respectively. Pellets were resuspended in 1 ml TRIzol (Invitrogen) with 0.1-mm glass/zirconia beads, and RNA was extracted as described previously (18). Total RNA (10 μg) of each sample was electrophoresed through a 1.5% agarose–0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA, pH 7.0), transferred to a Hybond-N+ membrane (Amersham), hybridized with gel-purified PCR fragments radiolabeled with [α-32P]dCTP by the random-priming method (Ready-To-Go Labeling kit; GE) under high-stringency conditions (21), washed, and autoradiographed. Primer extension experiments were performed with Moloney murine leukemia virus reverse transcriptase according to the manufacturer's instructions (Ambion) and 5′ UTR-specific sarA oligonucleotide primers (Table 2).
TABLE 2.
Primers used in primer extension and 3′ RACE-PCR
| Primer | Sequence |
|---|---|
| PS-1 | CATTTTTAGTGATAAAATTTTG |
| PS-2 | GGTAAATTATAAAAAATGCTG |
| PS-3 | GTTTTATAAACACTTTTTTG |
| PS-4 | GCAAAATTATGACTAACATATC |
| PA-11 | CTAAAGAGATTCTTTGTTATAGC |
| PA-12 | CAGCATTTTTTATAATTTACC |
| PA-13 | CAAAAAAGTGTTTATAAAAC |
| PA-14 | GATATGTTAGTCATAATTTTGC |
Western blotting.
Whole-cell extracts of S. aureus wild-type SH1000 or sarA mutants containing separate HP1 and HP2 stem and loop mutations grown to an OD650 of ∼0.7 were prepared. The concentrations of total proteins from clear lysates were determined with the Pierce BCA Protein Assay kit (Thermo Scientific, IL) with bovine serum albumin as the standard. Western blotting and detection were performed as described previously (22).
Biofilm formation.
Quantification of biofilm formation on abiotic surfaces was done as described elsewhere (23). Briefly, S. aureus grown overnight in TSB supplemented with 0.25% glucose (TSB-glucose) was diluted 1:40 in TSB-glucose. This cell suspension was used to inoculate sterile 96-well polystyrene microtiter plates (Iwaki Inc.) in triplicate. After 18 h of incubation at 37°C, wells were gently washed three times with 200 μl of sterile phosphate-buffered saline, air dried in an inverted position, and stained with 0.1% safranin for 30 s. Wells were rinsed again and solubilized with ethanol, and the absorbance at 550 nm was determined (FL800; BioTek Instruments). Each assay was repeated in five separate experiments. Colony morphology was studied on Congo red agar as previously described (24).
RNA-Seq.
teg48 and teg49 were previously discovered and reanalyzed in the whole transcriptome of S. aureus N315 (10). Double-stranded cDNAs were synthesized with random primers from DNase-treated RNA. cDNAs were fragmented by nebulization, ends were repaired, and fragments were ligated with Illumina genomic adapters. Size selection on agarose gel allowed the selection of inserts of approximately 30 to 150 bp that were used to construct the library by PCR amplification (see reference 10 for the detailed protocol). An sRNA orientation run was performed with total RNA purified with the MirVana isolation and MicrobExpress kits (Ambion) after 4 h of growth in rich medium. RNAs were prepared with a dir-mRNA-SEQ protocol. After end repair, RNAs were ligated with single-stranded sRNA adapters. After cDNA synthesis and PCR amplification for 15 cycles, the library was size selected on agarose gel to select inserts of 15 to 100 bp. Sequencing was performed with an Illumina GAII for 36 cycles. The reads obtained were mapped onto the annotated sequence of strain N315 (NC_002745) and analyzed with the Artemis genome viewer (25).
3′ RACE assay.
S. aureus strain N315 was grown for 4 h in Mueller-Hinton broth from a 0.1 McFarland suspension obtained from a diluted overnight culture. Total RNA from a cell suspension treated with lysostaphin (50-μg/ml final concentration) was purified with the RNeasy kit (Qiagen). RNA quality and quantity were assessed with the Bioanalyzer (Agilent) and NanoDrop ND-8000.
As the region upstream of sarA contains two RNA species, the transcripts were separated and purified on a 10% acrylamide gel. Two fractions with sizes of <400 and >400 nt were purified. RNA fractions were polyadenylated with the MessageAmp Bacteria-Prokaryotic RNA amplification kit (Ambion). Poly(A) RNA fractions were immediately used for nonspecific synthesis of cDNA with the SMARTer RACE cDNA amplification kit (Clontech). For 3′ RACE-PCR, cDNA synthesis was performed with 3′-CDS primer A (1 μM final concentration) for 90 min at 42°C, followed by 10 min at 70°C. cDNAs were diluted in 20 μl Tricine-EDTA buffer before proceeding to specific 3′ RACE-PCR with Advantage 2 Polymerase Mix (Clontech) in accordance with the manufacturer's instructions in 1× Universal Primer Mix A and teg49 GSP2 primer at a 0.2 mM final concentration. Samples were amplified by 30 cycles of 30 s at 94°C, 30 s at 62°C, and 3 min at 72°C. PCR products were purified by QIAquick columns (Qiagen) and quantified. Finally, 3′ RACE-PCR products were sequenced with the BigDye Terminator Cycle Sequencing v.3.1 kit (Life Technologies) with a 3130 XL device (Applied Biosystems). Sequences were aligned with the N315 annotated genome sequence (RefSeq accession no. NC_002745) with Artemis software (25).
RESULTS
Identification of sRNA within the sarA locus by RNA-Seq.
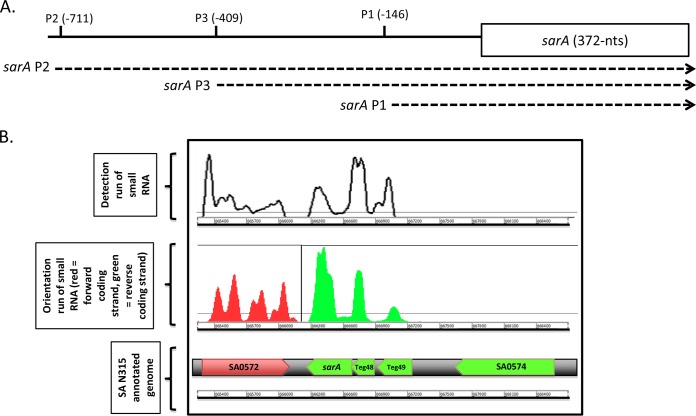
Beaume et al. (10) have previously described the use of RNA-Seq technology to obtain a representative map of the whole transcriptome of S. aureus strain N315. They identified 160 sRNA molecules in regions considered to be noncoding or intergenic. Some of the sRNAs are localized in biologically or clinically relevant regions, between key metabolic or virulence genes or within PIs, in strain N315 (10). Among these regions of interest, it was noticed that the sarA region contains two putative sRNAs. These data were reanalyzed, and we designated these sRNAs teg49 and teg48 (Fig. 1). The teg49 sRNA is located between the P3 and P1 promoters, while teg48 resides between the P1 promoter and the sarA translation start site (Fig. 2A), as confirmed by reanalysis by RNA-Seq and Northern blot assay.
FIG 1.
RNA-Seq analysis of the sarA locus. (A) Architecture of the sarA locus. Three promoters located between −710 and −146 upstream of the sarA translation start compose the topology of the sarA locus. (B) Schematic representation of the sarA region subjected to a transcriptomic study by RNA-Seq. An RNA-Seq detection run shows different transcript signals upstream of sarA (top). The orientation run (middle) shows the similar transcription orientation of the two smaller transcripts within the sarA locus and the transcription, in the opposite direction of sarA, of flanking gene SA0572, whereas the other flanking gene, SA0574, is not expressed under the conditions studied. The annotated genome is depicted at the bottom.
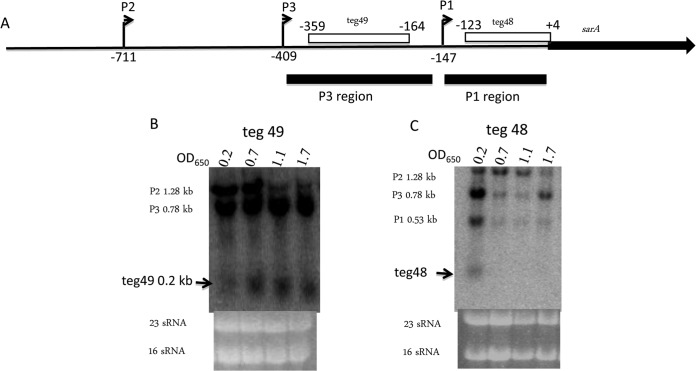
FIG 2.
Two sRNAs, teg49 and teg48, are detectable in the 5′ UTR of the sarA gene. (A) Map of 5′ UTR and sarA ORF. Two sRNAs, teg49 and teg48, are indicated in the P3-P1 and P1 promoter regions, respectively. The transcription start sites of P2, P3, and P1 are marked at −711, −409, and −149, with +1 corresponding to the sarA translational start site, as indicated by bent arrows. The P3 and P1 regions (black bars) were used as probes for Northern blot assays. The locations of teg49 and teg48 in the 5′ UTR of sarA are indicated at −356 to −164 and −123 to +4 (empty bars), respectively, with +1 corresponding to the sarA translational start site. (B, C) Northern blot assay detection of sRNAs teg49 (B) and teg48 (C). Total RNA (10 μg) from S. aureus Newman derivative ALC6094 in various growth phases (see Materials and Methods) were separated in a 1.5% agarose gel, transferred to an H+-bond membrane, and probed as described in Materials and Methods.
Northern blot analysis confirms the expression of two sRNAs, teg49 and teg48, within the 5′ UTR of sarA.
To verify the existence of both teg49 and teg48, we performed Northern blot assays with radiolabeled P3 and P1 fragments (Fig. 2A). Total cellular RNA of strain ALC6094, a derivative of strain Newman (Table 1), was extracted at various time points after inoculation. A probe from the sarA P3 region (−409 to −147, where +1 is the sarA translational start site) hybridized with three distinct transcripts of 1.15, 0.8, and 0.2 kb. The 1.15- and 0.8-kb transcripts correspond to the sarA P2 and P3 transcripts (6), while the ∼0.2-kb transcript is a sRNA corresponding to teg49 (Fig. 2B). Likewise, teg48 was also visualized with a P1 probe (−147 to −1) in Northern blot assays. More specifically, a labeled P1 probe (−147 to −1) yielded four bands, corresponding to sarA P2, P3, and P1 transcripts and also teg48 at ∼200 nt (Fig. 2C).
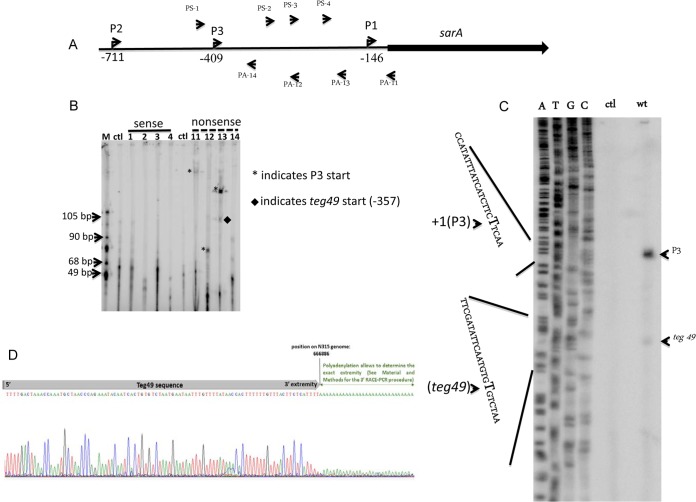
Mapping the 196-nt teg49 sRNA within the P3-P1 promoter region.
We initially focused on teg49 within the P3-P1 sarA promoter region. To map the start site and direction of transcription of teg49, we designed DNA primers from sense and antisense strands in the P3 promoter region (spanning −406 to −147 of the translational start site of sarA) for primer extension (Table 2) (Fig. 3A). Primer extension analysis with primer PA-13, representing the antisense strand, produced two clear transcripts, one larger transcript for P3 and a smaller transcript corresponding to the start site of teg49, while primers PS-1, -2, -3, and 4 of the sense strain (Table 2) did not yield any transcript (Fig. 3B). Primers PA-11 and PA-12 of the antisense strand both produced single transcripts, with one larger and one smaller than the corresponding sarA P3 transcriptional start site as identified by PA-13 (6). These results indicate that the sRNA teg49 exists and is transcribed in the same direction as the sarA P3 transcript.
FIG 3.
Primer extension studies of two transcripts in the sarA P3 promoter region. (A) Map of sarA and the 5′ UTR. The directions of transcription of P2, P3, and P1 are indicated by arrowheads. The oligomers used for primer extension are shown above and below the sarA genetic map. The directions of the oligomers are indicated by arrowheads. The oligomers were customized from the sense strand (PS-1, PS-2, PS-3, and PS-4) and the antisense strand (PA-11, PA-12, PA-13, and PA-14) (Table 2). (B) Primer extension analysis of teg49 containing putative primer extension products from the sense and nonsense strands, including a control reaction mixture (lane ctl) that contained only total RNA prepared from S. aureus ALC6094. The transcriptional start sites of P3 (at −409 from the translational start site of sarA) and teg49 (at −357 from the translational start site of sarA) are indicated by asterisks and a diamond, respectively. Lane M contained molecular size markers. (C) Primer extension with oligomer PA-13 and total RNA isolated from S. aureus ALC6094 grown to exponential phase (OD650 of 0.7) in TSB medium as the template. The transcriptional start sites of P3 and teg49 are indicated on the right. A part of the sequence of the P3 promoter deduced from the sequencing reaction is shown on the left; P3 and teg49 starting sites from T's (complementary nucleotides of A's) are indicated by bold letters and arrowheads. wt, wild type. (D) 3′ RACE DNA sequencing reactions indicating the 3′ end of teg49.
To determine the exact size of teg49, we mapped the 5′ end of the sRNA by primer extension with a sequencing ladder and the 3′ end by RACE-PCR assay. Using primer PA-13, which produced an additional band on the gel (arrow in Fig. 3C), we mapped the 5′ end of teg49 to nt 667081 of the S. aureus N315 genome by primer extension (Fig. 3C). We also mapped the 3′ end of teg49 to nt 666886 in the S. aureus N315 genome with the 3′ RACE-PCR assay (Fig. 3D). Thus, the teg49 sRNA, at 196 nt in length, is located within the sarA P3-P1 promoter region.
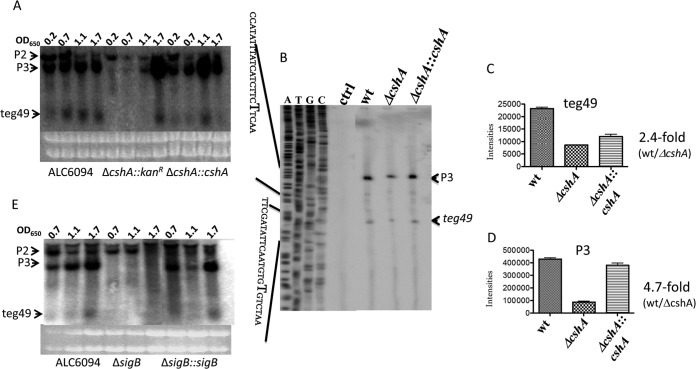
Expression of both P3 and teg49 transcripts is cshA and sigB dependent.
In recent studies, it has been reported that CshA, a DEAD box RNA helicase (26), can alter the stability of the agr mRNA in S. aureus. To assess if CshA can regulate the expression of sRNA teg49 and its associated sarA transcripts, we conducted Northern blot assays of parental strain ALC6094 and its isogenic cshA mutant and the complemented mutant with a teg49 probe. As shown in Fig. 4A, neither a sarA P3 transcript nor teg49 was readily detectable in the cshA mutant during the early, mid-log, and late log phases, in contrast to parental strain ALC6094 and the complemented mutant. In addition, the corresponding P2 transcript level was also lower in the cshA mutant than in the parental and complemented strains. Interestingly, both teg49 and sarA P3 transcripts, as detected by Northern blot assays, reemerged in cells grown to the stationary phase (OD650 of 1.7) (Fig. 4A), suggesting that factors other than CshA may be important in the generation of sRNA teg49 and the sarA P3 transcript during the stationary phase.
FIG 4.
Northern blot analysis of sRNA teg49 in S. aureus strains in various phases of growth. (A, E) Northern blot assay detection of sRNA teg49. Total RNAs (10 μg) from ΔcshA (A) and ΔsigB (E) mutants of strain Newman derivative ALC6094 grown in TSB medium were separated in a 1.5% agarose gel and then transferred to an H+-bond membrane for probing. sRNA teg49 was detected with a teg49 probe. (B) Primer extension analysis of sRNA teg49 in a cshA mutant of ALC6094. Primer extension assays were performed with RNAs purified from the parental (ALC6094), ΔcshA mutant, and complemented strains grown to exponential phase (OD650 of ∼0.7) in TSB medium with oligonucleotide PA-13 (Table 2). ctrl, control; wt, wild type. (C, D) Quantification of sarA P3 and teg49 transcripts in the cshA mutant, parental, and complemented strains from panel B. Transcripts sarA P3 (C) and sRNA teg49 (D) were quantified with ImageJ (NIH). These experiments were repeated at least three times, and one set of typical experiments is shown.
To confirm the Northern blot assay results for exponential-phase cells, we conducted primer extension studies of the cshA mutant, isogenic parental, and complemented strains at an OD650 of 0.7 with primer PA-13. We found weaker primer extension signals for both sarA P3 and teg49 in the cshA mutant than in the parental and complemented strains (Fig. 4B). More specifically, the cshA mutant exhibited notably lower levels of primer extension signals for teg49 (∼2.4-fold lower, as determined by densitometry) (Fig. 4C) and the sarA P3 transcript (∼4.7-fold less) (Fig. 4D) than the isogenic parent.
In previous studies, we and others have shown that sigB regulates sarA P3 transcript expression in S. aureus (27, 28). As SigB is involved in the postexponential stress response and teg49 somehow reemerges in a cshA mutant during the stationary phase, we evaluated whether sigB is involved in the expression of sRNA teg49. As expected, the sarA P3 transcript level was significantly lower in the sigB mutant than in parental strain ALC6094 and also the complemented mutant (Fig. 4E). Remarkably, sRNA teg49 was not observed in the sigB mutant, even during the stationary phase (OD650 of 1.7, Fig. 4E), suggesting that teg49 expression is dependent on sigB. Thus, the sigB mutant of ALC6094 cannot generate either a sarA P3 transcript or sRNA teg49 but the two transcripts reemerged in the complemented sigB mutant. In contrast to the cshA mutant, the sigB mutant did not yield any sarA P3 transcript or sRNA teg49 in any of the three growth phases, including the stationary phase (Fig. 4E).
sRNA teg49 has two hairpin structures and sequences conserved among S. aureus strains.
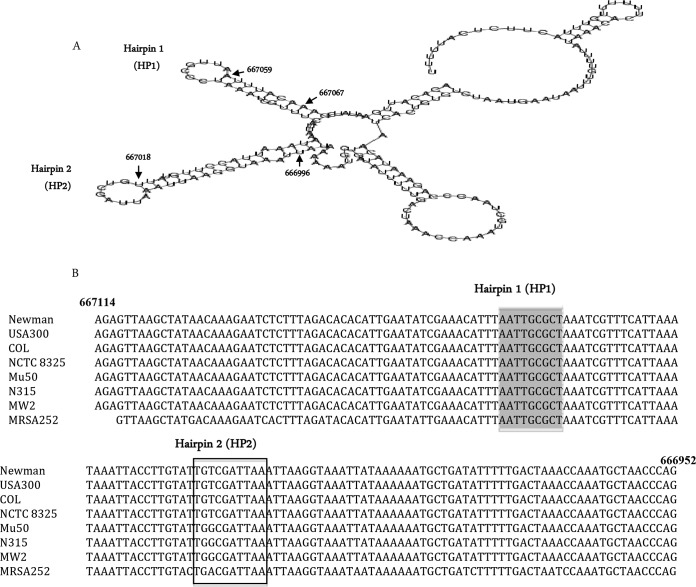
The RNA structure of teg49 was predicted by the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (Fig. 5A). The sRNA teg49 appears to have two stem-loop structures, each with a long stem and a short loop, designated HP1 and HP2. HP1 and HP2 are located at nt 667059 to 667067 and 667018 to 667027 of the S. aureus N315 genome (Fig. 5A), respectively. The sequences of teg49 are also highly conserved among methicillin-resistant and -susceptible S. aureus strains (Fig. 5B).
FIG 5.
Predicted secondary structure and comparison of the teg49 nucleotide sequences of S. aureus strains. (A) Predicted secondary structure of sRNA teg49. The structure of teg49 was predicted by the RNAfold web server (see Results). Two hairpin-loop structures, HP1 and HP2, within teg49 were found to be located at nt 667059 and 667018 of the S. aureus N315 genome, respectively. (B) Sequences upstream of the sarA coding region bearing putative sRNA teg49 are conserved. A partial alignment of the teg49 sequences of S. aureus strains, Newman, USA300, COL, NCTC 8325, Mu50, N315, MW2, and MRSA252 is shown. The HP1 and HP2 hairpin-loop sequences are shaded in gray and boxed, respectively.
HP1 loop mutation of teg49 has a prominent effect on sarA P3 expression.
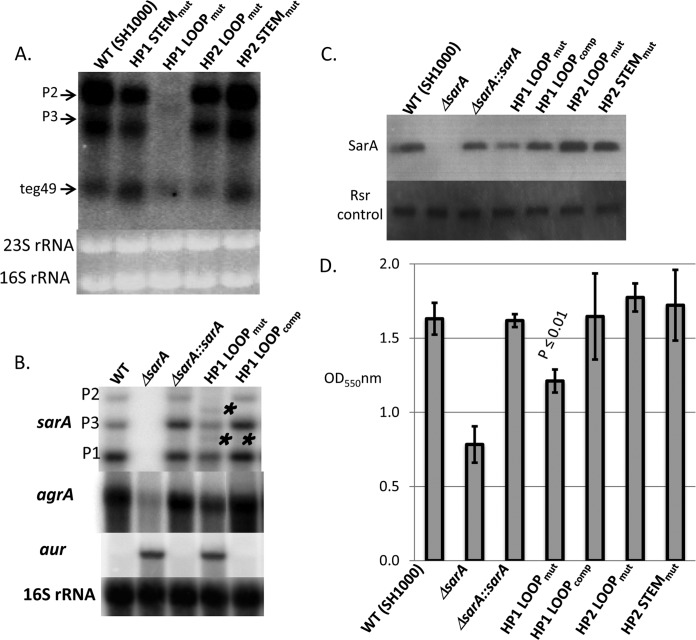
Previous experiments have shown that hairpins in noncoding sRNAs are important sites for posttranscriptional gene regulation (12, 13, 14). Given the conservation of teg49 among various S. aureus strains, we thus wanted to elucidate the contribution of the loop and stem sequences of HP1 and HP2 on sarA expression. In this instance, we elected to work with strain SH1000, which is a version of strain 8325-4 with rsbU restored that is more genetically amenable and has been extensively studied. To ensure that teg49 is also present within the sarA promoter region of strain SH1000, we conducted a Northern blot assay with a teg49 probe, which showed that teg49, along with P2 and P3 sarA transcripts, was easily detectable in this strain (Fig. 6A, first lane). We subsequently created chromosomal transversion mutations of teg49 in SH1000, yielding an HP1 stem mutant (ALC7288), an HP1 loop mutant (ALC7289), an HP2 loop mutant (ALC7290), and an HP2 stem mutant (ALC7291) (Table 1). The expression of teg49 with a teg49 probe appeared to be reduced in two loop mutants, ALC7289 (HP1 loop mutant) and ALC7290 (HP2 loop mutant), but not in two stem mutants (Fig. 6A). Surprisingly, in the HP1 loop mutant, where there is a 7-base transversion mutation in the chromosome, the P3 and P2 transcripts are almost undetectable or are significantly reduced (Fig. 6A). Thus, the HP1 loop in teg49 seems to be critical for sarA P3 and teg49 transcription.
FIG 6.
Analysis of HP1 loop mutant with respect to sarA P3 and teg49 transcription. (A) Northern blot assay (from a 1.5% agarose gel optimized for sRNA detection) of wild-type (WT) SH1000, HP1 stem and loop mutants (mut), and HP2 stem and loop mutants (from late-log-phase cells at an OD650 of 1.1) probed with a labeled teg49 probe. As expected, there are three transcripts from wild-type SH1000, sarA P2 and P3 transcripts, and teg49. The sarA P1 transcript was not expected to be detected by the teg49 probe. (B) Northern blot assays (from a 0.7% agarose gel to detect transcript larger than 500 bases) of isogenic sarA strains along with an HP1 loop mutant and a complemented (comp) HP1 loop mutant. The probes are the sarA coding region for sarA, agrA for RNAII, and aur for the aureolysin gene. 16S rRNAs were used as loading controls. We included wild-type SH1000, a sarA deletion mutant, and complemented mutants for sarA transcripts (P1, P3, and P2 for 0.5, 0.8, and 1.1 kb, respectively) as additional controls. The single and double asterisks represent altered sarA transcripts in the HP1 loop mutant. This defect was restored in the complemented HP1 loop mutant (HP1 LOOPcomp). (C) Western blot assay for SarA in isogenic sarA mutant strains with an HP1 loop mutant, a complemented mutant, and HP2 stem and loop mutants. The blot was probed with an anti-SarA monoclonal antibody at 1:1,000, followed by conjugate. Rsr, a cellular protein that was constant during growth, was used as a loading control and was detected by a mouse anti-Rsr antibody. (D) Biofilm formation by assorted sarA strains in microtiter wells containing TSB and glucose. Cells were inoculated into wells and grown overnight. Biofilms were stained with crystal violet and solubilized with ethanol, and OD550s were read. sarA mutant and complemented mutant strains were used as negative and positive controls, respectively.
To analyze the effect of the HP1 loop mutation of teg49 on sarA P2 and P3 transcription more clearly, we ran a lower-percentage agarose gel (0.7% in Fig. 6B versus 1.5% in Fig. 6A) for Northern blot assays with a sarA probe (sarA coding region) (Fig. 6B). As expected, compared to the parental and complemented strains, the HP1 loop mutant exhibited lower levels of sarA P1, P3, and P2 transcript expression. Remarkably, there were two additional bands, one located between the P2 and P3 transcripts (one asterisk in Fig. 6B) and the other located between the P3 and P1 transcripts (two asterisks in Fig. 6B) in the HP1 loop mutant. The origin of these two transcripts is not clear. However, it is plausible that the larger transcript is a truncated form or processed from the P2 transcript while the smaller transcript can be derived from either the P2 or the P3 transcript, indicating that the 7-base HP1 loop sequence is critical to processing of the sarA transcripts. More importantly, processing of the sarA transcripts is associated with reduced expression of teg49 (Fig. 6A). Notably, complementation of the 7-base mutation in the HP1 loop mutant in the chromosome restored the transcription of all three sarA transcripts in the mutant (Fig. 6B).
The effect of HP1 loop mutation on sarA transcription also includes lower SarA protein expression than in the parental and complemented mutant strains in whole-cell lysates of Western blot assays probed with an anti-SarA monoclonal antibody, but the level of expression was still higher than that of the sarA deletion mutant (Fig. 6C). Given that sarA is a positive regulator of agr expression (29), we conducted Northern blot assays with an agrA probe, which showed that RNAII (containing agrA) expression was lower in the HP1 loop mutant than in the parental and complemented mutant strains (Fig. 6B). However, the reduction was less than that of a true sarA deletion mutant. We next examined the aureolysin gene aur, which is negatively regulated by sarA (30). As expected, the expression of aur was upregulated in the HP1 loop mutant, similar to that in the sarA deletion mutant (Fig. 6B). Previous studies have described a critical role for SarA in biofilm formation by S. aureus (31, 32). As anticipated for a strain with reduced SarA expression, the HP1 loop mutant also exhibited less biofilm formation than the wild type and the complemented mutant (P ≤ 0.01 versus the parent by the Student t test), but the reduction in biofilm formation was less than that of the sarA deletion mutant (Fig. 6D).
Complementation of the HP1 loop mutant in trans with pEPSA5 carrying teg49.
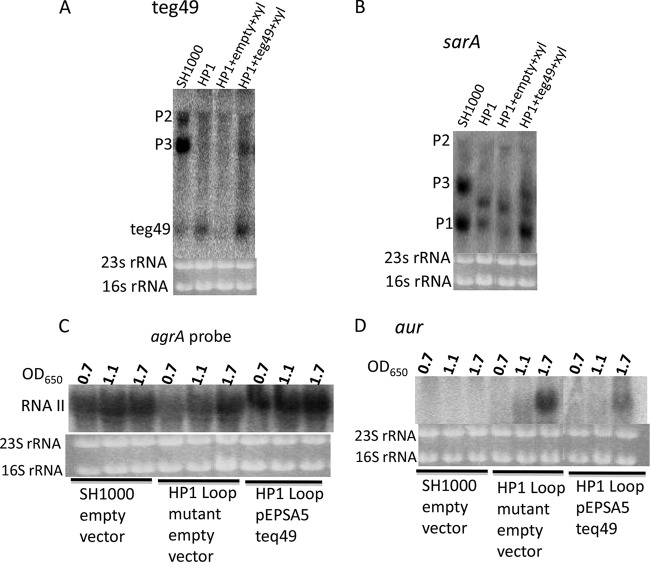
To complement the HP1 loop mutation in trans, we first cloned teg49 into pEPSA5, a shuttle plasmid containing a xylose-inducible promoter in E. coli; this was followed by transformation into the HP1 loop mutant. We were able to detect teg49 expression in the HP1 loop mutant upon induction with 1% xylose versus the empty-vector control (Fig. 7A). Using cells at an OD650 of 1.1 in the presence of 1% xylose, we discovered by Northern blot assays that the sarA P3 and P1 transcript levels were significantly enhanced in the HP1 loop mutant upon xylose induction while cells without xylose induction or those with the vector alone did not exhibit similar complementation. Interestingly, the cleaved transcript sizing between the P3 and P1 transcripts in the HP1 loop mutant persisted in the HP1 loop mutant carrying pEPSA5::teg49, consistent with the combination of defective and normal HP1 loops of teg49 in this strain. Importantly, the RNAII transcript, as detected by the agrA probe, was intensified in the HP1 loop mutant carrying pEPSA5::teg49 (Fig. 7C). Likewise, the aureolysin gene aur was more repressed in the HP1 loop mutant with pEPSA5::teg49 than in cells with only the vector control (Fig. 7D). Taken together, these data showed that teg49 is likely a trans-acting sRNA that is capable of complementing the defect in sarA transcription in the HP1 loop mutant.
FIG 7.
Northern blot analysis of S. aureus HP1 loop mutant containing pEPSA-teg49 in the presence of 1% xylose. An S. aureus HP1 loop mutant harboring pEPSA5-teg49 or control empty pEPSA5 was grown in TSB containing 1% xylose at 37°C, and cells were harvested at late exponential phase (OD650 of 1.1) (A, B) and at three different growth stages (OD650s of 0.7, 1.1, and 1.7) (C,D). Purified total RNAs (10 μg/lane) were separated in a 1.5% agarose gel, transferred to H+-bond membranes, and then hybridized with radiolabeled DNA probes for teg49 (A), sarA (B), agrA (C), and aur (D).
DISCUSSION
The sarA locus encompasses the 372-bp sarA coding region preceded by a long ∼800-bp 5′ UTR that adopts a three-promoter system responsible for modulating the abundance of the SarA protein. SarA is a pleiotropic regulator of virulence, oxidative stress, and biofilm formation in S. aureus (1, 2, 23, 31, 32, 33). In this study, we also detected two sRNAs in the 5′ UTR of the sarA locus and found that sRNA teg49 likely contributes to the virulence of S. aureus by modulating SarA expression. This report thus adds to the growing list of sRNAs involved in the control of bacterial virulence in S. aureus, including RNAIII, SprD, RsaE, SprA1, SSR42, and ArtR (12, 13, 14, 15, 34, 35). Unlike the orientation of many of the antisense sRNAs, teg49 is in the same direction as the sarA P1, P3, and P2 transcripts, as determined by RNA-Seq and primer extension studies (Fig. 1B). In the absence of any indication of a riboswitch, teg49 is likely a trans-acting sRNA. We were able to detect teg49 by Northern blot assays with a radiolabeled P3 (data not shown) and/or teg49 DNA probe but not with the sarA P1 and P2 and sarA ORF probes (data not shown), consistent with the location of teg49 between the sarA P3 and P1 promoter regions, as verified by mapping studies.
The expression of sRNA teg49 appears to be sigB dependent. However, we did not find a sigB-dependent promoter (GGGTAT at the −10 position) immediately upstream of the transcription start site. Notably, the absence of teg49 from a sigB mutant coincides with a lack of activation of the SigB-dependent sarA P3 promoter (27, 28) in a sigB mutant, while sarA P3 transcript restoration was accompanied by the reemergence of teg49 in a complemented sigB mutant (Fig. 4E). In addition, as expected with a sigB-dependent transcript (i.e., sarA P3 transcript) (36, 37), teg49 was maximally expressed during the stationary phase (Fig. 4E). Finally, erratic processing of the P2 and P3 sarA transcripts in the HP1 loop mutant also reduced teg49 expression (Fig. 6A). Taking our findings together, we conclude that teg49 does not have a promoter and is likely derived from the sigB-dependent sarA P3 transcript.
We have shown that the expression of teg49 is cshA dependent. CshA is an ATP-dependent DEAD box RNA helicase that unwinds duplex RNA. CshA has been known to be involved in ribosome biogenesis, ribosome assembly, and mRNA decay in a degradosome involving RNase J and Y and polynucleotide phosphorylase (PNPase) (26). However, the protection (or stability) of sRNAs such as teg49 by CshA has not been previously described. In addition, RNase E is absent from S. aureus. In a separate study, we recently analyzed the transcriptome of a cshA mutant of ALC6094, showing that CshA is indeed required for the preservation and/or stability of at least 15 sRNAs in the S. aureus Newman RNome (unpublished data). In contrast to the regulation of the sigB mutant, regulation of teg49 by cshA is active only from early to late log phase since teg49 expression reemerged in the cshA mutant during stationary phase (OD650 of 1.7) (Fig. 4A).
In silico analysis indicated that teg49 forms two hairpin structures with small loops, which we called HP1 and HP2 (Fig. 5A). These secondary structures are often the site of binding to other mRNAs or proteins. To confirm the importance of these secondary structures, we performed transversion mutations of the stem and loop sequences of HP1 and HP2. Mutational analysis of HP1 of teg49 indicated that the HP1 loop has a modulatory function important for controlling virulence gene expression by regulating SarA protein expression (Fig. 6C). In comparison to the parental strain, the HP1 loop mutant with a 7-bp replacement (ATTGCGC→CGGTATA) in the chromosome not only exhibited tapered expression of sarA P2, P3, and teg49 transcripts (Fig. 6A and B) but also led to the formation of two truncated transcripts. The larger truncated transcript is likely derived from the sarA P2 transcript, while the origin of the smaller truncated transcript is not clear and may be the P2 or P3 transcript (Fig. 6B, asterisks). A consequence of the modulation of sarA P2 and P3 transcripts as a result of the HP1 loop mutation is reduced synthesis of the SarA protein, resulting in the dysregulation of downstream genes, including increased aur but decreased RNAII expression (Fig. 6B). Importantly, this has also led to reduced biofilm formation in vitro in the HP1 loop mutant, but the level of reduction is less than that of the sarA deletion mutant.
We have noticed that expression of teg49 was lower in the HP1 loop mutant than in the parent (Fig. 6A). To verify that teg49 can act in trans to complement the HP1 loop mutant, we confirmed that sarA P3 and P1 transcription can be restored close to the parental level in this mutant by expressing teg49 from a xylose-inducible promoter in pEPSA5. Given that the orientation of teg49 is identical to that of the sarA transcripts, we ruled out teg49 as a cis-acting sRNA. In the absence of a riboswitch structure and the ability to complement in trans, we concluded that teg49 is likely a trans-acting sRNA. On the basis of the studies reported here, we propose that 5′-terminal stem-loop HP1 of teg49 is critical in contributing to the stability of sarA P3 and possibly other sarA transcripts. This hypothesis also implies that the HP1 loop sequence in teg49 may be important in preventing erratic processing by host RNases, presumably via an mRNA binding protein that confers protection from defective processing. One plausible candidate for this protection may be CshA, which is likely involved in the generation of teg49 (Fig. 4A) and possesses presumably RNA binding activity (38, 39, 40). Further investigations are needed to verify this possibility.
ACKNOWLEDGMENTS
This work was supported by grants AI106937 from the National Institutes of Health (to A.C.) and 31003A_153474/1 from the Swiss National Science Foundation (to P.F.).
Footnotes
Published ahead of print 4 August 2014
REFERENCES
- 1. Bronner S, Monteil H, Prevost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183–200. 10.1016/j.femsre.2003.09.003 [DOI] [PubMed] [Google Scholar]
- 2. Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9. 10.1016/S0928-8244(03)00309-2 [DOI] [PubMed] [Google Scholar]
- 3. Chien YT, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169–37176. 10.1074/jbc.274.52.37169 [DOI] [PubMed] [Google Scholar]
- 4. Sterba KM, Mackintosh SG, Blevins JS, Hurlburt BK, Smeltzer MS. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410–4417. 10.1128/JB.185.15.4410-4417.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheung AL, Projan SJ. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bayer MG, Heinrichs JH, Cheung AL. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pichon C, Felden B. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U. S. A. 102:14249–14254. 10.1073/pnas.0503838102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, Romby P. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 37:7239–7257. 10.1093/nar/gkp668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. 2010. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 88:565–575. 10.1007/s00109-010-0597-2 [DOI] [PubMed] [Google Scholar]
- 10. Beaume M, Hernandez D, Farinelli2 Deluen LC, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5(5):e10725. 10.1371/journal.pone.0010725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P. 2010. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 38:6620–6636. 10.1093/nar/gkq462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée Durant-E, Barbet R, Jacquets E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P. 2010. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 38:6620–6636. 10.1093/nar/gkq462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xue T, Zhang X, Sun H, Sun B. 2014. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5′ UTR of sarT mRNA. Med. Microbiol. Immunol. 203:1–12. 10.1007/s00430-013-0307-0 [DOI] [PubMed] [Google Scholar]
- 14. Chabelskaya S, Gaillot O, Felden B. 2010. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 6(6):e1000927. 10.1371/journal.ppat.1000927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morrison JM, Miller EW, Benson MA, Alonzo F, III, Yoong P, Torres VJ, Hinrichs SH, Dunmana PM. 2012. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J. Bacteriol. 194:2924–2938. 10.1128/JB.06708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 18. Fu Z, Tamber S, Memmi G, Donegan NP, Cheung AL. 2009. Overexpression of MazFSa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 191:2051–2059. 10.1128/JB.00907-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2002. σB modulates virulence determinants expression and stress resistance: characterization of a functional rsbU strain of Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamber S, Cheung AL. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 77:419–428. 10.1128/IAI.00859-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ballal A, Manna AC. 2009. Expression of the sarA family of genes in different strains of Staphylococcus aureus. Microbiology 155(Pt 7):2342–2352. 10.1099/mic.0.027417-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades LR. 2005. SarA positively controls Bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 187:5790–5798. 10.1128/JB.187.16.5790-5798.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888–2896. 10.1128/JB.183.9.2888-2896.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 26. Oun S, Redder P, Didier J, François P, Corvaglia A, Buttazzoni E, Giraud C, Girard M, Schrenzel J, Linder P. 2013. The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 10:157–165. 10.4161/rna.22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheung AL, Chien YT, Bayer AS. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oscarsson J, Kanth A, Tegmark-Wisell K, Arvidson S. 2006. SarA is a repressor of hla (α-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188:8526–8533. 10.1128/JB.00866-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heinrichs JH, Bayer MG, Cheung AL. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karlsson A, Arvidson S. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239–4246. 10.1128/IAI.70.8.4239-4246.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211. 10.1128/IAI.71.7.4206-4211.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valle J, Toledo-Arana A, Berasain C, Ghigo J, Amorena B, Penades JR, Lasa I. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075–1087. 10.1046/j.1365-2958.2003.03493.x [DOI] [PubMed] [Google Scholar]
- 33. Ballal A, Manna AC. 2010. Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J. Bacteriol. 192:336–345. 10.1128/JB.01202-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, Vandenesch F, Romby P. 2010. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 6(3):e1000809. 10.1371/journal.ppat.1000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sayed N, Jousselin A, Felden B. 2012. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 19:105–112. 10.1038/nsmb.2193 [DOI] [PubMed] [Google Scholar]
- 36. Manna AC, Bayer MG, Cheung AL. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171–5179. 10.1128/JB.183.17.5171-5179.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck VC, McKay DB. 2006. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA 12:959–967. 10.1261/rna.5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hardin JW, Hu Y, McKay DV. 2010. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J. Mol. Biol. 402:412–427. 10.1016/j.jmb.2010.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elles LM, Uhlenbeck OC. 2008. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 36:41–50. 10.1093/nar/gkm926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300–310. 10.1128/JB.01000-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fu Z, Donegan NP, Memmi G, Cheung AL. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 189:8871–8879. 10.1128/JB.01272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]