Abstract
Studies on the innate immune response against microbial infections in Drosophila melanogaster involve mutant strains and their reference strains that act as experimental controls. We used five standard D. melanogaster laboratory reference strains (Oregon R, w1118, Canton-S, Cinnabar Brown, and Yellow White [YW]) and investigated their response against two pathogenic bacteria (Photorhabdus luminescens and Enterococcus faecalis) and two nonpathogenic bacteria (Escherichia coli and Micrococcus luteus). We detected high sensitivity among YW flies to bacterial infections and increased bacterial growth compared to the other strains. We also found variation in the transcription of certain antimicrobial peptide genes among strains, with Oregon and YW infected flies showing the highest and lowest gene transcription levels in most cases. We show that Oregon and w1118 flies possess more circulating hemocytes and higher levels of phenoloxidase activity than the other strains upon infection with the nonpathogenic bacteria. We further observed reduced fat accumulation in YW flies infected with the pathogenic bacteria, which suggests a possible decline in physiological condition. Finally, we found that nitrite levels are significantly lower in infected and uninfected YW flies compared to w1118 flies and that nitric oxide synthase mutant flies in YW background are more susceptible to bacterial infection compared to mutants in w1118 background. Therefore, increased sensitivity of YW flies to bacterial infections can be partly attributed to lower levels of nitric oxide. Such studies will significantly contribute toward a better understanding of the genetic variation between D. melanogaster reference strains.
INTRODUCTION
Genetic variation in the immune response of animals to microbial infections is an important factor in host resistance to infectious disease. Variation in immune system function can directly increase or decrease host susceptibility to microbial challenges, as well as the persistence of microbes during the infection process. Differences between individuals in their ability to activate defense mechanisms against foreign microorganisms may also have significant effects on their fitness potential, such as their ability to survive or their competence to mate and reproduce. Insects are excellent organisms to investigate phenotypic variation of immune capacity among host populations because changes in the efficacy of immune reactions to microbial infections and the genetic/functional basis of phenotypic variation can be studied readily.
The fruit fly, Drosophila melanogaster, is an outstanding model to investigate host-pathogen interactions and innate immunity in eukaryotic organisms (1). Its major benefit is the development of a wide range of molecular and cellular tools, and applications for high-throughput forward and reverse genetic/genomic screens. Innate immunity in D. melanogaster can also be studied as an integrated system at the level of the whole organism. D. melanogaster has a multilayered immune response consisting of humoral and cellular mechanisms (2). The hallmark of the fly host defense is the definition of two main signaling pathways, Toll and immune deficiency (Imd), which lead to the activation of distinct members of the NF-κB family of transcription factors, and result in the expression of target genes, including those encoding antimicrobial peptides (AMPs) (3). The c-Jun N-terminal kinase (JNK) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathways can also act in either competing or cooperative modes to regulate immune effector genes in the fly (4, 5). The body cavities of D. melanogaster flies, like those of all arthropods, are filled with hemolymph that contains both free-floating and sessile hemocytes (blood cells). These are responsible for a number of cellular defenses, including formation of cell aggregates, nodulation, phagocytosis, and encapsulation (6). In addition, D. melanogaster has complex proteolytic cascades that regulate phenoloxidase (PO) activity, coagulation/melanization of hemolymph (7, 8), and defenses associated with the production of reactive oxygen and nitrogen species (9–11). The epithelial cells of the gut can also mount immune responses and have important roles in fighting microbial and parasite infections (12, 13).
Immunity studies in D. melanogaster involve direct comparison of the immune response between loss-of-function or gain-of-function mutant lines and their parental strains that serve as background controls. Interestingly, previous research has revealed extensive variability in susceptibility to bacterial and fungal infections among wild-derived D. melanogaster strains (14–16). Recent experimental evidence also suggests that certain D. melanogaster common laboratory strains exhibit variation in susceptibility to Listeria monocytogenes infection and substantial changes in AMP gene transcription after challenge with this intracellular bacterial pathogen (17). However, little was previously done to investigate in detail the similarities and differences in immune competence between D. melanogaster reference strains that are commonly used in immunity research.
In this study, we investigated the levels of immunocompetence of D. melanogaster laboratory stocks in response to bacterial infections. We used five common D. melanogaster laboratory strains to determine their survival ability against infection with four bacterial species, as well as the ability of the bacteria to proliferate within the infected flies. We then examined host immune capacity of these fly strains by estimating the transcriptional levels of AMP genes, the total number of hemocytes, the PO activity, and the nitrite levels in adult individuals. We found significant variation in the ability of the five laboratory strains to survive bacterial infection, as well as differences in their immune responses against the invading microbes. We finally show that nitric oxide (NO) synthase fly mutants generated in different genetic backgrounds exhibit changes in survival upon bacterial infection. These results are expected to further stress the importance of using the appropriate reference laboratory strains as background controls in D. melanogaster immunity research.
MATERIALS AND METHODS
Fly stocks.
All fly stocks were reared on instant Drosophila diet supplemented with yeast (Carolina Biological Supply) and maintained at 25°C. The following reference fly strains were used in the experiments: Oregon R (Bloomington stock 5), w1118 (Bloomington stock 107122), Canton-S (Bloomington stock 1), Cinnabar Brown (Cinn. Brown; Bloomington stock 103252), and Yellow White (YW; Bloomington stock 189). NOS mutants included the strains NOS24283 (background strain w1118, Bloomington stock 24283), and NOS53156 (background strain YW, Bloomington stock 53156). All stocks had been maintained in culture for 3 years prior to these experiments. Adult male and female flies aged 4 to 6 days old were used in all experiments. We used a diagnostic PCR assay to determine the presence of Wolbachia endosymbiotic bacteria in all fly strains (18). We detected the presence of Wolbachia in Oregon, w1118, and YW strains only. To eliminate Wolbachia endosymbionts, flies from all strains were reared for three generations on diet containing 50 μg of tetracycline/ml (19). After three generations of tetracycline treatment, Wolbachia was not detected in any of the fly strains, which were then returned to the standard diet without antibiotic for all subsequent generations.
Bacterial cultures.
The following bacteria were used in all experiments: Escherichia coli strain DH5a (nonpathogenic), Photorhabdus luminescens subsp. laumondii strain TT01, Micrococcus luteus strain (CIP A270), and Enterococcus faecalis strain ATCC 19433. All bacteria were maintained on petri dishes containing 2.5% Luria-Bertani (LB) and 1.5% agar (Difco Laboratories). For liquid cultures, bacteria were grown in sterile tubes containing 7.5 ml of 2.5% LB and incubated for 24 h on a rotary shaker at 265 rpm. E. coli and E. faecalis bacteria were incubated at 37°C, and M. luteus and P. luminescens at 30°C. Cultures were pelleted at 4°C, washed, and resuspended in phosphate-buffered saline (PBS). The bacterial density of the suspension was estimated with an optical density measurement (600 nm), using a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific) and a 10× serial dilution plating technique. Dead bacteria were prepared by resuspending the bacterial pellets in sterile PBS, and bacterial suspensions were placed on a heat block at 95°C for 30 min. The heat-killed bacteria were centrifuged at 10,000 × g for 5 min, and the dead bacterial pellet was resuspended in 5 ml of sterile PBS.
Fly infection and survival experiments.
A PBS suspension (18.4 nl) containing cells of each bacterial strain was injected into the hemocoel of adult flies at the lateral anterior aspect of the thorax through nanoinjection (Nanoject II apparatus; Drummond Scientific). Approximately 100 to 150 CFU of bacteria were injected per fly. A pure PBS injection was used as a negative control. Treated flies were kept at 30°C, and survival was monitored at 24-h intervals and up to 4 to 7 days after challenge. Thirty flies were used per treatment, and the results represent three independent experiments conducted on three different days.
Bacterial load in infected flies.
To estimate the bacterial load at various time points (0, 3, 6, 12, 24, and 48 h) after bacterial infection, flies from each treatment were individually crushed and homogenized using an electronic pestle in 1.5-ml Eppendorf tubes containing 1 ml of LB medium. The numbers of bacteria (CFU) within each fly were estimated by serial dilution and plating on LB agar plates. Plated bacteria were incubated at 37°C (E. coli and E. faecalis) or at 30°C (M. luteus and P. luminescens). Twenty flies were used for each combination of fly strain/time point/bacterial species, and the results represent the averages of three independent experiments.
Quantitative RT-PCR.
Flies were injected with the bacteria, and at various time points (0, 3, 6, 12, 24, and 48 h) they were frozen at −80°C. Total RNA, cDNA synthesis, and quantitative reverse transcription-PCR (RT-PCR) experiments using Diptericin and Defensin gene-specific primers were carried out as recently described (20). The data are presented as a ratio between infected versus PBS injected flies. The results represent mean values and standard deviations of relative values from three biological repetitions.
Number of hemocytes.
Flies from each strain were injected with the bacteria, and at 0, 3, 6, 12, 24, and 48 h after infection they were anesthetized, their thoraces were pierced, and approximately 2 to 3 μl of hemolymph was removed from each individual. Aliquots of hemolymph (50 μl) were immediately added to ice-cold Ringer solution supplemented with a protease inhibitor mixture (Sigma; 450 μl) in siliconized test tubes. Hemocytes were finally collected by centrifugation, and their numbers were counted at 100× using a Neubauer hemocytometer.
PO assays.
Flies were infected and hemolymph was extracted at 3, 6, 12, 24, and 48 h after bacterial challenge, and the protein concentration was estimated using the Bradford assay (21). PO activity in the hemolymph was quantified using a microplate enzyme assay. Briefly, a reaction mixture containing 10 μg of protein was suspended in 40 μl of 50 mM PBS buffer (pH 6.5) and protease inhibitors. The reaction started by adding a saturated solution of l-3-4-dihydroxyphenylalanine (L-DOPA; Sigma) to each sample. The change in absorbance was read at 490 nm for 5 min at room temperature. Twenty flies from each strain were used per treatment (time point and bacterial infection), and the experiment was repeated three times.
Fat quantification.
We used a colorimetric method to estimate the percentage of total fat in flies infected with bacteria (22). Briefly, following the homogenization of flies in 0.05% Tween 20 (Sigma), the samples were heated at 70°C for 5 min and centrifuged at 5,000 rpm to isolate the supernatants. Samples were then mixed with a Thermo Infinity Trig solution, followed by incubation at 37°C for 5 min. The absorbance of the produced dye, which corresponds to the concentration of the total fats in the sample, was measured at 570 nm on a plate reader (Biotek).
Nitrite assay.
Flies were homogenized in buffer (0.1 M phosphate buffer [pH 7.4], 0.015 M potassium chloride), followed by centrifugation for 10 min at 10,000 × g at 4°C. The supernatants were mixed in a 1:1 proportion with Griess reagent (Sigma) and incubated at room temperature for 15 min. The nitrite levels were estimated with an optical density measurement (595 nm), using a spectrophotometer (NanoDrop 2000c). Concentrations of nitrite were calculated against a silver nitrite-derived standard curve.
Statistical analysis.
All values were expressed as means ± the standard deviation. Statistics were performed using the GraphPad Prism version 5.0 software. For data analyses, means were compared using one-way analysis of variance (ANOVA) with a Tukey post hoc test for multiple comparisons or an unpaired two-tailed t test (bacterial load results). Comparison between survival curves (fraction death) was conducted using a log-rank (Mantel-Cox) test. P values less than 0.05 were considered statistically significant.
RESULTS
Differential survival response of D. melanogaster reference strains to bacterial infections.
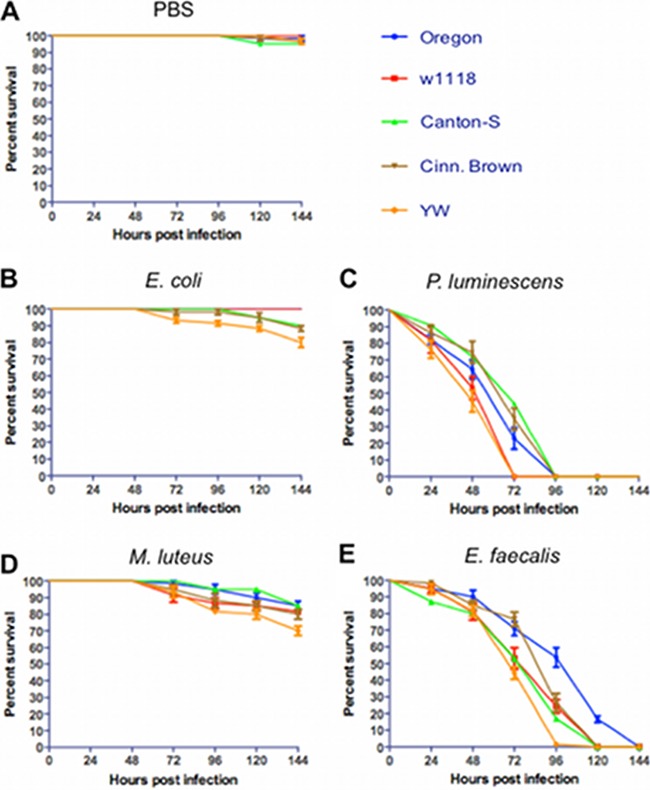
Injection of PBS buffer caused minimal or no fly mortality (Fig. 1A). We first found that all five fly strains survived infection with a nonpathogenic strain of E. coli (Fig. 1B). Although the YW strain was more sensitive compared to the other strains, percentage survival was never lower than 75% 6 days after infection, and there were no significant differences between survival curves (log-rank test, P > 0.05). However, survival curves for w1118 and YW flies infected with P. luminescens were different compared to the other strains (log-rank test, χ2 = 76.6, P < 0.001), and both strains succumbed to this pathogen 3 days after infection (Fig. 1C). We also found that survival curves varied among strains after infection with M. luteus (log-rank test, χ2 = 34.8, P < 0.01) with fly mortality ranging between 70 and 90% on day 6 after challenge (Fig. 1D). We further observed increased sensitivity of YW flies infected with E. faecalis bacteria compared to Canton-S, w1118, and Cinn. Brown flies and that Oregon flies showed the lowest susceptibility to this pathogen among all strains tested (log-rank test, χ2 = 66.4, P < 0.001; Fig. 1E). Although Cinn. Brown flies were less sensitive to E. faecalis for the first 72 h after infection compared to w1118 and Canton-S flies, they all succumbed on day 5, and there were no significant differences between their survival curves (log-rank test, P > 0.05). Injection with heat-killed bacteria caused no mortality to the flies (see Fig. S1 in the supplemental material).
FIG 1.
Survival results after injection of the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW with PBS buffer (negative controls) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Each replicate consisted of ten 4- to 6-day-old flies injected with PBS buffer containing approximately 100 to 150 bacterial cells. The survival of the flies was monitored every 24 h and up to 144 h after infection, and the data represent the percent survival of the infected flies. The averages from three separate experiments are shown. Survival curves were compared using a log-rank (Mantel-Cox) test.
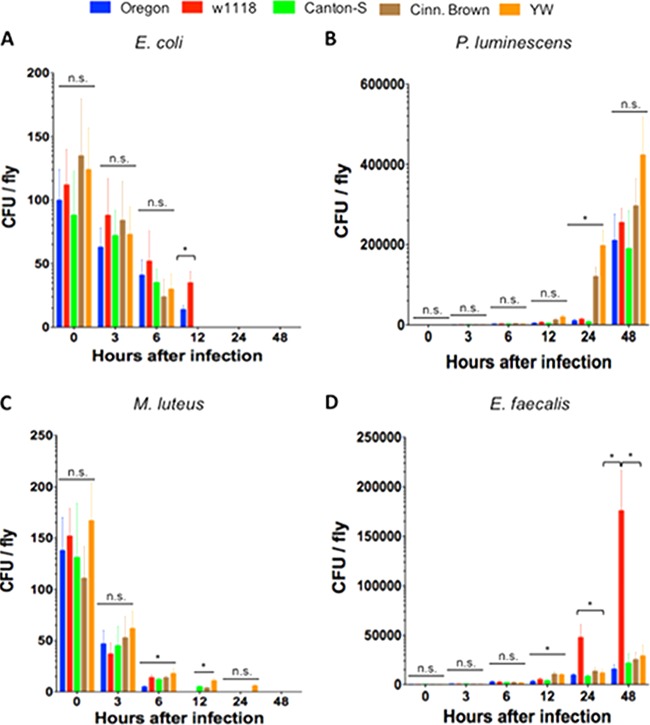
Pathogenic and nonpathogenic bacteria persist differently in D. melanogaster reference strains.
No bacterial growth was detected in flies injected with PBS. We found that numbers of E. coli bacteria decreased in the five D. melanogaster laboratory strains, and there were no bacterial cells detected 24 h after infection (Fig. 2A). There were also no significant differences (P > 0.05) in bacterial load between the five strains for the first two time points postinfection (3 and 6 h). However, we found that at 12 h, Canton-S, Cinn. Brown, and YW flies contained no E. coli bacteria, and the number of E. coli cells in w1118 strain was significantly higher (P < 0.05) compared to those recovered from Oregon flies.
FIG 2.
Bacterial load (CFU) in the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW after infection with the Gram-negative bacteria Escherichia coli (strain DH5a) (A) and Photorhabdus luminescens (strain TT01) (B) and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (C) and Enterococcus faecalis (strain ATCC 19433) (D). Flies (n = 20 per condition) were injected with PBS buffer containing approximately 100 to 150 bacterial cells. The numbers of recoverable bacteria were counted at 0, 3, 6, 12, 24, and 48 h after infection. Bars show means ± the standard deviations. The averages from three separate experiments are shown. Significant differences in bacterial load are indicated with asterisks (two-tailed t test). *, P < 0.05; n.s., nonsignificant differences between treatments.
In contrast, numbers of P. luminescens bacteria increased rapidly over time (Fig. 2B). There were no significant differences (P > 0.05) in the numbers of those bacteria among the five strains at early (3, 6 and 12 h) and late time points (48 h) after infection. At 24 h, we detected significantly lower (P < 0.05) numbers of P. luminescens cells in Oregon, w1118, and Canton-S flies compared to the other two strains.
For numbers of nonpathogenic M. luteus (Fig. 2C), we found no significant differences (P > 0.05) in CFU among the five strains at 3 h after infection and significantly lower (P < 0.05) numbers of those bacteria in Oregon flies compared to the other strains at 6 h after infection. Interestingly, we found no M. luteus cells in Oregon flies at 12 h, at which time point Canton-S and Cinn. Brown contained significantly fewer (P < 0.05) bacteria compared to YW flies. At 24 h postinfection, we detected M. luteus in YW flies only.
Finally, the numbers of E. faecalis bacteria started to increase rapidly at early time points after infection (Fig. 2D). We found no significant differences (P > 0.05) in E. faecalis CFU among the five strains at 3 and 6 h after injection with this pathogen. However, at 12 h after infection Oregon, w1118, and Canton-S strains carried significantly fewer (P < 0.05) E. faecalis bacteria compared to Cinn. Brown and YW flies, and at later time points (24 and 48 h) w1118 flies contained significantly more (P < 0.05) E. faecalis cells compared to the other four strains.
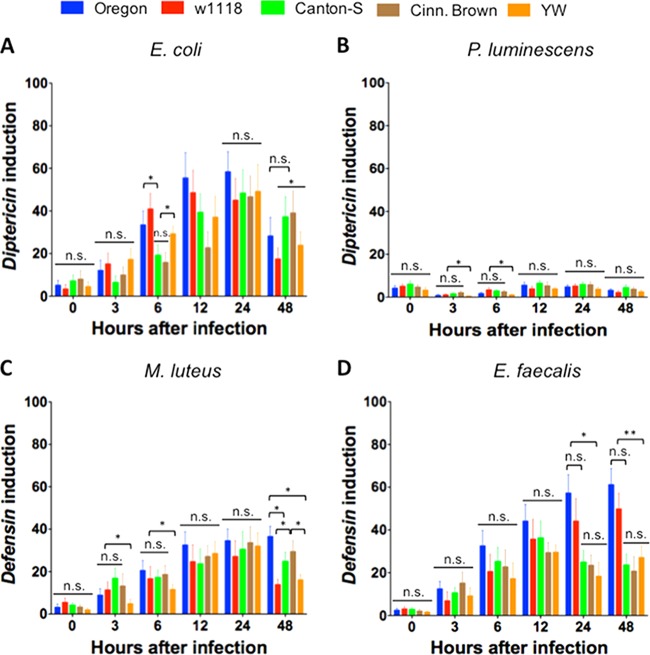
D. melanogaster reference strains transcribe AMP genes at different levels following bacterial infection.
We first tested the transcriptional induction of Diptericin in the five laboratory fly strains infected with the Gram-negative bacteria E. coli and P. luminescens. We found a delay in Diptericin upregulation in Canton-S and Cinn. Brown compared to the other three strains (Fig. 3A). In particular, Diptericin was upregulated at significantly lower levels (P < 0.05) in Canton-S and Cinn. Brown flies at 6 h after infection with E. coli, with no significant differences between those values (P > 0.05). Diptericin induction was also significantly lower (P < 0.05) in Cinn. Brown and YW flies infected with E. coli at 12 h compared to Oregon and w1118 infected individuals. We further found that at 48 h after infection with E. coli, Diptericin induction was significantly reduced (P < 0.05) in w1118 and YW flies compared to Canton-S and Cinn. Brown flies.
FIG 3.
Transcription of antimicrobial peptide genes Diptericin and Defensin in the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW after infection with the Gram-negative bacteria Escherichia coli (strain DH5a) (A) and Photorhabdus luminescens (strain TT01) (B) and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (C) and Enterococcus faecalis (strain ATCC 19433) (D). Flies (n = 20 per infection treatment) were injected with PBS buffer containing approximately 100 to 150 bacterial cells. Gene transcription was estimated at 0, 3, 6, 12, 24, and 48 h after infection. Diptericin and Defensin mRNA levels are shown as the relative abundance of transcripts normalized to RpL32 and are expressed as a ratio compared to flies injected with PBS only (negative controls). Values represent the means from three biological replicates, and error bars represent standard deviations. Asterisks show a value that is significantly different (one-way ANOVA with a Tukey post hoc test for multiple comparisons). **, P < 0.01; *, P < 0.05; n.s., nonsignificant differences.
For Diptericin induction in fly strains infected with the entomopathogen P. luminescens (Fig. 3B), we found that, overall, Diptericin was induced at lower levels in flies infected with P. luminescens compared to E. coli infected flies. We also noticed that Diptericin induction was significantly decreased (P < 0.05) in YW flies at early time points (3 and 6 h) after infection with the pathogen. Although there was variation in Diptericin upregulation among the five fly strains at later times postinfection, the differences were not statistically significant (P > 0.05).
We estimated the transcriptional activation of Defensin in laboratory fly strains following infection with the Gram-positive bacteria M. luteus. We found low (P < 0.05) Defensin induction in YW flies infected for 3 and 6 h with these bacteria and no significant differences (P > 0.05) among the other four strains (Fig. 3C). We also observed significantly reduced (P < 0.05) Defensin upregulation in w1118 and YW flies infected for 48 h with M. luteus compared to the other three strains.
We finally examined the transcriptional induction of Defensin in D. melanogaster reference strains infected with E. faecalis bacteria (Fig. 3D). We found no significant differences (P > 0.05) in Defensin upregulation among the five fly strains at early time points (3 to 12 h) postinfection with this pathogen. However, Defensin induction was significantly lower at 24 (P < 0.05) and 48 h (P < 0.01) in Canton-S, Cinn. Brown, and YW flies compared to Oregon and w1118 infected individuals. There were no significant differences in the transcriptional upregulation of Diptericin and Defensin (P > 0.05) between the fly strains 24 h after injection with heat-killed bacteria (see Fig. S2 in the supplemental material).
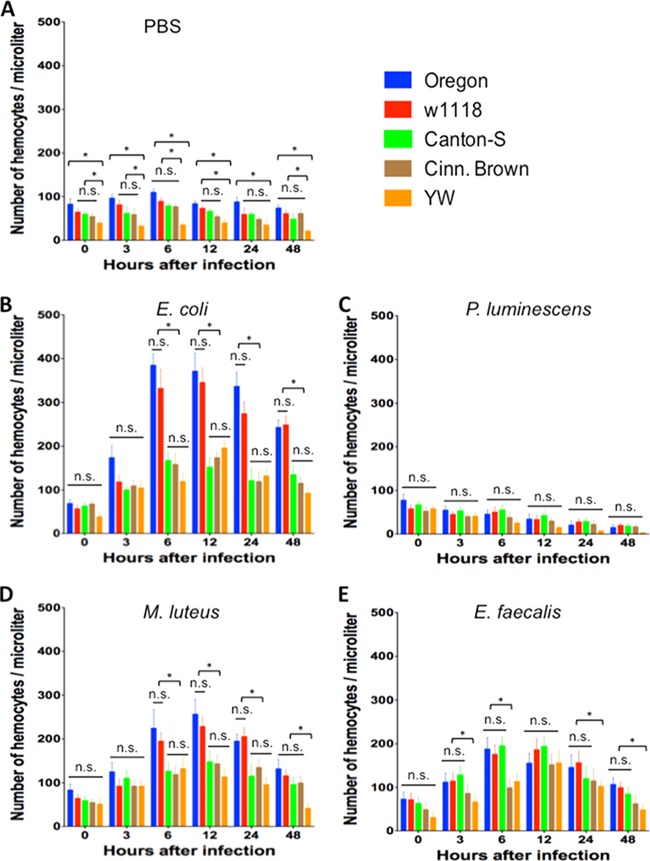
Variation in the number of hemocytes among D. melanogaster reference strains.
We first looked at the number of circulating hemocytes in control flies injected with PBS (Fig. 4A). We found that YW flies contained significantly fewer (P < 0.05) hemocytes compared to the other four strains at 6 and 48 h upon injection with the buffer. YW flies had consistently lower number (P < 0.05) of hemocytes compared to Oregon flies at all time points as well compared to w1118, Canton-S, and Cinn. Brown flies at the 0-, 3-, and 6-h time points. There were also reduced numbers (P < 0.05) of hemocytes in YW flies compared to w1118 flies at 12 h postinjection.
FIG 4.
Numbers of circulating hemocytes recovered from the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW after injection with PBS buffer (control treatment) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Flies (n = 20 per condition) were injected with PBS buffer containing approximately 100 to 150 bacterial cells. Hemocyte counts were determined per microliter of hemolymph at 0, 3, 6, 12, 24, and 48 h after fly injection. Bars represent the means from three separate experiments, and error bars represent the standard deviations. The asterisk shows a value that is significantly different from the other treatments (one-way ANOVA with a Tukey post hoc test for multiple comparisons). *, P < 0.05; n.s., nonsignificant differences between treatments.
There were no significant differences (P > 0.05) in the numbers of hemocytes among the five laboratory strains at 0 and 3 h after infection with E. coli bacteria (Fig. 4B). However, YW, Canton-S, and Cinn. Brown flies contained significantly fewer (P < 0.05) hemocytes compared to Oregon and w1118 flies at all later time points, and there were no significant differences (P > 0.05) among these three strains. Oregon and w1118 flies also contained similar numbers (P > 0.05) of hemocytes at all time points after E. coli infection.
We found that the numbers of hemocytes in all D. melanogaster strains were dramatically reduced over time after the injection of P. luminescens bacteria (Fig. 4C). There were no significant statistical differences (P > 0.05) in hemocyte numbers among the five reference strains at any time point after infection with the pathogen.
For flies injected with M. luteus bacteria, there were no significant differences (P > 0.05) in numbers of hemocytes among the five strains at 0 and 3 h postinfection (Fig. 4D). However, strains Canton-S, Cinn. Brown, and YW contained significantly fewer (P < 0.05) hemocytes compared to Oregon and w1118 flies at 6, 12, and 24 h after challenge with those bacteria. We further found that at 48 h after M. luteus infection YW flies had the lowest number (P < 0.05) of hemocytes among all fly strains.
For infections with E. faecalis bacteria, we recovered a significantly lower (P < 0.05) number of hemocyte cells from Cinn. Brown and YW flies at 6 h (Fig. 4E). Also, YW flies had significantly fewer hemocytes (P < 0.05) compared to Oregon and w1118 strains at 3, 6, 24, and 48 h after injection with this pathogen. We found no statistical differences (P > 0.05) in hemocyte numbers among Oregon, w1118, and Canton-S flies at any time point after infection with this pathogen.
There were no significant differences in the number of hemocytes between Canton-S, Cinn. Brown, and YW strains 24 h after injections with heat-killed E. coli or P. luminescens bacteria (see Fig. S3A and B in the supplemental material). However, Oregon and w1118 flies contained significantly higher number of hemocytes (P < 0.05) compared to the other three strains upon infection with E. coli or P. luminescens or compared to Canton-S flies only after infection with M. luteus bacteria (see Fig. S3C in the supplemental material). No significant changes in hemocyte numbers (P > 0.05) were found between flies injected with E. faecalis bacteria (see Fig. S3D in the supplemental material).
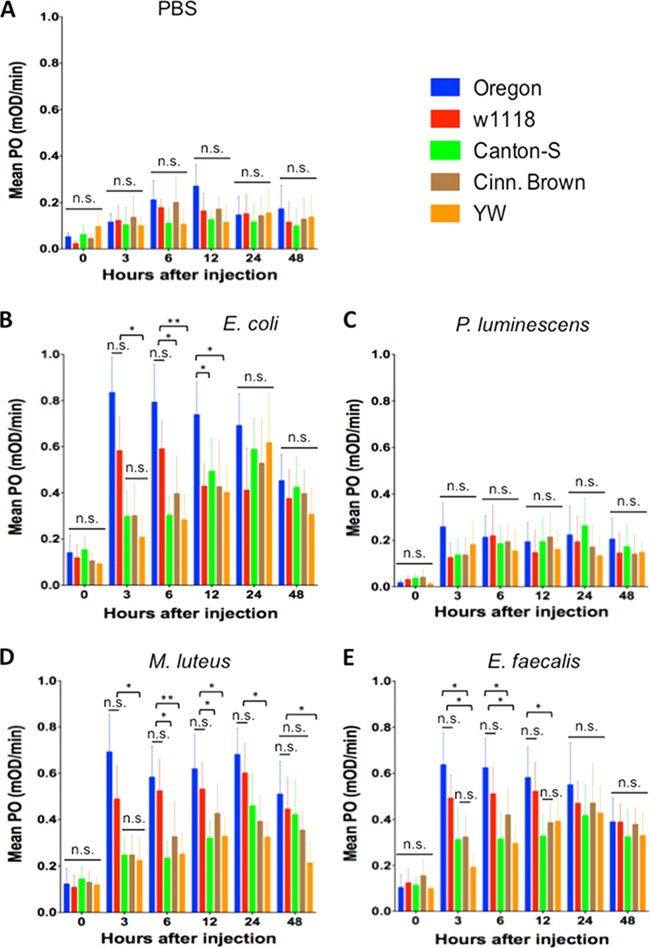
Variation in PO activity among D. melanogaster reference strains infected with bacteria.
We first measured PO activity in flies injected with PBS to estimate baseline enzyme levels in the five reference strains. We found no significant differences (P > 0.05) in PO levels among the five strains for any of the time points tested (Fig. 5A).
FIG 5.
Phenoloxidase (PO) activity in the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinnabar Brown (Cinn. Brown) and Yellow White (YW) after injection with PBS buffer (control treatment) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Flies (n = 20 per condition) were injected with PBS buffer containing approximately 100 to 150 bacterial cells. The PO activity in fly hemolymph samples was measured at 0, 3, 6, 12, 24, and 48 h after fly injection. Bars show means ± the standard deviations. The averages from three independent experiments are shown. Significant differences in PO activity are indicated with asterisks (one-way ANOVA with a Tukey post hoc test for multiple comparisons). **, P < 0.01; *, P < 0.05; n.s., nonsignificant differences between treatments.
For infections with E. coli bacteria, there were no significant differences (P > 0.05) in PO activity among strains at late (24 and 48 h) time points after infection (Fig. 5B). However, we found that PO activity levels in YW, Canton-S, and Cinn. Brown flies were significantly lower (P < 0.05) compared to PO levels in Oregon and w1118 flies at 3 h after infection. We also noticed that PO activity in YW and Canton-S flies was significantly reduced compared to Oregon and w1118 flies (P < 0.01 and P < 0.05, respectively) at 6 h after infection. Further, YW and w1118 flies had significantly lower (P < 0.05) PO activity at 12 h after E. coli challenge compared to Oregon flies.
Although PO values in all D. melanogaster strains injected with P. luminescens were notably lower than those for flies injected with E. coli, there were no significant differences (P > 0.05) in the PO activity among strains for any of the time points examined in our experiments (Fig. 5C).
For infections with M. luteus bacteria, the PO activity in Canton-S, Cinn. Brown, and YW flies at 3 h after injection was significantly lower than in Oregon and w1118 flies (P < 0.05) (Fig. 5D). The PO activity in YW flies infected with M. luteus was significantly than in Oregon and w1118 flies for the rest of the time points (P < 0.05). Also, the PO activity values in Canton-S flies were significantly lower than in Oregon and w1118 flies at 6 and 12 h postinfection (P < 0.01 and P < 0.05, respectively).
Finally, PO activity in YW flies was significantly lower (P < 0.05) than in Oregon and w1118 flies at 3 and 6 h after infection with E. faecalis bacteria (Fig. 5E). There were significantly lower PO values in Canton-S and Cinn. Brown flies than in Oregon flies at 3 and 12 h postinfection (P < 0.05). We also found significantly lower (P < 0.05) PO activity in Canton-S flies compared to Oregon flies at 6 h after E. faecalis challenge. There were no significant differences in PO levels among strains at the 24- and 48-h time points (P > 0.05).
We found no significant differences in PO activity (P > 0.05) between the five fly strains 24 h after injection of heat-killed E. coli, P. luminescens, or E. faecalis (see Fig. S4A, B, and D in the supplemental material). There was only significantly higher PO activity (P < 0.05) in Oregon and w1118 flies compared to YW flies 24 h after injection with heat-killed M. luteus bacteria (see Fig. S4C in the supplemental material).
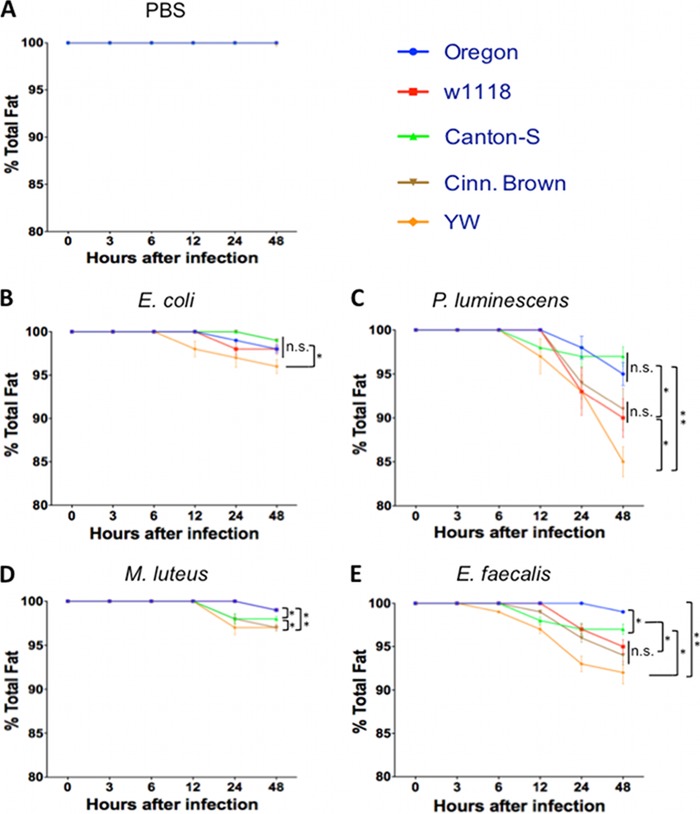
Total fat varies among reference fly strains following bacterial infection.
We injected the five D. melanogaster reference strains with pathogenic bacteria, nonpathogenic bacteria, or PBS and estimated total fat in the infected and uninfected control flies. We found no changes (P > 0.05) in total fat among strains injected with the buffer (Fig. 6A). We also found that YW flies started losing fat at 6 h after infection with E. coli bacteria, and fat loss gradually increased within the following 42 h (Fig. 6B). At 24 h after E. coli injection, YW flies had significantly less (P < 0.05) total fat compared to the other strains.
FIG 6.
Percentage of total fat in the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW after injection with PBS buffer (control treatment) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Flies (n = 20 per condition) were injected with PBS buffer containing approximately 100 to 150 bacterial cells. Total fat in flies was estimated using a colorimetric assay at 0, 3, 6, 12, 24, and 48 h after fly injection. Values represent means ± the standard deviations. The averages from three experiments conducted on different days are shown. Significant differences in total fat are indicated with a asterisks (one-way ANOVA with a Tukey post hoc test for multiple comparisons). **, P < 0.01; *, P < 0.05; n.s., nonsignificant differences between treatments.
For infections with P. luminescens bacteria, we observed an increase in total fat loss in all five fly strains compared to the other treatments (Fig. 6C). In particular, total fat significantly decreased (P < 0.05) in YW flies compared to Oregon, Canton-S (P < 0.01), and w1118, Cinn. Brown flies (P < 0.05). There were also no significant differences in total fat content between Oregon and Canton-S flies as well as between w1118 and Cinn. Brown individuals (P > 0.05). Oregon and Canton-S lines contained significantly higher (P < 0.05) total fat than w1118 and Cinn. Brown lines.
Although there were no dramatic changes in total fat in flies infected with M. luteus bacteria (Fig. 6D), we again found that YW and Cinn. Brown flies lost significantly more fat compared to Canton-S flies (P < 0.05) and Oregon and w1118 flies (P < 0.01). We also found that total fat in Canton-S flies was significantly lower (P < 0.05) than in Oregon and w1118 flies.
Infection with E. faecalis pathogenic bacteria also caused increased fat loss in the five laboratory strains, but at different rates (Fig. 6E). We found that total fat in Oregon flies was not highly affected by these bacteria, whereas YW flies lost significantly more fat compared to Oregon (P < 0.01) and Cantons-S flies (P < 0.05). At 48 h postinfection there were no significant changes (P > 0.05) in total fat between w1118 and Cinn. Brown, but total fat in these flies was significantly lower (P < 0.05) compared to Canton-S and Oregon individuals. The last two strains also contained significantly different (P < 0.05) amounts of total fat. Injection of heat-killed bacteria into flies of the five fly strains did not alter total fat (data not shown).
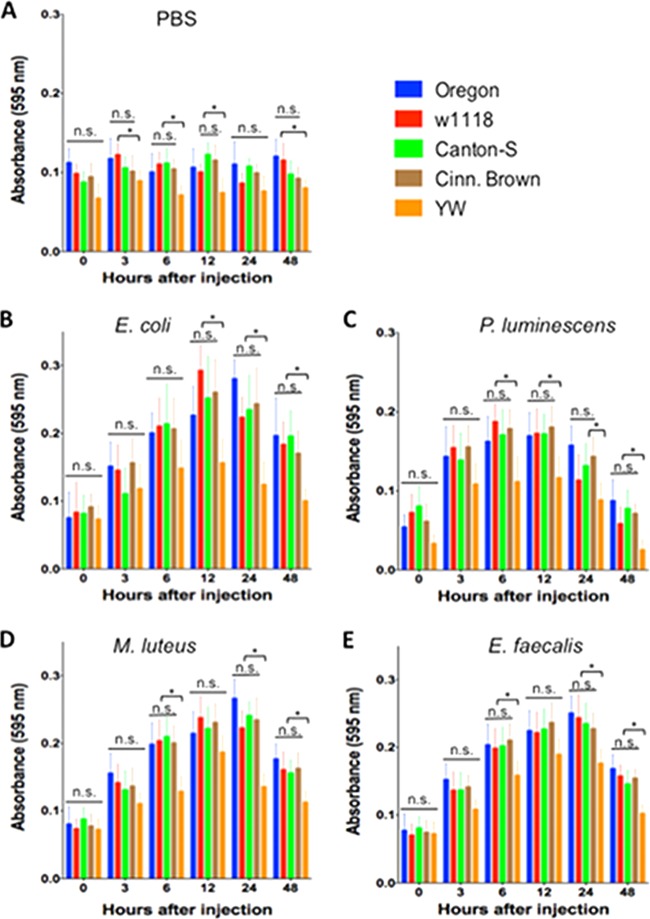
Differences in NO levels between fly strains upon bacterial challenge.
To evaluate NO levels in infected and uninfected flies, we quantified the levels of nitrites, the product of NO breakdown. For treatments involving PBS injections (Fig. 7A), we found significant lower nitrite concentration (P < 0.05) in YW flies compared to Oregon (48 h), w1118 (3, 6, 12, and 48 h), Canton-S and Cinn. Brown (6 h). For E. coli infections (Fig. 7B), there was significantly lower nitrite concentration (P < 0.05) in YW flies than in the other four strains at 12, 24, and 48 h after challenge. Infections with P. luminescens pathogens also resulted in significantly reduced (P < 0.05) nitrite levels in YW flies than in the other fours strains at 6, 12, 24, and 48 h (Fig. 7C). Similarly, nitrite levels were significantly lower (P < 0.05) in YW flies compared to flies of the other four strains at 6, 24, and 48 h after infection with either M. luteus or E. faecalis bacteria (Fig. 7D and E). For treatments with dead bacteria (see Fig. S5 in the supplemental material), we found only significantly lower nitrite concentration (P < 0.05) in YW flies 24 h after injection with heat-killed E. coli and M. luteus cells (see Fig. S5A and C in the supplemental material).
FIG 7.
Nitrite levels in the five Drosophila melanogaster laboratory strains Oregon, w1118, Canton-S, Cinn. Brown, and YW after injection with PBS buffer (control treatment) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Adult flies (n = 20 per condition) were injected with PBS containing 100 to 150 bacterial cells. Nitrite levels in flies were estimated using a spectrophotometer at 0, 3, 6, 12, 24, and 48 h after fly injection. Values represent means ± the standard deviations. The averages from three experiments conducted on different days are shown. Significant differences in nitrite levels are indicated with an asterisk (one-way ANOVA with a Tukey post hoc test for multiple comparisons). *, P < 0.05; n.s., nonsignificant differences between treatments.
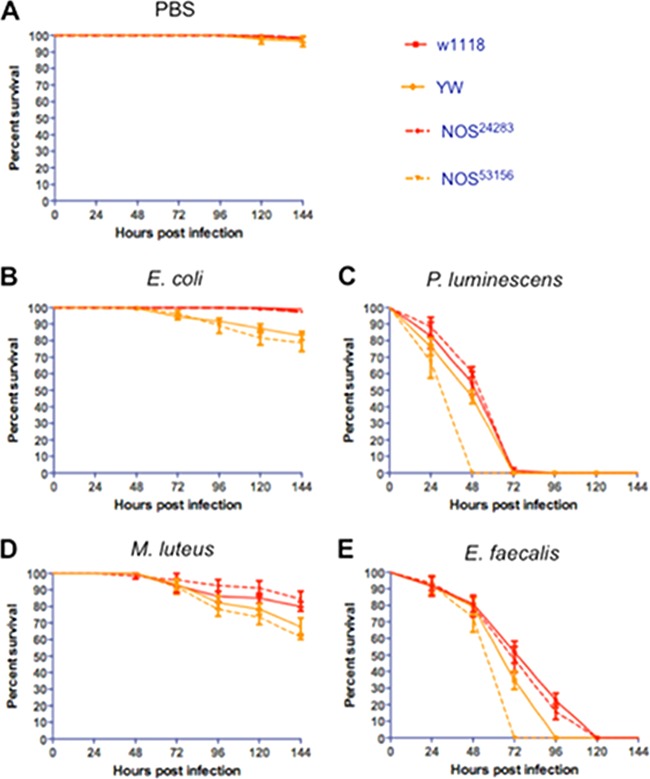
Mutant strains for NOS die at different rates upon bacterial infection.
To examine whether differences in nitrite levels between fly strains affects survival upon infection, we examined the susceptibility of NOS mutant flies to bacterial challenges (Fig. 8). We used mutant flies for NOS (NOS24283 and NOS53156) that were generated from two different reference strains (w1118 and YW, respectively). We found that injection of PBS did not affect the survival of NOS mutants or their background control strains (Fig. 8A). We also found that NOS53156 and YW flies were more sensitive (log-rank test, χ2 = 32.1, P < 0.01) than NOS24283 and w1118 flies to E. coli infection (Fig. 8B) and that NOS53156 mutants were significantly more susceptible (log-rank test, χ2 = 44.3, P < 0.01) compared to NOS24283 mutants upon infection with pathogenic P. luminescens (Fig. 8C). Similarly, infection with M. luteus bacteria resulted in higher sensitivity (log-rank test, χ2 = 22.5, P < 0.01) for NOS53156 and YW individuals (Fig. 8D), while NOS53156 mutant flies had the lowest survival rate (log-rank test, χ2 = 43.7, P < 0.01) after infection with E. faecalis bacteria (Fig. 8E). Injection of heat-killed bacteria into NOS mutant flies and their background controls did not affect survival (see Fig. S6 in the supplemental material).
FIG 8.
Survival results after injection of the Drosophila melanogaster laboratory strains w1118 and YW and the mutant strains for NOS, NOS24283, and NOS53156 with PBS buffer (negative controls) (A), the Gram-negative bacteria Escherichia coli (strain DH5a) (B) and Photorhabdus luminescens (strain TT01) (C), and the Gram-positive bacteria Micrococcus luteus (strain CIP A270) (D) and Enterococcus faecalis (strain ATCC 19433) (E). Each replicate consisted of ten 4- to 6-day-old flies injected with PBS buffer containing 100 to 150 bacterial cells. The survival of flies was monitored every 24 h and up to 144 h after infection, and data represent the percent survival of infected flies. The averages from three separate experiments are shown. Survival curves were compared by using a log-rank (Mantel-Cox) test.
DISCUSSION
D. melanogaster reference laboratory strains are commonly used in immunity studies as controls for the phenotypic characterization of mutant strains in response to pathogenic and nonpathogenic infections. Results in recent studies have indicated variation between certain reference laboratory lines in susceptibility to infection with bacterial pathogens, as well as differences in bacterial load and AMP gene transcription in the infected flies (17). To expand these studies, we used here Gram-negative and Gram-positive pathogenic and nonpathogenic bacteria, together with a range of functional assays, to investigate several aspects of the humoral and cellular immune response in five commonly used D. melanogaster laboratory strains.
The motivation for the present study was based on our recent observations that laboratory fly strains exhibit variation in their survival ability upon infection with bacteria. To document these observations, we initially tested the survival response of five laboratory strains (Oregon, w1118, Canton-S, Cinn. Brown, and YW) to the bacterial pathogens P. luminescens and E. faecalis and the nonpathogens E. coli and M. luteus. These bacteria are frequently used for understanding the insect immune system (23). Although the susceptibility of the five strains varied with the different types of infection, we consistently found that YW flies, and on one occasion w1118 flies, were particularly sensitive to all bacteria. A previous study involving infections with Salmonella enterica serovar Typhimurium, Listeria monocytogenes, and Staphylococcus aureus has also found variation in susceptibility to bacterial infections among wild-type fly strains (17). To determine whether variation in fly survival was due to increased bacterial burden, we estimated numbers of bacteria in the infected flies at several time points after infection. Although significant differences in bacterial load among strains were not observed in most cases, the general trend was that bacterial growth in YW flies was higher compared to the other strains for almost all time points. These results imply that increased susceptibility of YW flies to bacterial infections is at least in part due to the reduced ability of these flies to clear the bacteria from their bodies efficiently.
To understand the basis for the differences in the survival among the five fly strains, we investigated various aspects of their antibacterial immune response. We examined the transcriptional levels of two AMP genes (Diptericin and Defensin) representative of Toll and Imd immune pathway activation in D. melanogaster (24, 25). We chose to examine the transcriptional activation of those two AMP genes because diptericin is effective against Gram-negative bacteria and defensin is active against Gram-positive bacteria (26). Strikingly, we observed very low mRNA levels of Diptericin in all fly strains infected with the pathogen P. luminescens. These results agree with our previous findings and further support the notion that P. luminescens uses unknown mechanisms to suppress the transcriptional induction of AMP-encoding genes in insects (27). This hypothesis is also supported by the fact that injection of heat-killed P. luminescens bacteria upregulates transcription of AMP genes in adult flies. Infection of D. melanogaster strains with E. coli, M. luteus and E. faecalis results in quick upregulation of AMP gene transcription. Although Diptericin and Defensin mRNA levels were similar at early time points after bacterial infection for all five strains, there were distinct changes in AMP gene transcription among strains at late times. These results suggest that different genetic backgrounds among D. melanogaster reference strains probably play an important role in the AMP immune response against bacterial infections. Indeed, three of the five strains (w1118, YW, and Cinn. Brown) are known to carry known mutations (28–30). We also observed AMP gene upregulation upon injection with heat-killed E. coli, M. luteus, and E. faecalis bacteria; however, the level of gene induction was lower than that with live bacteria. These results imply that certain bacterial membrane components are able to activate the AMP transcriptional response and that bacterial replication further increases this effect.
We then assessed numbers of hemocytes in the five fly strains in the presence or absence of bacterial infection as an estimation of cellular immune efficacy (31). Previous studies have associated hemocyte load with immune system efficiency in D. melanogaster and lepidopteran caterpillars (32–35). Our data indicate that Oregon and YW uninfected flies possess the highest and lowest numbers of hemocytes, respectively. This indicates that adult flies from different laboratory fly strains contain variable numbers of circulating hemocytes, which derive during the embryonic and larval stages (36). Upon infection with E. coli or M. luteus, numbers of hemocytes in Oregon and w1118 strains dramatically increase at most time points after bacterial challenge. These results imply that Oregon and w1118 flies may rely mostly on the rapid activation of cellular immune defenses against infections with certain bacteria. Significant differences in the ability of w1118 and YW strains to phagocytose E. coli bacteria were also reported in a recent study (37). However, infection with the pathogen P. luminescens abolishes the differences in hemocyte numbers that are observed in control treatments and treatments involving infection with nonpathogenic E. coli or M. luteus. It was not surprising to find a drastic decrease in the numbers of hemocytes in all fly strains infected with P. luminescens. These bacteria produce virulence factors that aggressively attack and destroy insect hemocytes (38, 39). Nevertheless, injection of flies with dead P. luminescens increased the hemocyte numbers in all strains. We also noticed that infection with E. faecalis resulted in reduced numbers of hemocytes in all fly strains; this indicates an interaction between these bacteria and the fly cell-mediated immune response. Injection of dead E. coli, M. luteus, and E. faecalis bacteria into flies also increases hemocyte numbers, which suggests that bacterial proliferation is probably not a trigger for hemocyte proliferation in D. melanogaster adult flies.
Experiments estimating PO activity showed that most significant differences among laboratory strains occurred at early times-points postinfection, and that Oregon flies, and in some cases w1118 flies, exhibited the highest levels of enzyme activity. These results suggest that different D. melanogaster laboratory strains activate the PO system at different levels, which may in turn affect the immune response against bacterial infections (40). Injection of flies from the different strains with heat-killed bacteria did not affect PO activity, which implies that bacterial amplification does not augment the PO response. Again, we found that infection with live P. luminescens bacteria leads to strong decrease in PO activity in all fly strains. Previous studies in other insect models have reported P. luminescens molecules with potent PO inhibitory activity, which leads to a low melanization response and high levels of insect death (41–43). Future efforts will focus on the identification of P. luminescens effectors that interfere with the PO and coagulation cascades in D. melanogaster adult flies.
To monitor the physiological status of the five fly strains, we estimated the total fat in adults infected with pathogenic and nonpathogenic bacteria, as well as in uninfected controls. We found that fat stores in infected YW flies, especially those challenged with live P. luminescens and E. faecalis, were steadily reduced after infection with the pathogens. Low fat reserves in infected YW individuals can be attributed to starvation or overall decline in the metabolic state of the flies. These results, together with the bacterial load and survival data, suggest that high sensitivity of YW flies to bacterial infections is not only due to inefficient elimination of bacteria (resistance) but also due to a decline in metabolic fitness during the course of infection (tolerance) (44–46).
NO is an important molecule because it participates in several physiological and pathological processes, including immune response to microbes and parasites, in insects (47, 48). To examine the potential involvement of NO in the differential survival of background D. melanogaster strains to bacterial infections, we estimated the amount of the NO oxidation product nitrite. Measuring nitrite levels in whole insects or insect tissues is an accurate estimation of cellular nitric oxide production (49). We found here that uninfected or bacterium-infected YW flies contain substantially smaller amounts of nitrite compared to w1118 individuals and other fly strains. To test whether reduced nitrite quantities in YW flies reflect decreased survival to bacterial infections, we used NOS mutant flies in YW background that proved more sensitive to pathogenic and nonpathogenic bacteria compared to NOS mutants in w1118 background. These results indicate that the quantity of NO varies among different D. melanogaster reference strains and forms a crucial factor that regulates fly sensitivity to bacterial challenges. Indeed, previous studies have shown that ingestion of bacteria induces NO in the gut and hemocytes, which leads to the activation of antimicrobial peptides in the fat body (50). This effect is essential for the efficient immune response of Drosophila larvae against Gram-negative bacteria. Here, we found relatively low levels of NO in YW adult flies infected with Gram-negative bacteria, which might lead to reduced protection of these flies against bacterial challenge.
Because changes in single or multiple immune defenses are likely to be involved in the variation of antibacterial immune responses among fly strains, future experiments will investigate several aspects of hemocyte-related functions, as well as the interaction between humoral and cellular immune reactions in the five fly strains. Infections with a range of pathogenic and nonpathogenic bacteria, as well as with other microbes, will also provide significant clues on the specificity, selectivity, and efficiency of immune responses among the fly strains. Detailed analysis of the differences among strains at the transcriptome level will further clarify the molecular/genetic basis of the evolutionary events that shaped the antibacterial immune system variation in the fruit fly.
In conclusion, our findings indicate that D. melanogaster reference strains exhibit variation in sensitivity to infection by pathogenic and nonpathogenic bacteria. Variation in sensitivity to bacterial infections reflects significant changes in immune capacity and NO levels among the different fly strains. In turn, variation in antibacterial immune responses among fly strains can have serious consequences for the selection of suitable background controls in Drosophila immunity studies. Future research will concentrate on the identification of additional genetic/physiological factors that are responsible for conferring variation in the antibacterial immunity in D. melanogaster laboratory strains.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the George Washington University Columbian College of Arts and Sciences and a Harlan Fellowship from The Department of Biological Sciences at George Washington University to S.S.
We thank members of the Department of Biological Sciences for comments and providing feedback on the manuscript.
Footnotes
Published ahead of print 21 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02318-14.
REFERENCES
- 1. Bier A, Guichard A. 2012. Deconstructing host-pathogen interactions in Drosophila. Dis. Models Mechanisms 5:48–61. 10.1242/dmm.000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uvell H, Engström Y. 2007. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 23:342–349. 10.1016/j.tig.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 3. Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697–743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- 4. Delaney JR, Stoven S, Uvell H, Anderson KV, Engstrom Y, Mlodzik M. 2006. Cooperative control of Drosophila immune responses by the JNK and NF-κB signaling pathways. EMBO J. 25:3068–3077. 10.1038/sj.emboj.7601182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morin-Poulard I, Vincent A, Crozatier M. 2013. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT 2:e25700. 10.4161/jkst.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Honti V, Csordás G, Kurucz É Márkus R, ó I. 2014. The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy, and regulation. Dev. Comp. Immunol. 42:47–56. 10.1016/j.dci.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 7. Eleftherianos I, Revenis C. 2011. Role and importance of phenoloxidase in insect hemostasis. J. Innate Immun. 3:28–33. 10.1159/000321931 [DOI] [PubMed] [Google Scholar]
- 8. Theopold U, Krautz R, Dushay MS. 2014. The Drosophila clotting system and its messages for mammals. Dev. Comp. Immunol. 42:42–46. 10.1016/j.dci.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 9. Bae YS, Choi MK, Lee WJ. 2010. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 31:278–287. 10.1016/j.it.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 10. Dow JA, Romero MF. 2010. Drosophila provides rapid modeling of renal development, function, and disease. Am. J. Physiol. Renal Physiol. 299:F1237–F1244. 10.1152/ajprenal.00521.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies SA, Dow JA. 2009. Modulation of epithelial innate immunity by autocrine production of nitric oxide. Gen. Comp. Endocrinol. 162:113–121. 10.1016/j.ygcen.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 12. Charroux B, Royet J. 2010. Drosophila immune response: from systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly 4:40–47. 10.4161/fly.4.1.10810 [DOI] [PubMed] [Google Scholar]
- 13. Lee K-A, Lee W-J. 2014. Drosophila as a model for intestinal dysbiosis and chronic inflammatory diseases. Dev. Comp. Immunol. 42:102–110. 10.1016/j.dci.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 14. Lazzaro BP, Sceurman BK, Clark AG. 2004. Genetic basis of natural variation in Drosophila melanogaster antibacterial immunity. Science 303:1873–1876 [DOI] [PubMed] [Google Scholar]
- 15. Sackton TB, Lazzaro BP, Clark AG. 2010. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 6:e1000797. 10.1371/journal.pgen.1000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tinsley MC, Blanford S, Jiggins FM. 2006. Genetic variation in Drosophila melanogaster pathogen susceptibility. Parasitology 132:767–773. 10.1017/S0031182006009929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okado K, Shinzawa N, Aonuma H, Nelson B, Fukumoto S, Fujisaki K, Kawazu S, Kanuka H. 2009. Rapid recruitment of innate immunity regulates variation of intracellular pathogen resistance in Drosophila. Biochem. Biophys. Res. Commun. 379:6–10. 10.1016/j.bbrc.2008.11.097 [DOI] [PubMed] [Google Scholar]
- 18. Zhou W, Rousset F, O'Neil S. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265:509–515. 10.1098/rspb.1998.0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teixeira L, Ferreira A, Ashburner M. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e1000002. 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castillo JC, Shokal U, Eleftherianos I. 2013. Immune gene transcription in Drosophila adult flies infected by entomopathogenic nematodes and their mutualistic bacteria. J. Insect Physiol. 59:179–185. 10.1016/j.jinsphys.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 21. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 22. Ayyaz A, Giammarinaro P, Liégeois S, Lestradet M, Ferrandon D. 2013. A negative role for MyD88 in the resistance to starvation as revealed in an intestinal infection of Drosophila melanogaster with the Gram-positive bacterium Staphylococcus xylosus. Immunobiology 218:635–644. 10.1016/j.imbio.2012.07.027 [DOI] [PubMed] [Google Scholar]
- 23. Neyen C, Bretscher AJ, Binggeli O, Lemaitre B. 2008. Methods to study Drosophila immunity. Methods 68:116–128. 10.1016/j.ymeth.2014.02.023 [DOI] [PubMed] [Google Scholar]
- 24. Lindsay SA, Wasserman SA. 2014. Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 42:16–24. 10.1016/j.dci.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleino A, Silverman N. 2014. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42:25–35. 10.1016/j.dci.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanji T, Ip YT. 2005. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 26:193–198. 10.1016/j.it.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 27. Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. 2010. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 18:552–560. 10.1016/j.tim.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 28. Cook R. 1980. The extent of visual control in the courtship tracking of Drosophila melanogaster. Biol. Cybernet. 37:41–51. 10.1007/BF00347641 [DOI] [Google Scholar]
- 29. Biessmann H. 1985. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 82:7369–7373. 10.1073/pnas.82.21.7369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Warren WD, Palmer S, Howells AJ. 1996. Molecular characterization of the cinnabar region of Drosophila melanogaster: identification of the cinnabar transcription unit. Genetica 98:249–262. 10.1007/BF00057589 [DOI] [PubMed] [Google Scholar]
- 31. Gerritsma S, de Haan A, van de Zande L, Wertheim B. 2013. Natural variation in differentiated hemocytes is related to parasitoid resistance in Drosophila melanogaster. J. Insect Physiol. 59:148–158. 10.1016/j.jinsphys.2012.09.017 [DOI] [PubMed] [Google Scholar]
- 32. Lavine MD, Strand MR. 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32:1295–1309. 10.1016/S0965-1748(02)00092-9 [DOI] [PubMed] [Google Scholar]
- 33. Prévost G, Eslin P. 1998. Hemocyte load and immune resistance to Asobara tabida are correlated in species of the Drosophila melanogaster subgroup. J. Insect Physiol. 44:807–816. 10.1016/S0022-1910(98)00013-4 [DOI] [PubMed] [Google Scholar]
- 34. Eslin P, Prévost G. 1996. Variation in Drosophila concentration of haemocytes associated with different ability to encapsulate Asobara tabida larval parasitoid. J. Insect Physiol. 42:549–555. 10.1016/0022-1910(95)00134-4 [DOI] [Google Scholar]
- 35. Kraaijeveld AR, Limentani EC, Godfray HCJ. 2001. Basis of the trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Proc. Biol. Sci. 268:259–261. 10.1098/rspb.2000.1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chambers MC, Lightfield KL, Schneider DS. 2012. How the fly balances its ability to combat different pathogens. PLoS Pathog. 8:e1002970. 10.1371/journal.ppat.1002970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crozatier M, Meister M. 2007. Drosophila hematopoiesis. Cell. Microbiol. 9:1117–1126. 10.1111/j.1462-5822.2007.00930.x [DOI] [PubMed] [Google Scholar]
- 38. Waterfield NR, Ciche T, Clarke D. 2009. Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 63:557–574. 10.1146/annurev.micro.091208.073507 [DOI] [PubMed] [Google Scholar]
- 39. Rodou A, Ankrah DO, Stathopoulos C. 2010. Toxins and secretion systems of Photorhabdus luminescens. Toxins 2:1250–1264. 10.3390/toxins2061250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang H. 2009. Regulation and function of the melanization reaction in Drosophila. Fly 3:105–111. 10.4161/fly.3.1.7747 [DOI] [PubMed] [Google Scholar]
- 41. Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, ffrench-Constant RH, Reynolds SE. 2007. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Proc. Natl. Acad. Sci. U. S. A. 104:2419–2424. 10.1073/pnas.0610525104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eleftherianos I, Waterfield NR, Bone P, Boundy S, ffrench-Constant RH, Reynolds SE. 2009. A single locus from the entomopathogenic bacterium Photorhabdus luminescens inhibits activated Manduca sexta phenoloxidase. FEMS Microbiol. Lett. 293:170–176. 10.1111/j.1574-6968.2009.01523.x [DOI] [PubMed] [Google Scholar]
- 43. Crawford JM, Portmann C, Zhang X, Roeffaers MB, Clardy J. 2012. Small molecule perimeter defense in entomopathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:10821–10826. 10.1073/pnas.1201160109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schneider DS, Ayres JS. 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8:889–895. 10.1038/nri2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ayres JS, Schneider DS. 2012. Tolerance of infections. Annu. Rev. Immunol. 30:271–294. 10.1146/annurev-immunol-020711-075030 [DOI] [PubMed] [Google Scholar]
- 46. Medzhitov R, Schneider DS, Soares MP. 2012. Disease tolerance as a defense strategy. Science 335:936–941. 10.1126/science.1214935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivero A. 2006. Nitric oxide: an antiparasitic molecule of invertebrates. Trends Parasitol. 22:219–225. 10.1016/j.pt.2006.02.014 [DOI] [PubMed] [Google Scholar]
- 48. Nappi AJ, Vass E, Frey F, Carton Y. 2000. Nitric oxide involvement in Drosophila immunity. Nitric Oxide 4:423–430. 10.1006/niox.2000.0294 [DOI] [PubMed] [Google Scholar]
- 49. Ajjuri RR, O'Donnell JM. 2013. Novel whole-tissue quantitative assay of nitric oxide levels in Drosophila neuroinflammatory response. J. Vis. Exp. 82:e50892. 10.3791/50892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Foley E, O'Farrell PH. 2003. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17:115–125. 10.1101/gad.1018503 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.