Abstract
The Lyme disease spirochete, Borrelia burgdorferi, occupies both a tick vector and mammalian host in nature. Considering the unique enzootic life cycle of B. burgdorferi, it is not surprising that a large proportion of its genome is composed of hypothetical proteins not found in other bacterial pathogens. bb0238 encodes a conserved hypothetical protein of unknown function that is predicted to contain a tetratricopeptide repeat (TPR) domain, a structural motif responsible for mediating protein-protein interactions. To evaluate the role of bb0238 during mammalian infection, a bb0238-deficient mutant was constructed. The bb0238 mutant was attenuated in mice infected via needle inoculation, and complementation of bb0238 expression restored infectivity to wild-type levels. bb0238 expression does not change in response to varying culture conditions, and thus, it appears to be constitutively expressed under in vitro conditions. bb0238 is expressed in murine tissues during infection, though there was no significant change in expression levels among different tissue types. Localization studies indicate that BB0238 is associated with the inner membrane of the spirochete and is therefore unlikely to promote interaction with host ligands during infection. B. burgdorferi clones containing point mutations in conserved residues of the putative TPR motif of BB0238 demonstrated attenuation in mice that was comparable to that in the bb0238 deletion mutant, suggesting that BB0238 may contain a functional TPR domain.
INTRODUCTION
Borrelia burgdorferi, the causative agent of Lyme disease, is a zoonotic pathogen that utilizes an arthropod vector (i.e., Ixodes sp. hard ticks) for transmission between a vertebrate reservoir and humans (1–3). As the tick feeds, bacteria are introduced into a mammalian host to establish an initial localized infection at the bite site, after which the bacteria disseminate throughout the mammal and colonize other secondary tissues, such as the heart, joints, and central nervous system (4, 5). B. burgdorferi induces disease in the mammalian host, primarily due to its highly invasive nature and its ability to persist in various tissues. Therefore, B. burgdorferi virulence factors can be broadly defined as being encoded by genes that contribute to successful arthropod colonization, vector-mediated transmission, or vertebrate infection.
Considering the specialized enzootic life cycle of B. burgdorferi, it is not surprising that the B. burgdorferi genome contains many genes that are unique among bacterial pathogens (6). These hypothetical proteins are particularly interesting because they represent a group of borrelial proteins that might contribute to vector and/or mammalian colonization and, ultimately, may provide insight into molecular mechanisms contributing to host/vector-pathogen interactions. Although studying genes with unknown functions represents a challenge, a number of these unique B. burgdorferi proteins (i.e., BptA, BB0323, BBA52, BBA57, BBA62, and BBA64) are now known to play roles at different stages of the spirochete's enzootic cycle (7–12). Elucidation of the roles of individual hypothetical proteins in B. burgdorferi and identification of those necessary for mammalian infection would potentially provide new avenues of intervention or therapeutic targets.
BB0238 is a chromosomally encoded hypothetical protein unique to B. burgdorferi that is predicted to be 30 kDa in size and to contain a tetratricopeptide repeat (TPR) domain (6, 13). To evaluate the role of bb0238 during mammalian infection, a bb0238-deficient mutant of B. burgdorferi was generated. The bb0238 mutant was significantly attenuated in the murine borreliosis infection model. Specifically, spirochetes could not be cultured from ear punch or heart tissues recovered from any of the mice infected with the bb0238 mutant. The bb0238 mutant also exhibited a defect in its capacity to infect tibiotarsal and bladder tissues. Interestingly, multiple clones expressing bb0238 with individual point mutations engineered in the putative TPR domain exhibited attenuated infectivity similar to that observed with the bb0238 deletion mutant, thus suggesting that this domain may be essential to the function of BB0238 in the spirochete. However, it is not likely that BB0238 directly interacts with host cells and/or ligands, because BB0238 partitioned to the inner membrane of B. burgdorferi during cellular localization assays. While we were unable to determine the exact function of BB0238, these studies did identify a new protein that plays an important role during B. burgdorferi murine infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains and plasmids utilized in this study are described in Table 1. Escherichia coli strains TOP10F′ (Life Technologies, Carlsbad, CA) and MON1 (Monserate, San Diego, CA) were used for plasmid propagation and cloning. E. coli C41(DE3) (Lucigen, Middleton, WI) was used for expression of recombinant protein for purification. E. coli transformants were selected in Luria-Bertani medium supplemented with 100 μg/ml of ampicillin, 100 μg/ml of spectinomycin, or 50 μg/ml of kanamycin (Kan). The low-passage human isolate B. burgdorferi strain 297 (Bb297) was used in these studies (14). Unless noted, B. burgdorferi was grown in Barbour-Stoenner-Kelley-II (BSK-II) medium at 37°C with 3% CO2 and antibiotics when appropriate (150 μg/ml streptomycin [Strep] or 150 μg/ml kanamycin) (15). Transformation of B. burgdorferi was carried out as previously described (16).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Descriptiona | Source |
|---|---|---|
| Plasmids | ||
| pGEM-T Easy | TA cloning vector; Ampr | Promega |
| pJSB430A | pGEM-T Easy-bb0238::PflgB-Kan mutation construct; Kanr | This study |
| pJSB201 | pJD7 with the NdeI site in PflgB-aadA mutated; Specr Strepr | This study |
| pJD7 | Borrelial shuttle vector derived from pKFSS1 | 18 |
| pJSB445 | pJSB201::bb0238 ORF complementation vector; Specr Strepr | This study |
| pProEX-HTb | Expression construct; N-terminal, TEV-cleavable His6 tag; Ampr | Life Technologies |
| pJSB482 | pProEX-HTb::codon-adapted bb0238 ORF; no signal peptide; Ampr | This study |
| pJSB568 | pGEM-T Easy::flaB-actin qPCR standard; Ampr | This study |
| pJSB641 | pGEM-T Easy::flaB probe; Ampr | This study |
| pJSB643 | pGEM-T Easy::bb0238 probe; Ampr | This study |
| piRrp2(Kan) | Borrelial shuttle vector with IPTG-inducible rrp2; Kanr | 17 |
| pJSB499 | piRrp2(Kan) with rrp2 replaced by the ebfC ORF; Kanr | This study |
| pJSB598 | pJSB445 with BB0238Y43A mutation; Specr Strepr | This study |
| pJSB599 | pJSB445 with BB0238G47V mutation; Specr Strepr | This study |
| pJSB603 | pJSB445 with BB0238Y50A mutation; Specr Strepr | This study |
| pJSB604 | pJSB445 with BB0238A66G mutation; Specr Strepr | This study |
| pJSB602 | pJSB445 with BB0238P71A mutation; Specr Strepr | This study |
| Strains | ||
| E. coli | ||
| TOP10F′ | F′ [lacIqTn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 nupG ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 | Life Technologies |
| MON1 | F− endA1 recA1 mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15ΔlacX74 araΔ139 Δ(ara-leu)7697 galU galK nupG rpsL λ− | Monserate |
| C41(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Lucigen |
| B. burgdorferi | ||
| Bb297 | Strain 297; infectious human spinal fluid isolate | 14 |
| BbΔbb0238 | Bb297 with bb0238 interrupted with PflgB-Kan marker; Kanr | This study |
| Bbbb0238C | BbΔbb0238 complemented with pJSB445; Kanr Strepr | This study |
| BbiEbfC | Bb297 transformed with pJSB499; Kanr | This study |
| Bbbb0238Y43A | BbΔbb0238 complemented with pJSB598; Kanr Strepr | This study |
| Bbbb0238G47V | BbΔbb0238 complemented with pJSB599; Kanr Strepr | This study |
| Bbbb0238Y50A | BbΔbb0238 complemented with pJSB603; Kanr Strepr | This study |
| Bbbb0238A66G | BbΔbb0238 complemented with pJSB604; Kanr Strepr | This study |
| Bbbb0238P71A | BbΔbb0238 complemented with pJSB602; Kanr Strepr | This study |
Spec, spectinomycin; Strep, streptomycin; Amp, ampicillin; Kan, kanamycin.
Generation and confirmation of plasmids and clones used in this study.
The primers used in this study are summarized in Table S1 in the supplemental material. When PCR amplifying fragments for cloning, genomic DNA (gDNA) from Bb297 served as the template, and TaKaRa PrimeStar HS DNA Polymerase (Clontech, Mountain View, CA) was used for amplification. The amplicons were TA cloned into pGEM-T Easy (Promega Corp., Madison, WI) and confirmed by sequencing. Fermentas GeneRuler DNA ladder mix was the molecular weight standard used for DNA electrophoresis (Thermo Scientific, Rockford, IL). The endogenous plasmid content of each transformant was confirmed by PCR-based plasmid profiling (16).
The bb0238 mutant, BbΔbb0238, was created by first amplifying regions flanking bb0238 (accession no. NP_212372) with primers that introduced an AscI restriction site on the 3′ end of a 1,009-bp upstream flanking region (5′ F1-BB0238 and 3′ F1-BB0238-AscI) (see Table S1 in the supplemental material) and a 5′ AscI and a 3′ BssHII site in a 995-bp downstream flanking region (5′ F2-BB0238-AscI and 3′ F2-BB0238-BssHII) (see Table S1 in the supplemental material). To ligate the two flanking regions, the downstream F2 fragment was excised using BssHII and ligated into the vector containing the upstream F1 region linearized with AscI. A Kanr marker with a T7 transcriptional terminator fused to the end of the resistance gene, designated flgBp-aphI-T7t, was provided by Scott Samuels at the University of Montana (17). The Kanr marker was ligated into the unique AscI restriction site at the junction between the two flanking regions. The final mutagenesis construct, designated pJSB430A, contained a Kanr marker that replaced an internal region of the bb0238 open reading frame (ORF) between bp 259 and 500. The pJSB430A mutagenesis construct was electroporated into Bb297, and transformants were selected with kanamycin. Interruption of bb0238 in the chromosome by the Kanr marker was confirmed with bb0238 diagnostic primers (5′ BB0238-Diag and 3′ BB0238-Diag) (see Table S1 in the supplemental material).
To restore bb0238 expression in the BbΔbb0238 mutant, the bb0238 ORF, along with 420 bp upstream and 258 bp downstream of the ORF, was amplified using primers that introduced BamHI restriction sites on the 5′ and 3′ ends (5′ BB0238-Comp and 3′ BB0238-Comp) (see Table S1 in the supplemental material). This region was ligated into pJSB201, a derivative of the cp9-based pJD7 shuttle vector (18). The insert was confirmed by sequencing with vector-specific primers and internal primers, 5′ BB0238-Int and 3′ BB0238-Int (see Table S1 in the supplemental material). The resulting construct, designated pJSB445, contained the bb0238 ORF, along with a region containing its putative native promoter and a PflgB-aadA Strepr resistance marker. pJSB445 was electroporated into BbΔbb0238 with kanamycin selection during recovery and plated with kanamycin and streptomycin selection. The presence of pJSB445 in transformants was confirmed by plasmid recovery and PCR using bb0238 diagnostic primers (5′ BB0238-Diag and 3′ BB0238-Diag) (see Table S1 in the supplemental material). BbΔbb0238 transformed with pJSB445 was designated Bbbb0238C.
To produce recombinant BB0238 protein, the bb0238 ORF from Bb297 was first codon adapted to optimize expression in E. coli using GeneArt gene synthesis (Life Technologies). The codon-optimized ORF was PCR amplified using primers that introduced BamHI and EcoRI restriction sites on the 5′ and 3′ ends, respectively (5′ BB0238 Opt-SP/BamHI and 3′ BB0238 Opt/EcoRI) (see Table S1 in the supplemental material). These primers were designed to exclude the N-terminal 26 amino acids of BB0238, which was predicted by SignalP 4.1 to contain either a signal peptidase I cleavage site or a transmembrane domain (19). The ORF fragment was confirmed by sequencing and ligated into the pPROEX-HTb expression vector (Life Technologies) to generate pJSB482.
To generate copy number standards for quantitative PCR (qPCR) analysis of bacterial burdens in tissues, a region of the murine β-actin gene was amplified from C3H/HeN DNA using primers ActF-STD and ActR-STD (see Table S1 in the supplemental material) (20), which introduced a BamHI restriction site at one end. The amplicon was cloned into pGEM-T Easy. A region of flaB was then amplified from Bb297 gDNA using primers flaBF-STD and flaBR-STD (see Table S1 in the supplemental material). These primers introduced BamHI restriction sites on both ends of the fragment. The flaB amplicon was cloned into pGEM-T Easy containing the actin fragment using BamHI restriction enzyme sites. The resulting construct was designated pJSB568.
To generate hybridization probes for Northern blot analyses, regions corresponding to the genes of interest were amplified and cloned into pGEM-T Easy. The primers 5′ BB0238-probe and 3′ BB0238-probe amplified the DNA fragment probe for bb0238 (see Table S1 in the supplemental material). The primers for flaB (flaBd and flaBrc) (see Table S1 in the supplemental material) were based on those used by Bugrysheva et al. (21). The vectors were designated pJSB641 and pJSB643 for the flaB and bb0238 Northern hybridization probes, respectively.
To generate the B. burgdorferi clone expressing IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible ebfC, the ebfC ORF was amplified with primers that introduced NdeI and HindIII restriction enzyme sites at the 5′ and 3′ ends, respectively (5′ EbfC-NdeI and 3′ EbfC-HindIII) (see Table S1 in the supplemental material). The rrp2 ORF in piRrp2(Kan) (17) was then replaced by the ebfC ORF to generate pJSB499. pJSB499 was transformed into Bb297, and Kanr transformants were recovered. The presence of pJSB499 in clones was confirmed by plasmid recovery in E. coli. A single clone, designated BbiEbfC, was selected for characterization.
To generate B. burgdorferi clones expressing bb0238 with point mutations in the putative TPR domain, overlap extension PCR was used to mutate individual amino acids encoded in the bb0238 ORF of the complementation construct, pJSB445. Primers 5′ BB0238-Comp and 3′ BB0238-Comp (see Table S1 in the supplemental material) represent the outermost primers used for all amplifications. Internal primers used to mutate individual residues (i.e., Y43A, G47V, Y50A, A66G, and P71A) are indicated with their respective mutation denoted as sense or antisense in Table S1 in the supplemental material. Primers 5′ BB0238-Comp and 3′ BB0238-Comp were paired with the mutational sense and antisense primers, respectively. Amplicons from the individual reactions were gel purified and combined to serve as DNA templates in a second round of amplification using the outermost 5′ BB0238-Comp and 3′ BB0238-Comp primers. The mutated bb0238 alleles were ligated into pJSB201 to produce pJSB598 (bb0238Y43A), pJSB599 (bb0238G47V), pJSB603 (bb0238Y50A), pJSB604 (bb0238A66G), and pJSB602 (bb0238P71A). The inserts were confirmed by sequencing with vector-specific primers and internal primers, 5′ BB0238-Int and 3′ BB0238-Int. BbΔbb0238 was transformed with each plasmid, and single clones were recovered for genotyping. The presence of the shuttle vector in the transformants was confirmed by PCR using bb0238 diagnostic primers (5′ BB0238-Diag and 3′ BB0238-Diag) (see Table S1 in the supplemental material), as well as plasmid recovery from E. coli and sequencing. Confirmed clones were designated according to the BB0238 mutation that each transformant carried: Bbbb0238Y43A, Bbbb0238G47V, Bbbb0238Y50A, Bbbb0238A66G, or Bbbb0238P71A.
Expression and purification of recombinant BB0238 protein.
Expression from pProEX-HTb generates a recombinant protein with an N-terminal His6 tag followed by a tobacco etch virus (TEV) protease cleavage site. To produce recombinant BB0238, pJSB482 was transformed into E. coli C41(DE3), and cultures were induced for 3 h with 0.5 mM IPTG. The recombinant protein was affinity purified by the batch method with HisPur Ni-nitrilotriacetic acid (NTA) resin (Thermo Scientific) under nonnative conditions according to the manufacturer's instructions, except 8 M urea was used to solubilize the inclusion body pellet and included in all purification buffers. The concentration of purified recombinant BB0238 was determined using the DC protein assay kit (Bio-Rad Laboratories, Hercules, CA).
Generation of BB0238-specific antisera.
To generate polyclonal antisera, 25 μg of recombinant protein in 200 μl of sterile phosphate-buffered saline (PBS) was combined with complete Freund's adjuvant (Sigma, St. Louis, MO) at a 1:1 ratio. After emulsification of the antigen-adjuvant mixture, 3- to 4-week-old female Sprague-Dawley rats (Harlan, Indianapolis, IN) were injected intraperitoneally. The rats were then boosted twice with 25 μg of recombinant protein, mixed, and emulsified as described above with incomplete Freund's adjuvant (Sigma) at 4-week intervals. Sera were collected 2 weeks following the final immunization. The University of Arkansas for Medical Sciences (UAMS) is accredited by the International Association for Assessment and Accreditation of Laboratory Animals Care (AAALAC), and all immunization protocols were approved by the UAMS Institutional Animal Care and Use Committee (IACUC).
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses were performed as previously described (16). A volume of cell lysate equivalent to 2 × 107 bacteria was loaded in each gel lane of an SDS-12.5% PAGE gel and separated by electrophoresis. The electrophoresed proteins were either stained with Sypro Ruby stain (Bio-Rad) or transferred to nitrocellulose membranes for colorimetric immunoblot detection, using 4-chloro-1-naphthol as the substrate. For chemiluminescence detection with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific), proteins were transferred to polyvinylidene difluoride (PVDF) membranes. Antibodies and antisera used for detection of FlaB, BB0796, OspA, DbpA, and OspC were previously described (16, 22, 23). BB0238-specific antiserum production is described above. The molecular mass standard used for all immunoblots was the All Blue Precision Plus marker (Bio-Rad). Bio-Rad Low Range Unstained SDS-PAGE standards were used for Sypro Ruby-stained SDS-PAGE gels.
Infection of mice by needle inoculation.
All murine infection experiments were approved by the UAMS institutional IACUC. Four- to 5-week-old C3H/HeN mice (Harlan) were used for mouse infection studies. Four- to 5-week-old CBYSmn.CB17-Prkdcscid/J mice (Jackson Laboratories, Bar Harbor, ME) were used for immunocompromised scid mouse infection studies. Clones were grown to the mid-log phase with antibiotic selection, at which time the bacterial density in each culture was enumerated using dark-field microscopy and diluted appropriately. Mice were infected via intradermal injection in the sternal region of the thorax as previously described (16). To determine the 50% infectious dose (ID50) for Bb297, BbΔbb0238, and Bbbb0238C, mice were injected with serial 10-fold dilutions containing 102 to 105 bacteria. For all other infection studies, a dose of 105 bacteria was used. At 2 weeks postinfection, tissue samples (i.e., ear punch, back skin, brachial and inguinal lymph nodes, heart, tibiotarsal tissue, and bladder) were harvested and cultured in BSK-II medium supplemented with Borrelia antibiotic mixture (BAM) (Monserate). Additional heart, tibiotarsal, and back skin samples were harvested and frozen for qPCR and quantitative real-time PCR (qRT-PCR) analysis (see below). Heart and knee tissues were also collected for histopathology (see below). Cultures containing tissue samples were examined by dark-field microscopy for the presence of spirochetes 1 to 2 weeks after collection. Aliquots of cultures with positive growth were passed to BSK-II supplemented with BAM and confirmed to contain the respective genotype of the isolated bacteria.
Histological assessment.
Heart and knee tissues were fixed in 10% neutral buffered formalin, paraffin embedded, routinely processed, and stained with hematoxylin and eosin (H&E). Knees were decalcified in 5% formic acid prior to processing. A blind histological assessment was then performed on stained tissue sections to evaluate the presence and severity of carditis and arthritis in each mouse (24). Sagittal sections through the heart, which included the aorta, were scored for severity of inflammation (0 [absent] to 4 [marked]), aortic involvement (0 [present] or 1 [absent]), pericardial inflammation (0 [present] or 1 [absent]), and endocardial inflammation (0 [present] or 1 [absent]). Sagittal sections through the knee were scored for severity of inflammation (0 [absent] to 4 [marked]), synovial proliferation (0 [present] or 1 [absent]), significance of sheath thickening (0 [absent] to 4 [marked]), and significance of involvement of adjacent tissues (0 [absent] to 4 [marked]). Individual criterion scores were then totaled for a maximum cumulative score of 7 or 13 for carditis and arthritis, respectively.
qPCR analyses of spirochete tissue burden.
DNA was extracted from back skin, heart, and tibiotarsal samples from infected mice as previously described by Maruskova et al. (20). DNA was extracted using a High Pure PCR template preparation kit (Roche Applied Sciences, Indianapolis, IN) according to the manufacturer's instructions, except that 200 μg of collagenase (5 mg/ml stock; Sigma) was added during lysis. Purified DNA was analyzed via qPCR on a StepOnePlus Real Time PCR System using TaqMan Fast Advanced master mix (Life Technologies). The primer set and probe used to detect flaB were FlaB-ABI-F, FlaB-ABI-R, and FlaB-ABI-Probe. The primer set and probe used to detect the β-actin gene were Act-ABI-F, Act-ABI-R, and Actin-ABI-Probe. The flaB and β-actin probes were labeled on the 5′ end with 6-carboxyfluorescein (FAM) and MAX, respectively (Integrated DNA Technologies, Coralville, IA). To quench the probe fluorophores, probes were also labeled at the 3′ end with Iowa Black FQ and internally quenched with ZEN (Integrated DNA Technologies). The number of flaB copies was used to quantify spirochetal burdens based on a copy number standard curve using the pJSB568 flaB/β-actin vector as a standard. Bacterial burdens were reported as numbers of flaB copies per 106 copies of mouse β-actin.
Cultivation of B. burgdorferi for in vitro analyses.
For growth curve analyses, cultures of Bb297, BbΔbb0238, and Bbbb0238C were inoculated to an initial density of 1 × 103 bacteria/ml in BSK-II medium containing appropriate antibiotic selection. Beginning at 3 days postinoculation, culture densities were determined daily by enumerating spirochetes using dark-field microscopy; 10 fields were counted per culture, and analyses were performed in duplicate. For room temperature growth curves, samples were inoculated at 1 × 103 bacteria/ml, left at room temperature, and counted once a week. BbiEbfC was grown as described above, except cultures were divided at 4 days postinoculation and 1 mM IPTG was added to one portion of the culture. BbiEbfC cultures were then grown for 48 h posttreatment, and cells were collected for analysis.
Transcriptional analysis of bb0238.
Total RNA was isolated from in vitro-cultivated bacteria or murine tissues using TRIzol reagent (Life Technologies) according to the manufacturer's protocol. RNA was purified and DNase treated using the RNeasy minikit and RNase-free DNase I, respectively (Qiagen Inc., Valencia, CA). The presence of contaminating DNA was tested for amplification using TaKaRa EmeraldAmp GT PCR master mix (Clontech) and primers specific for flaB.
For qRT-PCR analyses, purified RNA was converted to cDNA using an iScript cDNA synthesis kit according to the manufacturer's instructions. qRT-PCR analyses were performed with the StepOnePlus Real Time PCR System using TaqMan Fast Advanced master mix (Life Technologies). Control reactions prepared without reverse transcriptase were included in the analysis. The primer set and probe used to detect flaB transcripts were FlaB-F, FlaB-R, and FlaB-Probe. The primer set and probe used to detect the bb0238 transcripts were BB0238-F, BB0238-R, and BB0238-Probe. The primer set and probe used to detect ospC transcripts were OspC-F, OspC-R, and OspC-Probe. The probes were labeled at the 5′ and 3′ ends with FAM and Black Hole Quencher-1, respectively (Integrated DNA Technologies).
For quantitation of ebfC transcripts, qRT-PCR analyses were performed using Power SYBR green (Life Technologies). The primer set used to detect ebfC was EbfC-ABI-F and EbfC-ABI-R, and flaB was detected using primers FlaB-ABI-F and FlaB-ABI-R. flaB transcripts were used as an internal control to normalize qRT-PCR data, and differential expression was determined using the 2−ΔΔCT calculation. Results are expressed relative to flaB.
To identify the transcriptional start site (TSS) of bb0238 in Bb297, 5′ rapid amplification of cDNA ends (RACE) was performed using the 5′ RACE system v2.0 (Life Technologies) according to the manufacturer's directions. bb0238-specific primers 5′ RACE-GSP1 and 5′ RACE-GSP2 were used for cDNA synthesis and amplification, respectively (see Table S1 in the supplemental material). Amplified fragments were cloned into pGEM-T Easy, and single clones were sequenced with vector-specific primers to determine the TSS.
Reverse transcription-PCR (RT-PCR) was used to evaluate transcriptional linkage of bb0238 and bb0239. The iScript cDNA Synthesis kit (Bio-Rad) was used to synthesize cDNA from Bb297 RNA via random priming. A control reaction mixture lacking reverse transcriptase was also included. cDNA was used as the template for primer pairs corresponding to the bb0238 ORF (5′ BB0238-Diag and 3′ BB0238-Diag) and the bb0238-bb0239 intergenic region (5′ BB0238-link and 3′ BB0238-link) (see Table S1 in the supplemental material). Each PCR with cDNA was accompanied by a positive-control reaction that utilized the same set of primers and Bb297 gDNA. All PCR products were analyzed by electrophoresis on a 0.8% agarose gel stained with ethidium bromide.
The Ambion NorthernMax-Gly kit (Life Technologies) was used for Northern blot analyses; 5 μg of denatured RNA from Bb297 and BbΔbb0238 was loaded per lane, electrophoresed, and transferred to an Ambion BrightStar-Plus positively charged nylon membrane (Life Technologies) according to the manufacturer's instructions. To generate Northern hybridization probes, the DNA fragments corresponding to flaB and bb0238 were excised from pJSB641 and pJSB643, respectively, using EcoRI and gel purified. The fragments were labeled with digoxigenin (DIG) via random priming using the DIG High Prime DNA labeling and detection starter kit II (Roche) according to the manufacturer's directions. Hybridization was performed according to the conditions provided in the NorthernMax-Gly kit, and detection was carried out according to the protocol outlined in the DIG High Prime DNA labeling and detection starter kit II. The DIG-labeled RNA Molecular Weight Marker I, 0.3 to 6.9 kb (Roche), was included as a standard on all Northern blots.
Subcellular localization studies.
To evaluate localization of BB0238 to the bacterial surface, proteinase K accessibility assays were performed as previously described (25). In brief, 2 × 109 spirochetes were incubated in PBS with 5 mM MgCl2 (PBS-MgCl2) in the absence or presence of 400 μg/ml proteinase K (Life Technologies) for 60 min. Digestion was stopped using 5 mM phenylmethylsulfonyl fluoride (Thermo Scientific). Cells were then prepared for SDS-PAGE and immunoblot analysis for BB0238, FlaB (periplasmic control), and OspC (outer membrane control). To determine whether BB0238 was intrinsically resistant to proteinase K degradation, proteinase K accessibility assays were repeated in the presence and absence of 0.05% Triton X-100 (Promega).
Immunofluorescence assays (IFA) were also used to evaluate surface accessibility of BB0238 on the spirochete. For IFA with permeabilized (fixed) cells, bacteria were prepared as described by Revel et al. (7). Bb297 or BbΔbb0238 spirochetes were collected at mid-logarithmic growth phase, washed with PBS-MgCl2, and resuspended in PBS-MgCl2 to 1.5 × 107 spirochetes/ml. Cells were spotted on a CSS-100 silylated slide (CEL Associates, Pearland, TX) and allowed to air dry. The slides were fixed at 65°C, followed by incubation in acetone. The fixed cells were washed three times in cold PBS-MgCl2 and incubated with blocking buffer (PBS-MgCl2 with 0.05% Tween 20 and 5% bovine serum albumin [BSA]). The slides were then washed with cold PBS-MgCl2 and incubated with primary antibodies recognizing FlaB, OspA, and BB0238 diluted in blocking buffer at 1:1,000, 1:1, and 1:500, respectively. Finally, the slides were washed with cold PBS-MgCl2 and incubated with Alexa Fluor-conjugated secondary antibodies diluted 1:600 in blocking buffer (i.e., Alexa Fluor 488 anti-rat, Alexa Fluor 594 anti-mouse, and Alexa Fluor 647 anti-chicken; Life Technologies). The slides were then washed with cold PBS-MgCl2, and the coverslip was affixed with Mowiol (Sigma) and prepared for imaging. Nonpermeabilized (unfixed) cells were prepared as previously described (25). Briefly, 5 × 107 spirochetes of Bb297 or BbΔbb0238 were resuspended in 100 μl of blocking buffer (PBS-MgCl2 with 10% fetal bovine serum [FBS] [Life Technologies]). Primary antibodies were applied to the cells as described above and incubated at room temperature. The cells were washed with blocking buffer, resuspended in blocking buffer, and spotted on a silylated slide. The cells were fixed with PBS-MgCl2 containing 4% formaldehyde, washed, and incubated with blocking buffer containing secondary antibodies at the concentrations listed above. The slides were washed and fixed with PBS-MgCl2 containing 4% formaldehyde, and the coverslip was affixed with Mowiol. Fluorescence microscopy was performed using a Ti-U microscope with a 60× oil immersion objective, and images were taken using a D5-QilMc digital camera (Nikon, Melville, NY).
To assess localization of BB0238 to the bacterial membrane, Triton X-114 phase partitioning was performed as described by Brooks et al. (26). Protein in the detergent (i.e., membrane) and aqueous fractions were precipitated, prepared for SDS-PAGE, and transferred to nitrocellulose and PVDF membranes. The presence of BB0238, OspC, and BB0796 in each fraction was determined by immunoblot analysis.
Additional whole-cell fractionation was performed as previously described (27, 28) to further separate the protoplasmic cylinder from the outer membrane and periplasmic fractions. In brief, 5 × 109 spirochetes were washed, resuspended in PBS with 1% Triton X-114, and incubated overnight at 4°C. Centrifugation at 20,000 × g for 30 min at 4°C resulted in protoplasmic cylinder isolation; the supernatant was then warmed to 37°C and centrifuged at 15,000 × g for 15 min at room temperature. The aqueous upper phase was separated from the lower, detergent phase, and the two phases were washed. The protoplasmic cylinders and the aqueous (periplasmic) and detergent (outer membrane) fractions were then prepared for SDS-PAGE and immunoblot analyses.
Proteomic analysis of native BB0238.
Gel electrophoresis combined with liquid chromatography-tandem mass spectrometry (GeLC–MS-MS) analysis of SDS-PAGE protein bands was performed as previously described (29). Volumes of cell lysate equivalent to 5 × 107 bacteria for Bb297 or BbΔbb0238 were loaded in a gel lane of a 12.5% SDS-PAGE gel in duplicate. One portion of the gel was transferred to nitrocellulose, and BB0238 was detected by immunoblotting as described above. The second portion was stained with GelCode Blue Stain Reagent (Thermo Scientific) and prepared for proteomic analysis. To generate gel slices, the PAGE gel was overlaid on the immunoblot to localize the regions of the lanes that corresponded to BB0238. Peptides and proteins were identified from tandem MS (MS-MS) spectra by database searching using the Mascot search engine (Matrix Science, Boston, MA).
Statistical analyses.
The ID50 values for Bb297, BbΔbb0238, and Bbbb0238C were calculated as described previously (30). Comparisons between the clones' ID50 values were made by using a probit regression model, where we assumed identical slopes in the response/log-dose relationship but different intercepts for each clone. Graphically, they manifest as dose-response curves with lateral shifts corresponding to the changes in intercept. Additionally, an overdispersion parameter was fitted to accommodate greater homogeneity in infection rates than would normally be permitted by the model.
Tissue infection rates were analyzed using Fisher's exact tests. Analysis of variance (ANOVA) models were used to compare clones with respect to qPCR measurements, and Tukey's procedure was used to perform pairwise comparisons. Student unpaired t tests were used to analyze qRT-PCR measurements. Finally, histopathologic scores were compared using Wilcoxon rank sum tests with adjustments for multiple comparisons made using Sime's procedure. For all analyses, P values of less than 0.05 were considered statistically significant. All analyses were performed using R software version 3.0.1 (R Foundation for Statistical Computing, Vienna, Austria) and SAS 9.3 (SAS Institute Inc., Cary, NC).
RESULTS
Inactivation of bb0238 in B. burgdorferi.
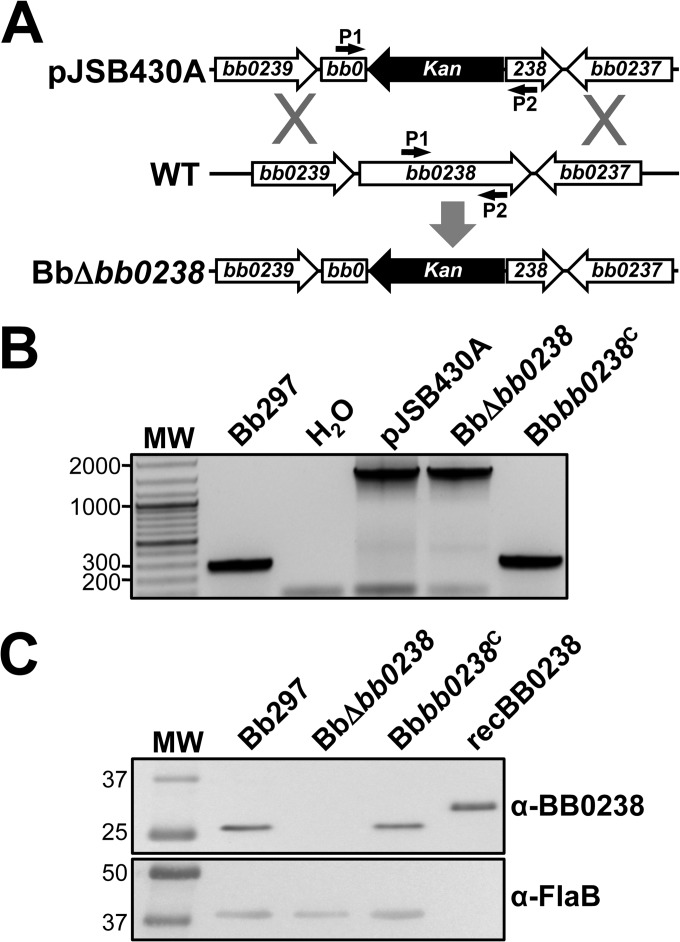
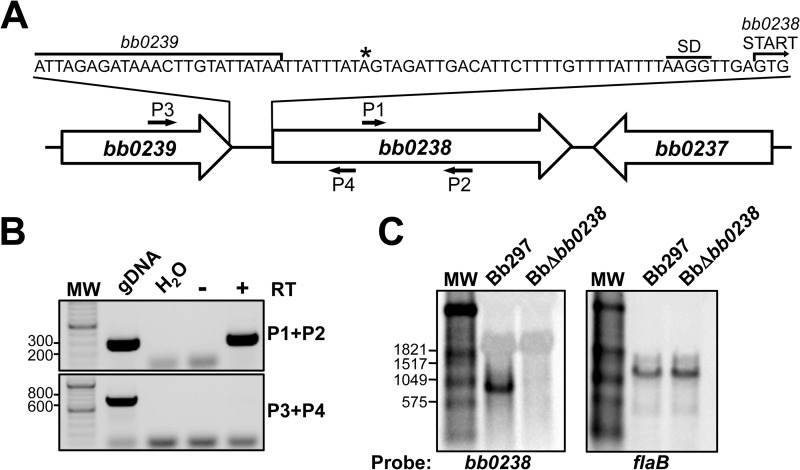
bb0238 encodes a hypothetical protein on the chromosome of B. burgdorferi (6). BB0238 is approximately 30.4 kDa in size (i.e., 262 amino acids) and is predicted to contain a TPR structural motif. While a BLASTP search identified homologs of BB0238 in both Lyme disease (99 to 90% identity) and relapsing fever (54 to 56% identity) Borrelia spp., no homologs were identified in other bacterial species (31). To evaluate the contribution of this novel protein during mammalian infection, allelic exchange was used to create a bb0238-deficient mutant in which a portion of the bb0238 ORF was replaced with a PflgB-aphI resistance marker. Wild-type Bb297 was electroporated with pJSB430A, a suicide vector containing regions flanking bb0238 with a Kanr cassette interrupting the ORF (Fig. 1A). Kanr transformants were recovered, integration of PflgB-aphI into bb0238 was confirmed via PCR (Fig. 1B), and PCR-based plasmid profiling confirmed the endogenous plasmid content of BbΔbb0238 (see Fig. S1 in the supplemental material). The BbΔbb0238 mutant failed to produce BB0238 protein when whole-cell lysates were probed with BB0238-specific rat antisera (Fig. 1C). To restore bb0238 expression in BbΔbb0238, the mutant was transformed with pJSB445A, a shuttle plasmid that contained bb0238, the 420-bp upstream region with the putative bb0238 promoter, and a PflgB-aadA Strepr resistance marker. PCR confirmed that the Bbbb0238C clone contained an intact bb0238 ORF (Fig. 1B) and that its endogenous plasmid profile was similar to that of Bb297 (see Fig. S1 in the supplemental material). Complementation of BbΔbb0238 with pJSB445A restored production of the BB0238 protein (Fig. 1C), suggesting that the 420-bp region upstream of the ORF contains the bb0238 promoter.
FIG 1.
Generation and confirmation of BbΔbb0238 and the Bbbb0238C complemented clone. (A) Diagram illustrating the approach to inactivating bb0238 in Bb297. pJSB430A, relevant region of the bb0238::Kan mutagenesis construct; WT, bb0238 and flanking regions from the B. burgdorferi chromosome; BbΔbb0238, genomic arrangement of the bb0238::Kan mutant. The small arrows denote the relative positions of the bb0238 P1 and P2 diagnostic primer pair. (B) PCR confirmation of the BbΔbb0238 mutant and complemented Bbbb0238C. PCR with primers P1 and P2 results in different amplicon sizes, 287 bp (wild type) and 1,475 kb (bb0238::Kan). Bb297, wild-type B. burgdorferi; H2O, negative control for amplification; pJSB430A, bb0238::Kan suicide vector used to interrupt bb0238. DNA size standards (lane MW), indicated on the left, are shown in kilobases. (C) Immunoblot analysis of BB0238 and FlaB in Bb297, BbΔbb0238, and Bbbb0238C. Antibodies used to detect the respective proteins are indicated on the right. recBB0238 denotes the lane containing His6-tagged BB0238 recombinant protein used to generate antisera. The relevant molecular masses (kDa) of the standard (lane MW) are shown on the left.
To assess total protein differences in Bb297 and BbΔbb0238, whole-cell lysates were separated by SDS-PAGE and stained with Sypro Ruby. Although two low-molecular-mass bands (i.e., <31 kDa) appeared to be altered between the parent strain and the bb0238 mutant (see Fig. S2 in the supplemental material), the comparison revealed no major global differences in protein expression profiles. The identities of the proteins represented by these two bands are currently unknown, but the upper band, which has an approximate mass of 30 kDa and was present in Bb297, may possibly represent BB0238. In vitro growth curve analyses showed that BbΔbb0238 grew similarly to Bb297 at 37°C, and no gross morphological defects were noted when BbΔbb0238 was visualized by dark-field microscopy (data not shown). Additionally, swarm plate motility assays suggested that BbΔbb0238 exhibited no general motility defects (data not shown).
Requirement for bb0238 during infection of mice.
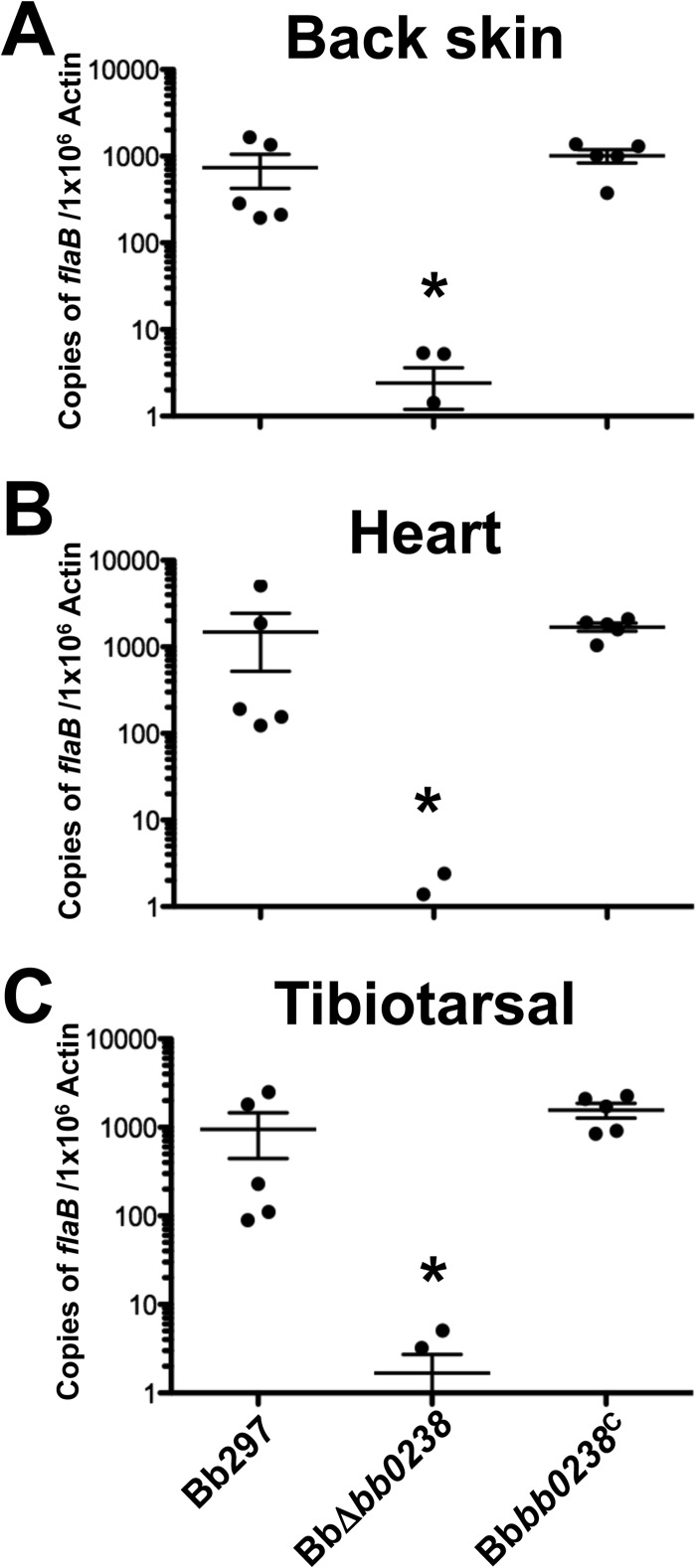
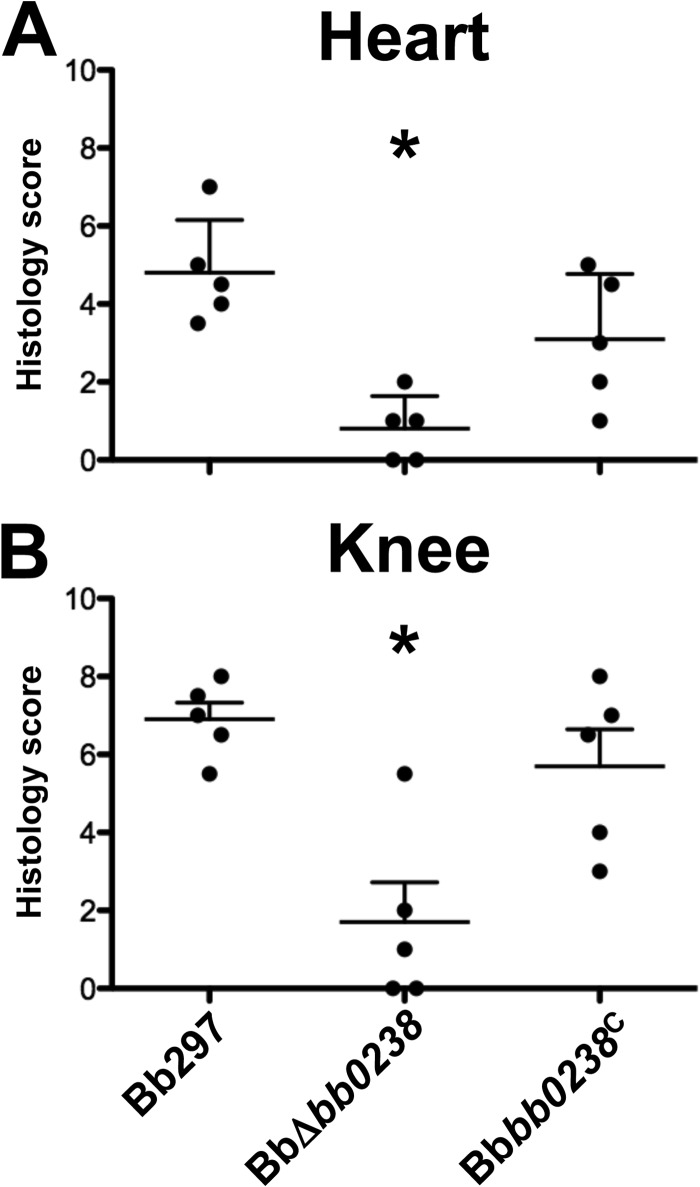
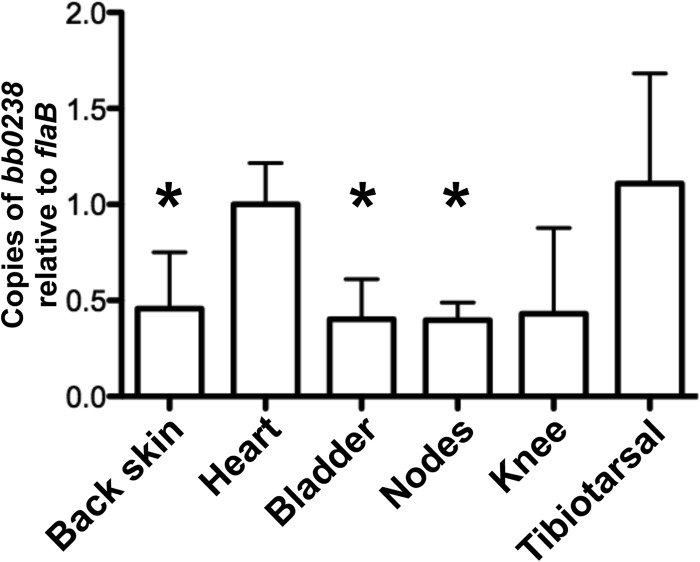
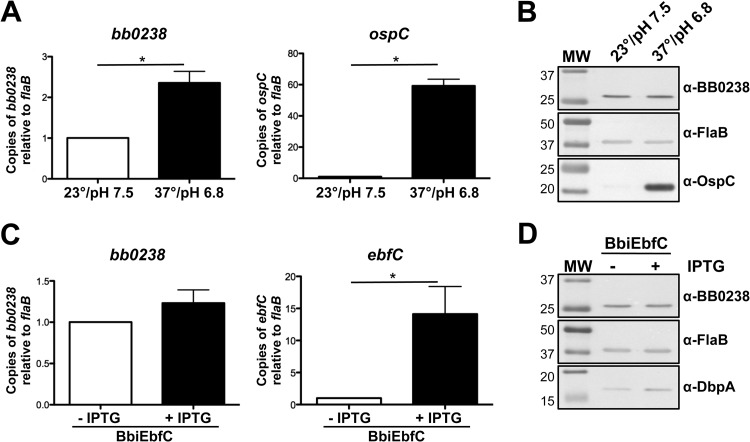
To determine the contribution of BB0238 to B. burgdorferi infection and pathogenesis, C3H/HeN mice were challenged via intradermal needle inoculation with increasing concentrations of in vitro-cultivated Bb297, BbΔbb0238, or Bbbb0238C (i.e., serial 10-fold dilutions from 102 to 105 bacteria). Ear punch, back skin, heart, bladder, tibiotarsal tissue, and lymph nodes were collected from mice at 2 weeks postinfection, and the tissues were cultured in BSK-II medium. The cultures were then examined by dark-field microscopy for the presence of spirochetes. Mice infected with BbΔbb0238 demonstrated a significant reduction in total numbers of culture-positive tissues, and this defect was not readily overcome by increasing the inoculum (Table 2). Interestingly, bacteria could not be recovered from ear punch biopsy specimens or heart tissues isolated from any of the BbΔbb0238-infected mice. BbΔbb0238 also exhibited a defect in its capacity to infect tibiotarsal and bladder tissues, although this deficiency was partially alleviated upon inoculation of mice with higher doses of BbΔbb0238 (a maximum infection rate of 30% was observed at doses of 104 or 105 spirochetes). The infection rates in mice challenged with Bbbb0238C were comparable to those of Bb297, suggesting the attenuation noted in BbΔbb0238 was specifically due to lack of BB0238. The ID50 of the BbΔbb0238 mutant was 1,507 spirochetes, which was comparable to those of Bb297 and Bbbb0238C (i.e., 378 and 234 bacteria, respectively) (Table 2). However, this calculation was based on the number of infected mice and does not reflect the tissue-specific infection defect observed in mice challenged with BbΔbb0238. To further assess the infectivity phenotype of BbΔbb0238, a subset of tissue samples were analyzed via qPCR to measure spirochetal burdens in BbΔbb0238-infected tissues (Fig. 2). qPCR results, reported as copies of flaB per 106 copies of mouse actin, showed significantly reduced spirochetal burdens in tissues from mice infected with BbΔbb0238, even in culture-positive samples, compared to tissues isolated from mice infected with Bb297 and Bbbb0238C. In agreement with these findings, histological scoring of heart and knee tissues also showed that the severity of carditis and arthritis, respectively, was significantly reduced in tissues collected from mice infected with BbΔbb0238 (Fig. 3). To confirm that bb0238 is expressed during mammalian infection, five mice were infected with Bb297 at a dose of 105 bacteria, and total RNA was extracted from tissues at 14 days postinfection. bb0238 transcripts, which were quantified using qRT-PCR and normalized to flaB (Fig. 4), were found to be similar between the six tissues tested. Although the elevated levels of bb0238 in bacteria colonizing the heart and tibiotarsal tissue suggested that effective infection of these sites may require bb0238, the increase was relatively modest (i.e., only 2-fold higher) compared to other tissues.
TABLE 2.
Infectivity of the BbΔbb0238 mutant in C3H/HeN mice
| Strain and dose (spirochetes) | No. of positive cultures/total |
No. of infected mice/total | ID50 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ear punch | Back skin | Lymph nodesa | Heart | Tibiotarsal tissue | Bladder | All sites | |||
| Bb297 | |||||||||
| 102 | 1/10 | 1/10 | 1/10 | 1/10 | 2/10 | 1/10 | 7/60 | 2/10 | |
| 103 | 5/10 | 6/10 | 7/10 | 7/10 | 9/10 | 7/10 | 41/60 | 9/10 | 378b |
| 104 | 7/10 | 8/10 | 10/10 | 8/10 | 10/10 | 10/10 | 53/60 | 10/10 | |
| 105 | 7/10 | 10/10 | 10/10 | 7/10 | 10/10 | 10/10 | 54/60 | 10/10 | |
| BbΔbb0238 | |||||||||
| 102 | 0/10 | 2/10 | 2/10 | 0/10 | 1/10 | 1/10 | 6/60 | 2/10 | |
| 103 | 0/10e | 4/10 | 4/10 | 0/10e | 2/10e | 0/10e | 10/60e | 4/10 | 1507c,e |
| 104 | 0/10e | 8/10 | 8/10 | 0/10e | 3/10e | 2/10e | 21/60e | 9/10 | |
| 105 | 0/10e | 6/10 | 9/10 | 0/10e | 2/10e | 3/10e | 20/60e | 10/10 | |
| Bbbb0238C | |||||||||
| 102 | 1/10 | 1/10 | 1/10 | 1/10 | 1/10 | 1/10 | 6/60 | 1/10 | |
| 103 | 3/10 | 4/10 | 4/10 | 4/10 | 7/10 | 3/10 | 25/60 | 7/10 | 234d |
| 104 | 9/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 59/60 | 10/10 | |
| 105 | 7/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 57/60 | 10/10 | |
A single brachial and inguinal lymph node were collected from each mouse and cultured together.
95% confidence interval (CI); range, 148 to 971 spirochetes.
95% CI; range, 617 to 3,690 spirochetes.
95% CI; range, 91 to 607 spirochetes.
Statistically significant difference relative to Bb297 (P < 0.05).
FIG 2.
Spirochete burdens in tissues from mice infected with BbΔbb0238. Tissues were collected 2 weeks postinfection from mice needle inoculated with Bb297, BbΔbb0238, or Bbbb0238C at a dose of 105 spirochetes. DNAs from tissue samples were analyzed in triplicate by qPCR to measure spirochete burdens. The results from back skin, heart, and tibiotarsal tissue are representative of a single infection experiment; n = 5 mice for each strain. The error bars represent standard errors of the mean (SEM). Statistical significance was determined by ANOVA and Tukey's procedure. The asterisks indicate a statistically significant difference relative to Bb297 or Bbbb0238C (P < 0.05).
FIG 3.
Histological evaluation of infected tissues. Heart and knee samples were collected 2 weeks postinfection from mice needle inoculated with Bb297, BbΔbb0238, and Bbbb0238C at a dose of 105 spirochetes. A blind histological assessment was performed, and disease severity scores from heart and knee are shown. The scores are representative of a single infection experiment; n = 5 mice for each strain. The error bars represent SEM. Statistical significance was determined using a Wilcoxon rank sum test with adjustments using Sime's procedure. The asterisks indicate a statistically significant difference relative to Bb297 or Bbbb0238C (P < 0.05).
FIG 4.
Expression of bb0238 by spirochetes in infected murine tissues. RNA was isolated from tissues collected 2 weeks postinfection from mice needle inoculated with Bb297 at a dose of 105 spirochetes. Samples were analyzed in triplicate by qRT-PCR to quantify bb0238 expression. The results are representative of a single infection experiment; n = 5 mice. The error bars represent SEM. Statistical significance was determined using an unpaired Student t test. The asterisks indicate a statistically significant difference relative to heart or tibiotarsal tissue (P < 0.05).
It is well established that B. burgdorferi must evade the adaptive immune response to infect and disseminate in mice (32–38). To assess whether the attenuation of BbΔbb0238 is due to the mutant's inability to avoid clearance by adaptive immunity, scid mice were challenged via intradermal needle inoculation with 105 spirochetes of Bb297, BbΔbb0238, or Bbbb0238C. At 2 weeks postinoculation, tissues (i.e., ear punch, back skin, heart, bladder, and tibiotarsal) were isolated from infected mice and cultured in BSK-II. Examination of cultures inoculated with tissues from the scid mice infected with BbΔbb0238 revealed a reduction in culture-positive tissues (Table 3) comparable to that observed with BbΔbb0238-infected C3H/HeN mice. Though all of the scid mice inoculated with BbΔbb0238 were infected, similar tissue-specific trends in attenuation were observed in C3H/HeN mice. Specifically, all of the ear punch cultures were negative, while only one heart was culture positive. In contrast to the results observed with the C3H/HeN mice, viable spirochetes were not recovered from any of the back skin cultures from the scid mice. In addition, BbΔbb0238 was able to infect the tibiotarsal tissues of scid mice more efficiently than those of C3H/HeN mice. The mice challenged with Bbbb0238C were fully infected, confirming that the defect noted in BbΔbb0238-infected mice is due to the lack of bb0238. Given that comparable reductions in infectivity were observed in both C3H/HeN and scid mice challenged with BbΔbb0238, the attenuation of BbΔbb0238 does not appear to be solely due to the inability of the BbΔbb0238 spirochetes to avoid clearance via the adaptive immune response.
TABLE 3.
Infectivity of the BbΔbb0238 mutant in scid mice
| Strain | No. of positive cultures/total |
No. positive mice/total | |||||
|---|---|---|---|---|---|---|---|
| Ear punch | Back skin | Heart | Tibiotarsal tissue | Bladder | All sites | ||
| Bb297 | 5/5 | 3/5 | 5/5 | 5/5 | 3/5 | 21/25 | 5/5 |
| BbΔbb0238 | 0/5a | 0/5 | 1/5a | 5/5 | 1/5 | 7/25a | 5/5 |
| Bbbb0238C | 5/5 | 3/5 | 5/5 | 5/5 | 5/5 | 23/25 | 5/5 |
Statistically significant difference relative to Bb297 (P < 0.05).
Characterization of bb0238 gene architecture.
Published annotation for bb0238 provided limited information regarding the putative function of BB0238. However, the genes adjacent to bb0238, bb0237 and bb0239, are predicted to encode an apolipoprotein, N-acyltransferase, and a deoxyguanosine/deoxyadenosine kinase, respectively (6). Since bb0238 and bb0237 are convergently transcribed, it is likely they do not comprise an operon (Fig. 5A). bb0239 and bb0238, on the other hand, are oriented in the same direction on the chromosome with a relatively small intergenic region (45 bp), suggesting they may be cotranscribed. This is especially relevant considering that bb0239 is predicted to be involved in B. burgdorferi purine salvage, and this pathway is essential for mammalian infection (39–41).
FIG 5.
Analysis of the bb0238 transcript. (A) Diagram illustrating bb0238 linkage primer sites and TSS mapped by 5′ RACE. The small arrows denote the relative positions of bb0238 diagnostic primers (P1 and P2) and bb0238-bb0239 linkage primers (P3 and P4). The asterisk denotes the TSS predicted by 5′ RACE. (B) RT-PCR results with the bb0238 internal control primers (P1 and P2) and linkage primers spanning the bb0238 and bb0239 intergenic region (P3 and P4). gDNA, Bb297 DNA; H2O, negative control for amplification; for Bb297 RNA, − and + designate the reactions prepared without and with reverse transcriptase (RT), respectively. DNA size standards (lane MW), indicated to the left of the panel, are shown in base pairs. (C) Northern blot analysis of Bb297 and BbΔbb0238 RNAs. The probes and transcripts detected are indicated beneath the gels. Relevant sizes of the DIG-labeled RNA standard (lanes MW) are shown on the left in base pairs.
RT-PCR with RNA extracted from Bb297 was used to determine whether bb0239 and bb0238 comprised an operon. Primers that amplified a section internal to the bb0238 ORF were employed, as well as a primer set that spanned the intergenic region (Fig. 5B). The bb0238 internal primers generated an amplicon of the appropriate size, confirming the presence of bb0238 transcripts, while the primer set spanning the bb0239-bb0238 intergenic region did not yield a PCR product. Northern blot analysis was used to confirm the RT-PCR results and the size of the bb0238 mRNA. Total RNA was extracted from Bb297 and BbΔbb0238 and hybridized with DIG-labeled probes recognizing flaB or bb0238 transcripts. flaB transcripts were detected in both strains (Fig. 5C) at the correct size of ∼1,100 bp (42, 43), thus confirming equal loading of sample. bb0238 mRNA was detected only in Bb297, confirming the specificity of the probe. The bb0238 transcript migrated at a size slightly smaller than the 1,059-bp band in the molecular weight standard. This agrees with an estimated size of a monocistronic bb0238 transcript of ∼800 bp. Finally, 5′ RACE was utilized to identify the transcription start site for the bb0238 gene. Using a bb0238-specific primer that recognized a region 525 bp downstream of the start codon, 33 clones were isolated and sequenced. The amplified fragments from 16 of these 33 clones stopped within the bb0238 ORF, suggesting they arose from premature termination of the extension reaction. Of the remaining 17 clones that mapped to a region upstream of the bb0238 ORF, 12 mapped to a common TSS that was 37 bp upstream from the predicted start codon (Fig. 5A). The start sites in the remaining five fragments did not map to a common consensus residue. Based on this predicted start site, the bb0238 transcript would be approximately 830 bp in length. Taken together, these data indicate that the promoter for bb0238 lies within the predicted deoxyguanosine/deoxyadenosine gene bb0239, but these two genes are not operonic. Furthermore, these findings, in conjunction with the genetic complementation studies described above, confirm that the attenuated-infection phenotype observed in BbΔbb0238 is not likely due to a polar mutational effect on bb0239 that arose from the insertion of the resistance marker in this mutant.
In vitro characterization of bb0238 regulation.
To determine whether bb0238 expression might differ during the B. burgdorferi enzootic life cycle, wild-type Bb297 was cultured in vitro under conditions that mimic the conditions with the mammal/fed tick (pH 6.8 and 37°C) and unfed tick (pH 7.5 and 23°C) (44–47). bb0238 expression was analyzed via qRT-PCR (Fig. 6A) and immunoblotting (Fig. 6B). Although there was a modest change in bb0238 mRNA expression under the two conditions, there was no demonstrable difference in the levels of BB0238 protein. To date, only one other study that described differential regulation of bb0238 has been published. In this study, the authors detected an increase in bb0238 transcription by comparative RNA sequencing when the EbfC (YbaB) nucleoid-associated DNA-binding protein was overproduced (48). To confirm these results, the borrelial IPTG-inducible expression vector was adapted to express ebfC by replacing the rrp2 ORF in piRrp2(Kan) with the ebfC ORF to create pJSB499. pJSB499 was transformed into Bb297, and the transformants, designated BbiEbfC, were recovered. A concentration of 1 mM IPTG was selected for induction because growth curve analyses determined that treatment of cultures of BbiEbfC with this concentration did not adversely affect growth (data not shown). Samples from the induced and uninduced BbiEbfC cultures were analyzed via qRT-PCR and immunoblotting for changes in expression. As expected, qRT-PCR showed over 10-fold increase in ebfC transcription (Fig. 6C). However, there was no significant increase in bb0238 expression (Fig. 6C) or BB0238 protein levels (Fig. 6D). Prior studies showed that dbpBA transcription was elevated when ebfC was induced (48), and in agreement with these findings, DbpA production was higher in the samples from the IPTG-induced BbiEbfC cultures (Fig. 6D). Taken together, these data suggest that bb0238 is not differentially expressed by B. burgdorferi during its enzootic cycle, nor is bb0238 regulated by ebfC.
FIG 6.
Regulation of bb0238. (A and B) qRT-PCR (A) and immunoblot analysis (B) of bb0238 expression in Bb297 cultivated to mid-exponential growth phase at 23°C, pH 7.5, or grown to the late exponential growth phase at 37°C, pH 6.8. (C and D) qRT-PCR (C) and immunoblot analysis (D) of bb0238 expression in BbiEbfC without IPTG or induced with 1 mM IPTG for 48 h. Detection of OspC protein and ospC transcript was used as a positive control to confirm rpoS activation. Detection of DbpA protein and ebfC transcript was included as a positive control to confirm ebfC expression. Relevant protein molecular masses (lane MW) are shown on the left in kDa. The immunoblot results are representative of two independent experiments. The qRT-PCR results are based on data from three biological replicates. The error bars represent SEM. Statistical significance was determined using an unpaired Student t test. The asterisks indicate a statistically significant difference between the respective conditions (P < 0.05).
Determination of BB0238 cellular localization.
Prediction using SignalP 4.1 suggested the N terminus of BB0238 contains either a transmembrane domain or a signal peptidase I cleavage site between amino acids 26 and 27 (19). Therefore, several assays were employed to determine where BB0238 is localized within the B. burgdorferi cell. First, a proteinase K accessibility assay was conducted to determine if BB0238 is exposed on the bacterial cell surface (Fig. 7A). When intact Bb297 cells were incubated with proteinase K, the OspC positive control was degraded due to surface localization. On the other hand, the FlaB negative control showed little to no degradation due to its protected location within the periplasmic space. Similar to FlaB, BB0238 was not degraded upon incubation of spirochetes with proteinase K. To confirm that BB0238 was not intrinsically resistant to degradation by proteinase K, spirochetes were permeabilized with Triton X-100 and then treated with proteinase K. BB0238, along with OspC and FlaB, were all readily degraded, confirming that BB0238 was sensitive to digestion by proteinase K. These results were supported by microscopic IFAs with fixed (permeabilized) and unfixed (nonpermeabilized) spirochetes (Fig. 7B). The outer surface protein OspA could be detected in assays with both fixed and unfixed cells, while the periplasmic FlaB could be detected only in cells that had been fixed prior to incubation with antibody. BB0238 could not be detected in unfixed cells, suggesting subsurface localization, supporting the proteinase K results. The absence of a signal with BbΔbb0238 cells confirmed the specificity of the BB0238-specific antisera.
FIG 7.
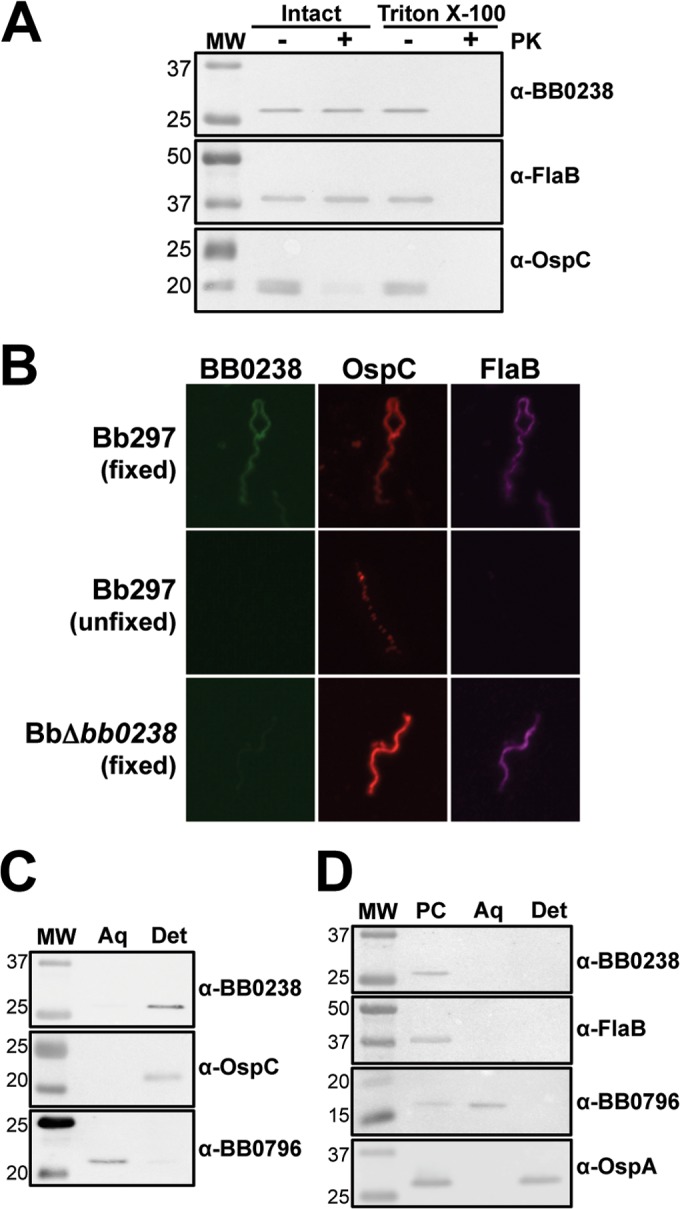
Cellular localization of BB0238. (A) Immunoblot analysis of the proteinase K accessibility assay. Bb297 cells were incubated with or without proteinase K (PK) in the presence or absence of Triton X-100. Antibodies to detect the respective proteins are listed on the right, and the relevant molecular masses (lane MW) are shown on the left in kDa. FlaB, negative control; OspC, positive control. (B) IFA of Bb297 and BbΔbb0238. Fixed, cells were fixed to glass slides by air drying, heated, and then incubated in acetone; the slides were blocked and incubated with primary antibody and then secondary antibody. Unfixed, cells were incubated with primary antibody and then washed and fixed to glass slides with formaldehyde; the slides were then incubated with secondary antibody. Secondary antibodies for detection of BB0238, OspA, and FlaB were anti-rat Alexa Fluor 488, anti-mouse Alexa Fluor 594, and anti-chicken Alexa Fluor 647, respectively. (C) Immunoblot analysis of whole-cell phase partitioning. Bb297 cells were phase separated into aqueous (Aq) and detergent (Det) fractions. Antibodies to detect the respective proteins are listed on the right, and the relevant molecular masses (lane MW) are shown on the left in kDa. OspC was used as a control for the membrane-localized, detergent-enriched fraction, and detection of BB0796 was used as an aqueous control. (D) Immunoblot analysis of cellular fractionation. Bb297 cells were incubated with Triton X-114. The protoplasmic cylinder (PC) was pelleted, and samples were incubated at 37°C for phase separation. The aqueous (Aq) and detergent (Det) fractions were then separated. Antibodies to detect the respective proteins are listed on the right, and the relevant molecular masses (lane MW) are shown on the left in kDa. FlaB was used as a protoplasmic cylinder control, periplasmic BB0796 as an aqueous control, and outer-membrane-localized OspA as a detergent control. The data shown are representative blots from a minimum of two independent experiments.
To further examine localization, BB0238 was analyzed by whole-cell phase partitioning, which separates fractions based on their amphiphilic properties (i.e., membrane localization). Bb297 was sonicated, incubated in Triton X-114, and separated into the detergent (membrane-enriched) and aqueous (non-membrane-enriched) fractions. Immunoblot analysis was then used to detect the presence of the proteins of interest (Fig. 7C), OspC, BB0796, and BB0238. BB0796 is a soluble aqueous-phase control described by Lenhart et al. (23). As expected, OspC, a known outer surface lipoprotein, was found in the detergent fraction, and BB0796 was detected in the aqueous fraction. BB0238 was enriched in the detergent fraction, which suggested association of BB0238 with the membrane. Cellular fractionation was utilized to more finely assess localization to the outer membrane, periplasmic space, or protoplasmic cylinder fractions (inner membrane and cytoplasm) (Fig. 7D). Fractions prepared from Bb297 were analyzed via immunoblot analyses. As expected, OspA was found in the outer membrane and protoplasmic cylinder fractions, and FlaB was detected in the protoplasmic cylinder. Like FlaB, BB0238 was also found in the protoplasmic cylinder fraction, suggesting either inner membrane or cytoplasmic localization. Given that the phase-partitioning data suggest BB0238 is membrane associated but cellular fractionation shows that BB0238 is localized to the protoplasmic cylinder, it is likely that BB0238 is associated with the inner membrane. However, these techniques do not distinguish whether BB0238 is anchored in the inner membrane or secreted into the periplasmic space and associated with the outer leaflet of the inner membrane. In an attempt to distinguish between these two possibilities, the mature BB0238 protein in a whole-cell lysate of Bb297 was analyzed by GeLC–MS-MS. A total of 217 proteins were identified in the excised gel slice, and BB0238 was the protein with the second highest relative abundance in the sample (data not shown). A total of 274 spectra that mapped to BB0238 were identified, with 81% coverage of the protein, but no peptides that mapped to the N-terminal 26 amino acids of the predicted BB0238 ORF were identified (Fig. 8). Since this portion of BB0238 seems to be absent in the mature protein, it is likely that BB0238 is secreted into the periplasm and is physically associated with the outer leaflet of the inner membrane directly or via interaction with another inner-membrane-associated protein.
FIG 8.

Analysis of mature BB0238 in Bb297 by GeLC–MS-MS. The amino acid sequence for BB0238 is shown, with underlining indicating regions of the protein for which peptides were detected. Boldface indicates amino acids that were oxidized or pryomodified. The arrow indicates the signal peptide cleavage site predicted by SignalP. The findings represent the results obtained from analysis of two biological replicates.
Contribution of the putative TPR domain to BB0238 function.
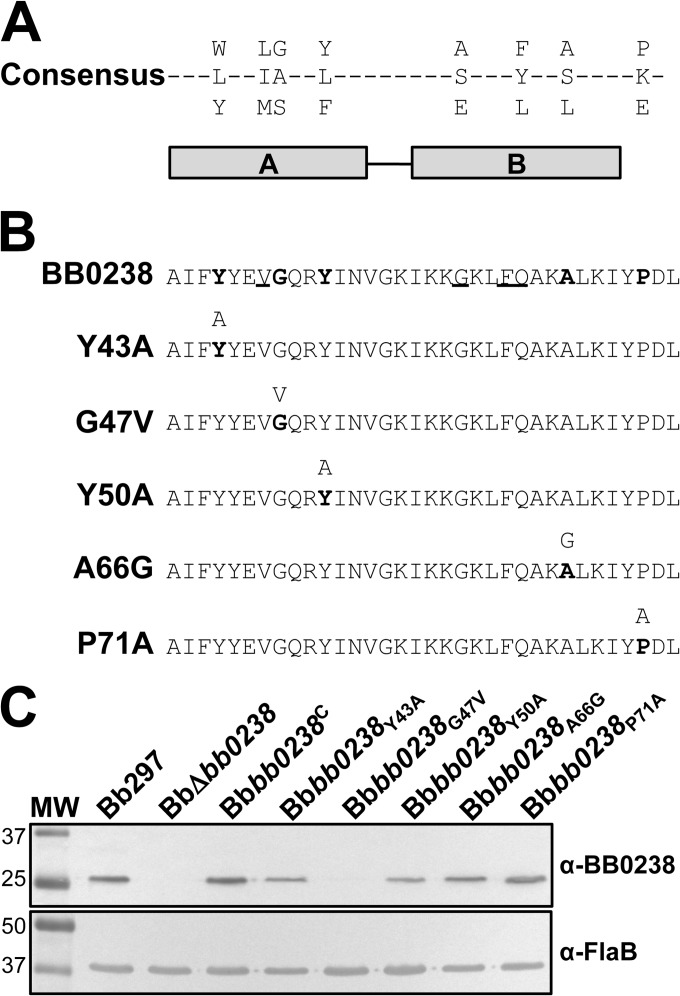
The fact that bb0238 encodes a conserved hypothetical protein with no homology to proteins with known functions provides limited information regarding the mechanism by which it may contribute to mammalian infection. The original annotation of BB0238 on the Comprehensive Microbial Resource website (49) identified a putative TPR domain as the only functional motif within BB0238. The TPR motif is a structural domain that is associated with protein-protein interaction for proteins such as chaperones or protein transporters (13, 50, 51). The TPR domain consists of two α-helices that fold in an antiparallel fashion. Most TPR-containing proteins have multiple TPR domains that combine to create a binding cleft, though proteins with as few as one motif have been identified, as seen in BB0238. TPRpred (52) identified a single TPR domain between amino acids 40 and 73 (P = 7.2 × 10−5). Although the probability that this region functions as a TPR had limited predicted probability (<1%), comparison of the predicted TPR domain in BB0238 with a consensus TPR sequence shows alignment of key residues needed for secondary-structure formation and interaction (Fig. 9A) (13). To determine whether this domain contributed to BB0238 function, five bb0238 mutants were generated with single point mutations of conserved residues in the TPR domain, three residing in helix A (Y43A, G47V, and Y50A) and two in helix B (A66G and P71A) (Fig. 9B). Point mutations were introduced in the TPR domain by overlap extension PCR, and the mutated bb0238 was cloned into the pJSB201 borrelial shuttle vector to generate complementation vectors expressing each mutated ORF under the control of the native bb0238 promoter. Each complementation construct was then transformed into BbΔbb0238, and BB0238 production was evaluated in the mutants by immunoblotting. BB0238 was detectable in Bbbb0238Y43A, Bbbb0238Y50A, Bbbb0238A66G, and Bbbb0238P71A and present at levels comparable to those in Bb297 and Bbbb0238C. Bbbb0238G47V, on the other hand, did not produce BB0238 (Fig. 9C). qRT-PCR showed that bb0238 transcript levels for all TPR mutants were equivalent to those measured in Bbbb0238C (data not shown); therefore, the lack of BB0238 in Bbbb0238G47V is likely due to protein instability.
FIG 9.
Characterization of BB0238 TPR point mutants. (A) Consensus sequence of the TPR motif predicted from sequence alignment of known TPR-containing proteins (13). The horizontal bars below the alignment indicate helices A and B. (B) BB0238 predicted TPR region (residues 40 to 73), with consensus residues in boldface and conserved residues underlined. The TPR mutations engineered in BB0238 are shown below the parental sequence, with the targeted residue(s) in boldface and the mutated residue above. (C) Immunoblot analysis of BB0238 expression in Bb297, BbΔbb0238, Bbbb0238C, Bbbb0238Y43A, Bbbb0238G47V, Bbbb0238Y50A, Bbbb0238A66G, and Bbbb0238P71A assessing protein expression of native and mutated BB0238. Detection of FlaB was included as a loading control. The relevant molecular weight markers (lane MW) are noted on the left. The immunoblot results are representative data from two independent experiments.
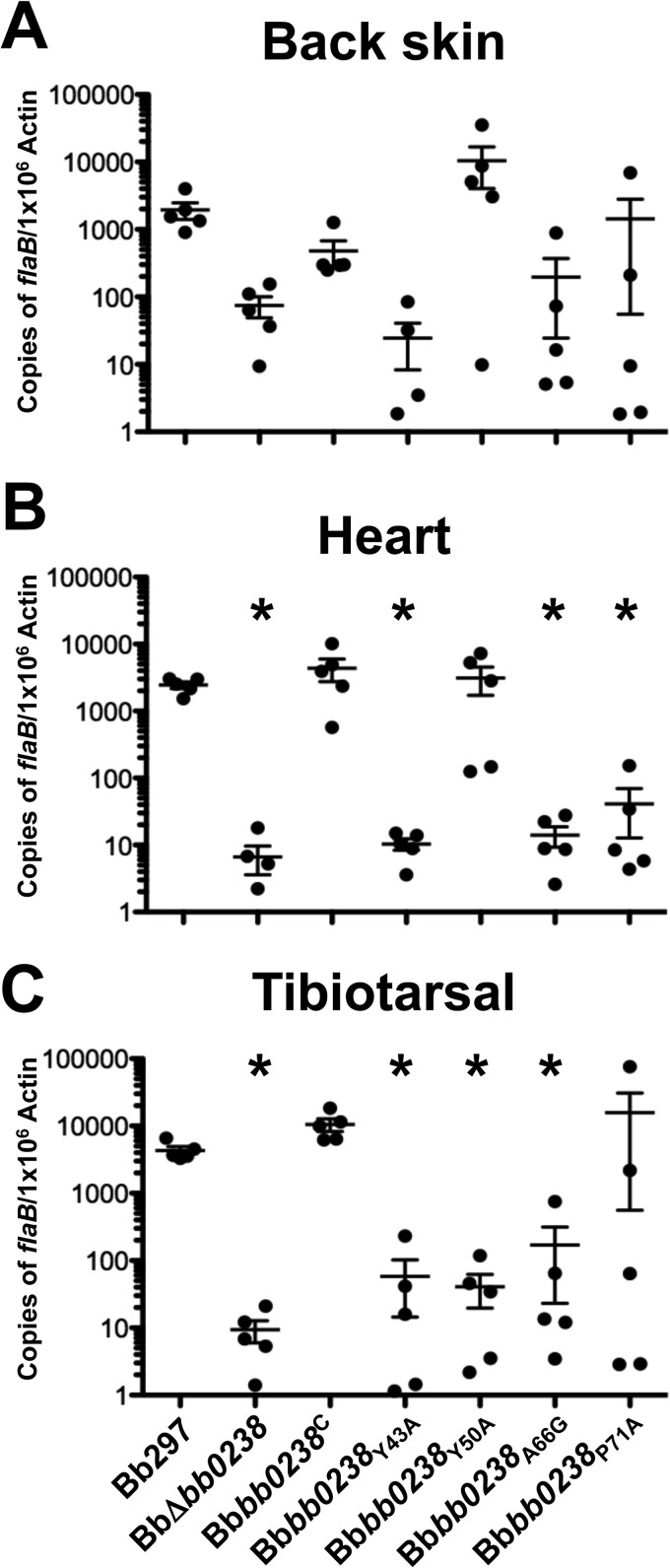
To evaluate the impacts of these TPR point mutations on BB0238 function during infection, C3H/HeN mice were challenged with Bbbb0238Y43A, Bbbb0238Y50A, Bbbb0238A66G, and Bbbb0238P71A via intradermal needle inoculation with 105 bacteria. Because Bbbb0238G47V did not produce the BB0238 protein, the clone was excluded from the infection experiments, as it would presumably have the same phenotype as BbΔbb0238. The Bbbb0238Y43A, Bbbb0238Y50A, Bbbb0238A66G, and Bbbb0238P71A TPR mutants were confirmed to contain all Bb297 endogenous plasmids known to be important for infection; Bbbb0238Y50A and Bbbb0238A66G were missing lp38 (see Fig. S1 in the supplemental material), but lp38 is not required by B. burgdorferi during the enzootic cycle (53). Ear punch, back skin, heart, bladder, tibiotarsal tissue, and lymph nodes were collected from mice at 2 weeks postinfection, and the tissues were cultured in BSK-II medium. The cultures were then examined by dark-field microscopy for the presence of spirochetes. Relative to Bb297 and Bbbb0238C, Bbbb0238Y43A, Bbbb0238A66G, and Bbbb0238P71A demonstrated reduced tissue infection rates and attenuation comparable to that of BbΔbb0238 (Table 4). As noted in earlier experiments (Table 2), the total numbers of infected mice in each group were similar, but tissue-specific attenuation was observed in mice infected with BbΔbb0238, Bbbb0238Y43A, Bbbb0238A66G, and Bbbb0238P71A. Interestingly, there was an increase in the incidence of positive heart tissues for Bbbb0238Y50A compared to BbΔbb0238 and the other three TPR mutant clones. The results from the qPCR analysis of spirochetal burdens in tissues from infected mice showed trends similar to those in the culture data (Fig. 10). Tissues from mice infected with Bbbb0238Y43A, Bbbb0238A66G, and Bbbb0238P71A showed bacterial burdens comparable to those of BbΔbb0238 in all tissues tested. In agreement with the infectivity results, Bbbb0238Y50A was present in the heart tissues at nearly wild-type levels. In addition, the mutant also showed bacterial numbers equivalent to those of Bb297 and Bbbb0238C in back skin tissues. However, bacterial burdens in tibiotarsal tissues from mice infected with Bbbb0238Y50A were reduced to levels similar to those detected in BbΔbb0238. While the exact function of BB0238 remains unknown, these data suggest that several of these conserved residues in the putative TPR domain are important for BB0238 function.
TABLE 4.
Infectivity of B. burgdorferi bb0238 TPR mutants in C3H/HeN mice
| Strain | No. of positive cultures/total |
No. positive mice/total | ||||||
|---|---|---|---|---|---|---|---|---|
| Ear punch | Back skin | Lymph nodesa | Heart | Tibiotarsal tissue | Bladder | All sites | ||
| Bb297 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 30/30 | 5/5 |
| BbΔbb0238 | 0/5b | 5/5 | 5/5 | 1/5b | 3/5 | 0/5b | 14/30b | 5/5 |
| Bbbb0238C | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 30/30 | 5/5 |
| Bbbb0238Y43A | 0/5b | 3/5 | 1/5b | 1/5b | 3/5 | 0/5b | 8/30b | 4/5 |
| Bbbb0238Y50A | 0/5b | 3/5 | 5/5 | 5/5 | 3/5 | 2/5 | 18/30b | 5/5 |
| Bbbb0238A66G | 0/5b | 2/5 | 2/5 | 0/5b | 3/5 | 1/5b | 8/30b | 3/5 |
| Bbbb0238P71A | 0/5b | 3/5 | 2/5 | 0/5b | 3/5 | 1/5b | 9/30b | 4/5 |
A single brachial and inguinal lymph node were collected from each mouse and cultured together.
Statistically significant difference relative to Bbbb0238C (P < 0.05).
FIG 10.
Spirochete burdens in tissues from mice infected with BB0238 TPR point mutants. Tissues were collected 2 weeks postinfection from mice needle inoculated with a dose of 105 spirochetes of Bb297, BbΔbb0238, Bbbb0238C, Bbbb0238Y43A, Bbbb0238Y50A, Bbbb0238A66G, or Bbbb0238P71A. DNAs from tissue samples were analyzed in triplicate by qPCR to measure spirochete burdens. The results from back skin, heart, and tibiotarsal tissue are representative of a single infection experiment (n = 5 mice for each strain). The error bars represent SEM. Statistical significance was determined using an unpaired Student t test. The asterisks indicate a statistically significant difference relative to Bbbb0238C (P < 0.05).
DISCUSSION
Given the high percentage of hypothetical proteins present in the genome of B. burgdorferi (6), characterization of genes of unknown function could help define the molecular mechanisms of host-pathogen interactions and identify new targets for disease intervention. While we were unable to define the function of BB0238 in B. burgdorferi and its specific role in virulence remains unknown, this study provides sufficient evidence to conclude that it is required for efficient infection in the murine infection model. The bb0238 mutant exhibited significant attenuation during experimental mammalian infection. This defect, characterized by reduced infection in specific tissues, most notably the heart and ear tissues (Table 2 and Fig. 2), could not be diminished even when the inoculum was increased significantly above the parental ID50. Though there were substantial decreases in tissue spirochete burdens and a large number of culture-negative tissues, the total number of mice infected by BbΔbb0238 remained similar to the total number of mice infected by Bb297. As a result, only a modest increase in the ID50 for BbΔbb0238 was observed. Reduced infection rates in the tibiotarsal joints of BbΔbb0238-infected mice were not a consistent phenotype across infection experiments and models. However, in experiments where higher infection rates were detected, the spirochete burden in these tissues was always significantly lower than for the wild type. The inability of BbΔbb0238 to infect immunocompromised scid mice suggests that the bb0238 mutant is not attenuated solely because it was unable to evade the adaptive immune response (Table 3). It is worth noting that all infection analyses in mice were conducted at 2 weeks postchallenge, and it is possible that the observed defect may be overcome during long-term infections. Additionally, the mammalian infection model evaluates only one side of the B. burgdorferi life cycle. Given that bb0238 is expressed under conditions mimicking the unfed tick and fed tick (Fig. 6A and B), one would expect bb0238 to be expressed by bacteria during both tick colonization and transmission to the mammal. Upon evaluation of BbΔbb0238 in the tick and during vector-mediated transmission, it may be found that bb0238 also plays a role during these events.
Because the bb0238 mutant was specifically unable to infect the heart and ear tissue (Table 2), we wanted to determine if bb0238 was differentially regulated by bacteria within different mammalian tissues (Fig. 4). In agreement with these findings, a minor increase in the levels of bb0238 transcripts was detected in heart tissues. A similar increase in bb0238 expression was observed in the tibiotarsal tissues, which also correlated with reduced infection rates in BbΔbb0238-infected mice. It is possible that variations within tissue microenvironments (i.e., unknown host stimuli or nutrient availability) trigger a change in bb0238 expression, but the changes among tissues were relatively modest (approximately 2-fold). Although the in vitro expression data determined that bb0238 was not differentially regulated (Fig. 6A and B), it is likely that the in vitro culture conditions do not fully recapitulate the complex environments within the tick or vertebrate host. Therefore, comparison of bb0238 transcription in the vector and host during each stage of the enzootic cycle is necessary to fully confirm that bb0238 is constitutively expressed by B. burgdorferi.
Cellular localization assays suggest that BB0238 fractionates to the inner membrane (Fig. 7), and SignalP predicted that the N terminus of BB0238 may contain either a transmembrane domain or a signal peptidase I cleavage site. Since the proteomic analysis of mature BB0238 was unable to identify a peptide mapping to the region containing the predicted signal peptide (Fig. 8), it is likely that BB0238 is secreted into the periplasm and is associated with some component of the inner membrane. In addition, the subsurface localization of BB0238 suggests that it does not directly interact with mammalian proteins. Therefore, the attenuated infectivity in the bb0238 mutant is not due to the inability of BbΔbb0238 to interact with extracellular matrix host proteins, unless BB0238 aids in export of such proteins to the cell surface. Because B. burgdorferi encodes other proteins that contain TPR domains, it is possible that one of these other TPR-containing proteins may compensate to a lesser degree for the lack of BB0238 in certain tissues of the mammalian host (i.e., lymph nodes and back skin) during infection with the BbΔbb0238 mutant, thus explaining the tissue-specific attenuation in infection and bacterial burden. Results from swarm assays suggest that basic bacterial motility is not compromised in BbΔbb0238 and that it does not contribute to the infectivity defect (data not shown), though functionality of the chemosensory pathways has not been ruled out. Similarly, no growth defects were identified for BbΔbb0238 in vitro (data not shown), but the attenuation could be due to the bb0238 mutant's inability to obtain nutrients from the mammalian host. Considering that BSK-II is a nutrient-rich medium (15), it is possible that cultivation in this medium in vitro could likely mask growth defects that may be more apparent in the mammal. Cultivation of BbΔbb0238 within dialysis membrane chambers (DMC) implanted in rat peritoneal cavities could provide insight into a possible role for BB0238 in nutrient acquisition in the host. Though the DMC experiments do not address tissue-specific differences in nutrient requirements or availability, the experiments may identify a general in vivo growth defect that could result in the attenuated-infectivity phenotype similar to that seen with pncA (54). As noted above, the gene adjacent to bb0239 is predicted to encode a deoxyguanosine/deoxyadenosine kinase involved in B. burgdorferi purine salvage, and this pathway is essential for mammalian infection (39, 40). Due to the physical proximity of bb0238 and bb0239 in the genome, it is possible that bb0238 may play an ancillary role in purine salvage. Interestingly, attempts to disrupt bb0239 in Bb297 were unsuccessful, suggesting bb0239 may be essential for B. burgdorferi growth (data not shown).
Despite our limited knowledge regarding the function of bb0238, the original annotation of BB0238 on the Comprehensive Microbial Resource database identified several residues in the N-terminal region that matched the consensus for a TPR domain (13, 50). This putative TPR domain also appears to be functionally important for BB0238, as clones carrying point mutations engineered in the more highly conserved consensus residues either (i) did not express BB0238, presumably due to destabilization and degradation of the G47V mutant protein, or (ii) exhibited attenuated infectivity comparable to that observed in the bb0238 deletion mutant (Table 4 and Fig. 9 and 10). BB0238 contains only one TPR motif, which is unusual among TPR motif-containing proteins, though examples of proteins in which a single TPR motif mediates protein-protein interactions exist (13, 50, 51). At this time, it still remains to be determined whether this region functions as an actual TPR domain, and the contribution of the domain to the physiological role of BB0238 remains unclear. It is possible that the point mutations generated in BB0238 could have altered the overall protein structure; however, attempts to generate soluble recombinant BB0238 protein for the purposes of structure-function analyses and nuclear magnetic resonance (NMR) structure characterization of the TPR mutants have been unsuccessful. If the TPR motif of BB0238 does indeed function as a prototypical protein-protein interaction domain, then BB0238 may be a chaperone, a common role for TPR-containing proteins. If BB0238 does act as a chaperone, it could be implicated in a number of physiological pathways, such as protein folding, protein transport, or chaperone-protease interactions.
Future studies will focus on delineating the function of BB0238. Given the presence and presumed importance of the TPR domain, attempts to identify a putative binding partner for BB0238 have high priority. Identification of a binding partner would not only shed light on a possible role for BB0238 in B. burgdorferi function, but would allow use of our TPR mutants in identifying relevant residues for protein-protein interactions. Additionally, all the infection analyses reported here were performed at 2 weeks postinfection. Analysis of BbΔbb0238 during long-term infections may determine whether the infectivity defect seen with the bb0238 knockout is alleviated during chronic infection. Finally, the bb0238 mutant should be evaluated throughout the rest of the experimental enzootic cycle to determine if BbΔbb0238 is able to colonize ticks and is competent for tick-to-mouse transmission. In conclusion, these studies have demonstrated that bb0238 is required for efficient B. burgdorferi mammalian infection, but further experimentation is required to determine the precise contribution(s) of BB0238 during infection.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by funding to Jon S. Blevins through the Arkansas Biosciences Institute (a major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000) and NIH/NIAID R01-AI087678, as well as support through the Translational Research Institute (UL1-TR000039; NIH National Center for Research Resources and National Center for Advancing Translational Sciences) and the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (P20-GM103625).
We thank Daniel Voth for his assistance with microscopy and for the use of the microscope. We also thank Ricky Edmondson, Alan Tackett, and Samuel Mackintosh in the UAMS Proteomics Core and Allen Gies in the UAMS Sequencing Core Facility for their technical assistance. Finally, we thank Darrin Akins for providing the BB0796-specific antiserum used in these studies.
Footnotes
Published ahead of print 28 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01977-14.
REFERENCES
- 1. Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. 10.1126/science.7043737 [DOI] [PubMed] [Google Scholar]
- 2. Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10:87–99. 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733–740. 10.1056/NEJM198303313081301 [DOI] [PubMed] [Google Scholar]
- 4. Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473. 10.1016/S0140-6736(11)60103-7 [DOI] [PubMed] [Google Scholar]
- 5. Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Invest. 113:1093–1101. 10.1172/JCI200421681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- 7. Revel AT, Blevins JS, Almazan C, Neil L, Kocan KM, de la Fuente J, Hagman KE, Norgard MV. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. U. S. A. 102:6972–6977. 10.1073/pnas.0502565102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang X, Yang X, Kumar M, Pal U. 2009. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J. Infect. Dis. 200:1318–1330. 10.1086/605846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X, Hegde S, Shroder DY, Smith AA, Promnares K, Neelakanta G, Anderson JF, Fikrig E, Pal U. 2013. The lipoprotein La7 contributes to Borrelia burgdorferi persistence in ticks and their transmission to naive hosts. Microbes Infect. 15:729–737. 10.1016/j.micinf.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar M, Yang X, Coleman AS, Pal U. 2010. BBA52 facilitates Borrelia burgdorferi transmission from feeding ticks to murine hosts. J. Infect. Dis. 201:1084–1095. 10.1086/651172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. 2009. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol. Microbiol. 74:112–125. 10.1111/j.1365-2958.2009.06853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patton TG, Dietrich G, Dolan MC, Piesman J, Carroll JA, Gilmore RD., Jr 2011. Functional analysis of the Borrelia burgdorferi bba64 gene product in murine infection via tick infestation. PLoS One 6:e19536. 10.1371/journal.pone.0019536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blatch GL, Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939 [DOI] [PubMed] [Google Scholar]
- 14. Hughes CA, Kodner CB, Johnson RC. 1992. DNA analysis of Borrelia burgdorferi NCH-1, the first north central U.S. human Lyme disease isolate. J. Clin. Microbiol. 30:698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pollack RJ, Telford SR, Spielman A. 1993. Standardization of medium for culturing Lyme disease spirochetes. J. Clin. Microbiol. 31:1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blevins JS, Hagman KE, Norgard MV. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 8:82. 10.1186/1471-2180-8-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groshong AM, Gibbons NE, Yang XF, Blevins JS. 2012. Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J. Bacteriol. 194:3336–3342. 10.1128/JB.00253-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blevins JS, Revel AT, Smith AH, Bachlani GN, Norgard MV. 2007. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 73:1501–1513. 10.1128/AEM.02454-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 20. Maruskova M, Esteve-Gassent MD, Sexton VL, Seshu J. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect. Immun. 76:391–402. 10.1128/IAI.01118-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bugrysheva JV, Godfrey HP, Schwartz I, Cabello FC. 2011. Patterns and regulation of ribosomal RNA transcription in Borrelia burgdorferi. BMC Microbiol. 11:17. 10.1186/1471-2180-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang X, Popova TG, Hagman KE, Wikel SK, Schoeler GB, Caimano MJ, Radolf JD, Norgard MV. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenhart TR, Kenedy MR, Yang X, Pal U, Akins DR. 2012. BB0324 and BB0028 are constituents of the Borrelia burgdorferi beta-barrel assembly machine (BAM) complex. BMC Microbiol. 12:60. 10.1186/1471-2180-12-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blevins JS, Revel AT, Caimano MJ, Yang XF, Richardson JA, Hagman KE, Norgard MV. 2004. The luxS gene is not required for Borrelia burgdorferi tick colonization, transmission to a mammalian host, or induction of disease. Infect. Immun. 72:4864–4867. 10.1128/IAI.72.8.4864-4867.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bryksin AV, Tomova A, Godfrey HP, Cabello FC. 2010. BmpA is a surface-exposed outer-membrane protein of Borrelia burgdorferi. FEMS Microbiol. Lett. 309:77–83. 10.1111/j.1574-6968.2010.02020.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brooks CS, Vuppala SR, Jett AM, Akins DR. 2006. Identification of Borrelia burgdorferi outer surface proteins. Infect. Immun. 74:296–304. 10.1128/IAI.74.1.296-304.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cunningham TM, Walker EM, Miller JN, Lovett MA. 1988. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170:5789–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H, Raji A, Theisen M, Hansen PR, Marconi RT. 2005. bdrF2 of Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J. Bacteriol. 187:175–184. 10.1128/JB.187.1.175-184.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 86:1183–1196. 10.1111/mmi.12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358–1374. 10.1111/j.1365-2958.2007.05746.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101–5109. 10.1111/j.1742-4658.2005.04945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barthold SW, Sidman CL, Smith AL. 1992. Lyme borreliosis in genetically resistant and susceptible mice with severe combined immunodeficiency. Am. J. Trop. Med. Hyg. 47:605–613 [DOI] [PubMed] [Google Scholar]
- 33. McKisic MD, Barthold SW. 2000. T-cell-independent responses to Borrelia burgdorferi are critical for protective immunity and resolution of Lyme disease. Infect. Immun. 68:5190–5197. 10.1128/IAI.68.9.5190-5197.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bockenstedt LK, Kang I, Chang C, Persing D, Hayday A, Barthold SW. 2001. CD4+ T helper 1 cells facilitate regression of murine Lyme carditis. Infect. Immun. 69:5264–5269. 10.1128/IAI.69.9.5264-5269.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schaible UE, Kramer MD, Museteanu C, Zimmer G, Mossmann H, Simon MM. 1989. The severe combined immunodeficiency (scid) mouse. A laboratory model for the analysis of Lyme arthritis and carditis. J. Exp. Med. 170:1427–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McKisic MD, Redmond WL, Barthold SW. 2000. Cutting edge: T cell-mediated pathology in murine Lyme borreliosis. J. Immunol. 164:6096–6099. 10.4049/jimmunol.164.12.6096 [DOI] [PubMed] [Google Scholar]
- 37. Schaible UE, Gay S, Museteanu C, Kramer MD, Zimmer G, Eichmann K, Museteanu U, Simon MM. 1990. Lyme borreliosis in the severe combined immunodeficiency (scid) mouse manifests predominantly in the joints, heart, and liver. Am. J. Pathol. 137:811–820 [PMC free article] [PubMed] [Google Scholar]
- 38. Fikrig E, Barthold SW, Chen M, Grewal IS, Craft J, Flavell RA. 1996. Protective antibodies in murine Lyme disease arise independently of CD40 ligand. J. Immunol. 157:1–3 [PubMed] [Google Scholar]
- 39. Jain S, Sutchu S, Rosa PA, Byram R, Jewett MW. 2012. Borrelia burgdorferi harbors a transport system essential for purine salvage and mammalian infection. Infect. Immun. 80:3086–3093. 10.1128/IAI.00514-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jewett MW, Lawrence KA, Bestor A, Byram R, Gherardini F, Rosa PA. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 191:6231–6241. 10.1128/JB.00450-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lawrence KA, Jewett MW, Rosa PA, Gherardini FC. 2009. Borrelia burgdorferi bb0426 encodes a 2′-deoxyribosyltransferase that plays a central role in purine salvage. Mol. Microbiol. 72:1517–1529. 10.1111/j.1365-2958.2009.06740.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ge Y, Old IG, Girons IS, Charon NW. 1997. The flgK motility operon of Borrelia burgdorferi is initiated by a sigma 70-like promoter. Microbiology 143:1681–1690. 10.1099/00221287-143-5-1681 [DOI] [PubMed] [Google Scholar]
- 43. Noppa L, Burman N, Sadziene A, Barbour AG, Bergstrom S. 1995. Expression of the flagellin gene in Borrelia is controlled by an alternative sigma factor. Microbiology 141:85–93. 10.1099/00221287-141-1-85 [DOI] [PubMed] [Google Scholar]
- 44. Revel AT, Talaat AM, Norgard MV. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 99:1562–1567. 10.1073/pnas.032667699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brooks CS, Hefty PS, Jolliff SE, Akins DR. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371–3383. 10.1128/IAI.71.6.3371-3383.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689–1705. 10.1128/IAI.71.4.1689-1705.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470–1479. 10.1046/j.1365-2958.2000.02104.x [DOI] [PubMed] [Google Scholar]
- 48. Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. 2012. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J. Bacteriol. 194:3395–3406. 10.1128/JB.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, Sutton G. 2010. The comprehensive microbial resource. Nucleic Acids Res. 38:D340–D345. 10.1093/nar/gkp912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 51. Main ER, Jackson SE, Regan L. 2003. The folding and design of repeat proteins: reaching a consensus. Curr. Opin. Struct. Biol. 13:482–489. 10.1016/S0959-440X(03)00105-2 [DOI] [PubMed] [Google Scholar]
- 52. Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34:W335–W339. 10.1093/nar/gkl217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dulebohn DP, Bestor A, Rego RO, Stewart PE, Rosa PA. 2011. Borrelia burgdorferi linear plasmid 38 is dispensable for completion of the mouse-tick infectious cycle. Infect. Immun. 79:3510–3517. 10.1128/IAI.05014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Purser JE, Lawrenz MB, Caimano MJ, Howell JK, Radolf JD, Norris SJ. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753–764. 10.1046/j.1365-2958.2003.03452.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.