Abstract
Malaria is a major public health problem in tropical and subtropical countries, including India. This study elucidates the cause of chloroquine treatment failure (for Plasmodium falciparum infection) before the introduction of artemisinin combination therapy. One hundred twenty-six patients were randomized to chloroquine treatment, and the therapeutic efficacy was monitored from days 1 to 28. An in vitro susceptibility test was performed with all isolates. Parasitic DNA was isolated, followed by PCR and restriction digestion of different codons of the pfcrt gene (codons 72 to 76) and the pfmdr1 gene (N86Y, Y184F, S1034C, N1042D, and D1246Y). Finally, sequencing was done to confirm the mutations. Forty-three (34.13%) early treatment failure cases and 16 (12.69%) late treatment failure cases were observed after chloroquine treatment. In vitro chloroquine resistance was found in 103 isolates (81.75%). Twenty-six (60.47%) early treatment failure cases and 6 (37.5%) late treatment failure cases were associated with the CVMNK-YYSNY allele (the underlined amino acids are those that were mutated). Moreover, the CVIEK-YYSNY allele was found in 8 early treatment failure (18.60%) and 2 late treatment failure (12.5%) cases. The presence of the wild-type pfcrt (CVMNK) and pfmdr1 (YYSNY) double mutant allele in chloroquine-nonresponsive cases was quite uncommon. In vivo chloroquine treatment failure and in vitro chloroquine resistance were strongly correlated with the CVMNK-YYSNY and CVIEK-YYSNY haplotypes (P < 0.01).
INTRODUCTION
The emergence and subsequent spread of chloroquine (CQ)-resistant Plasmodium falciparum in areas that are endemic for malaria are the greatest challenges to malaria control programs (1). Knowledge about the molecular mechanisms of drug-resistant malaria is most important for designing new drugs and providing molecular markers to monitor drug activity and its efficacy in treatment (2). Some definite mutations in parasitic proteins and enzymes allow the parasite to survive under high drug pressure conditions. The selected parasite populations with these mutations proliferate in the environment. Indeed, these mutations can be used as molecular markers to detect the drug-resistant parasite (3–6). It is suggested that the substitution of lysine to threonine (K76T) at codon 76 of the pfcrt gene is associated with in vivo and in vitro CQ resistance in Africa, South America, and Southeast Asia (7, 8). Now, it has been proven with precision that the entire 72 to 76 amino acid residues (haplotypes) play a key role in chloroquine resistance (9). The pfmdr1 gene, located on chromosome 5 containing P-glycoprotein homologue 1 (Pgh 1), has been implicated in making the parasite less susceptible to CQ and other antimalarials (9–11). Studies conducted in different geographical areas of the world have suggested that the point mutation of tyrosine at codon 86 (N86Y) is related to CQ resistance (10, 12–15). Several other pfmdr1 polymorphisms at 184F, 1034C, 1042D, and 1246Y are being implicated in altering the degree of CQ resistance (12, 13). In India, 86Y and 184F are commonly observed pfmdr1 mutant alleles with high in vitro 50% inhibitory concentrations (IC50s) for CQ (9, 16). In some parts of the world, CQ resistance occurs due to polymorphisms in both the pfcrt and pfmdr1 genes (17). In India, in vivo CQ treatment failure and in vitro CQ resistance are solely linked with pfcrt 76T mutations but not in the pfmdr1 gene (18–20).
Jangalmahal is one of the zones in eastern India that is endemic for malaria, where transmission intensity is high (21). CQ has been used here for >6 decades against P. falciparum malaria. The present study was designed to evaluate the molecular basis of CQ resistance in this part of India just before the launch of a new national drug policy, i.e., artemisinin combination therapy (ACT). In this study, the presence of the wild-type pfcrt (CVMNK) and pfmdr1 double mutant (YYSNY) alleles in CQ-nonresponsive cases was reported for the first time in India. A large number of the early treatment failure and late treatment failure cases were associated with these CVMNK-YYSNY and CVIEK-YYSNY alleles. This finding was new in the field of malarial epidemiology in eastern India.
MATERIALS AND METHODS
Study site.
This study was conducted from March 2008 to June 2009 in the Jangalmahal area of eastern India. It is one of the largest hilltop forest areas situated on the border of three states: West Bengal, Orissa, and Jharkhand. This rural forest area is one of the largest zones endemic for malaria in eastern India. The experimental design and protocol of this study were duly approved by the Vidyasagar University Ethical Committee.
Assessment of clonality in clinical isolates.
Clonality was defined as the highest number of alleles detected in either of the two loci and was used to classify isolates as monoclonal or polyclonal and to distinguish recrudescence from new infection for all patients failing therapy after day 7. The clonality of an infection was estimated by using an allelic family-specific nested-PCR, as described previously (22). All PCR amplifications contained both positive controls (genomic DNA from P. falciparum strains Dd2 and 3D7) and a negative control (no target DNA).
Patients and in vivo study.
Patients suffering from fever with headache, shivering, and vomiting tendency during the previous 24 h were sent for an evaluation of possible malaria diagnosis. Five milliliters of intravenous blood was collected from each of 171 patients using EDTA-coated Vacutainer tubes. The aliquots were stored at −20°C for further molecular analysis. The identification of P. falciparum monoinfection was based on the microscopic examination of Giemsa-stained thin and thick blood smears, as well as rapid diagnostic tests to detect Plasmodium-specific lactate dehydrogenase (OptiMAL-DT; Biomed, Indonesia). Finally, patients with malaria infection with a parasite density of 1,000 to 100,000 asexual parasites/μl blood and no recent history of self-medication with antimalarial drugs (according to the expressed verbal concern of the patients) were randomized to CQ treatment, as it was the first-line drug treatment against uncomplicated malaria in India during 2008 and early 2009 (23). The patients with signs and symptoms of severe complicated malaria and age of <1 year were excluded (24). Informed consent was taken from the patients, and the consent of the guardian was taken for child patients. The patients carrying P. falciparum infections were treated with 10 mg/kg of body weight CQ on days 1 and 2 and 5 mg/kg of body weight CQ on day 3. The clinical conditions and parasite densities were monitored on days 0, 1, 2, 3, 7, 14, 21, and 28. The therapeutic responses were classified as adequate clinical and parasitological response (ACPR), early treatment failure (ETF), late treatment failure (LTF), and late parasitological failure (LPF), according to the criteria adopted by the WHO. The patients who did not respond to CQ treatment were treated with ACT (artesunate plus sulfadoxine-pyrimethamine).
Separation of red blood cells.
Red blood cells (RBC) were separated using a Histopaque-1077 density gradient, followed by centrifugation at 350 × g for 45 min at 4°C. An aliquot of 3 to 4 ml of red blood cell pellet was found. Finally, RBC were washed with folate and p-aminobenzoic acid-free RPMI 1640 medium three times, as described previously (25).
In vitro drug sensitivity assay.
An in vitro drug sensitivity assay was performed with the clinical isolates, as described previously (26). P. falciparum isolates were grown and maintained through culture, using some modifications of the method of Trager and Jensen (27). The clinical isolates with a parasite density of ≥0.1% were suspended in the complete folate and p-aminobenzoic acid-free RPMI 1640 medium consisting of 0.5 g% AlbuMAX II, 25 mM HEPES, 25 mM NaHCO3, 25 μg/ml gentamicin, and 0.25% hypoxanthine at a hematocrit of 1.5%. If required, a dilution was done by adding uninfected O+ erythrocytes to obtain a parasite density of 0.5% and 1.5% hematocrit.
The IC50s were routinely checked on fresh isolates after culture adaptation as a part of the surveillance of the antimalarial susceptibility test. Before CQ treatment, a synchronized parasite culture was maintained over three life cycles. RPMI 1640 was used to prepare stock solutions and dilutions of CQ ranging from 6 nM to 3,072 nM. Two hundred microliters per well of parasitized erythrocyte suspensions was distributed in 96-well tissue culture plates. Twenty-five microliters of each concentration of the drug was distributed in each well. Three drug-free wells were used as controls for each 96-well plate. Each concentration was studied in duplicate or triplicate. The plates were incubated for 48 h at 37°C in an atmosphere of 5% CO2 and a relative humidity of 95%. The IC50 was defined as the drug concentration corresponding to 50% of hypoxanthine uptake by the parasites in the drug-free control wells, determined by probit/logit regression analysis. The CQ-sensitive 3D7 and CQ-resistant Dd2 strains were used as controls.
DNA extraction, PCR amplification, and restriction digestion.
Parasite DNA was extracted from 2 ml of infected blood using the phenol-chloroform extraction method, as described before (26). DNA was quantified by 1.2% agarose gel electrophoresis, and the purity of DNA was checked spectrophotometrically by calculating the A260/A280 ratio.
Primers were designed on the basis of the complete P. falciparum Dd2/Indochina strain sequence (GenBank accession no. AF030694 for pfcrt and KJ690043.1 for pfmdr1) (26, 28). Approximately 200 ng of genomic DNA, 15 pmol of primers, 1× reaction buffer (10 mM Tris, 50 mM KCl [pH 8.4]), 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), and 1 unit of Taq DNA polymerase were used to prepare a 25-μl reaction mixture (master mix). The PCR cycle conditions varied in the different genes. The detailed primer sequences and cycle conditions are presented in Table 1. In the nest I reaction, the pfcrt gene was amplified by the 1F and 1R primer pair. The 2F and 2R primer pair was used to amplify the pfmdr1 gene, particularly codons 86 to 184. An additional nest reaction with 3F-3R was designed to amplify the entire codons 1034 to 1246 of the pfmdr1 gene.
TABLE 1.
PCR primer sequences and reaction conditions used for the amplification of sequences containing P. falciparum pfcrt and pfmdr1 genes
| Gene | Primer name (positions) | Primer sequence | Product size (bp) | Restriction endonuclease | PCR cycling conditions |
|---|---|---|---|---|---|
| Nest I | |||||
| pfcrt | 1F | 5′-CCGTTAATAATAAATACAGGC-3′ | 261 | NDa | 95°C for 5 min; (95°C for 35 s; 56°C for 40 s; 72°C for 1 min) × 35 cycles; 72°C for 10 min |
| 1R | 5′-CTTTAAAAATGGAAGGGTGT-3′ | ||||
| pfmdr1 | 2F (86–184) | 5′-TGTTGAAAGATGGGTAAAGAGCAGAAAGAG-3′ | 780 | ND | 95°C for 5 min; (95°C for 40 s; 58°C for 50 s; 72°C for 1 min) × 36 cycles; 72°C for 10 min |
| 2R (86–184) | 5′-TACTTTCTTATTACATATGACACCACAAACA-3′ | ||||
| pfmdr1 | 3F (1034–1246) | 5′-AGAAGATTATTTCTGTAATTTGATACAAAAAGC-3′ | 880 | ND | 95°C for 5 min; (95°C for 40 s; 60°C for 42 s; 72°C for 1 min 20 s) × 38 cycles; 72°C for 10 min |
| 3R (1034–1246) | 5′-ATG ATT CGA TAA ATT CAT CTA TAGCAG CAA-3′ | ||||
| Multiplex | |||||
| pfcrt | K76T F | 5′-TGTGCTCATGTGTTTAAACTTAT-3′ | 134 | ApoI | 95°C for 5 min; (95°C for 30 s; 56°C for 40 s; 72°C for 1 min) × 32 cycles; 72°C for 10 min |
| K76T R | 5′-CAAAACTATAGTTACCAATTTTG-3′ | ||||
| pfmdr1 | N86Y F | 5′-TTTACCGTTTAAATGTTTACCTGC-3′ | 310 | AflIII | 95°C for 5 min; (95°C for 35 s; 58°C for 40 s; 72°C for 1 min) × 38 cycles; 72°C for 10 min |
| N86Y R | 5′-CCATCTTGATAAAAAACACTTCTT-3′ | ||||
| Y184F F | 5′-AAA GAT GGT AAC CTC AGT ATC AAA GAA GAG-3′ | 560 | DraI | 95°C for 5 min; (95°C for 35 s; 58°C for 40 s; 72°C for 1 min) × 34 cycles; 72°C for 8 min | |
| Y184F R | 5′-GTC AAA CGT GCA TTT TTT ATT AAT GAC CAT TTA-3′ | ||||
| S1034C F | 5′-AGAATTATTGTAAATGCAGCTTTATGGGGAC TC-3′ | 230 | DdeI | 95°C for 5 min; (95°C for 38 s; 60°C for 42 s; 72°C for 1 min 20 s) × 35 cycles; 72°C for 10 min | |
| S1034C R | 5-′AATGGATAATATTTCTCAAATGATAACTTAGCA-3′ | ||||
| N1042D F | 5′-TATGTCAAGCGGAGTTTTTGC-3′ | 337 | AsnI | 95°C for 5 min; (95°C for 40 s; 58°C for 40 s; 72°C for 1 min) × 38 cycles; 72°C for 10 min | |
| N1042D R | 5′-TCTGAATCTCCTTTTAAGGAC-3′ | ||||
| D1246Y F | 5′-GTGGAAAATCAACTTTTATGA-3′ | 499 | EcoRV | 95°C for 5 min; (95°C for 42 s; 60°C for 40 s; 72°C for 1 min) × 42 cycles; 72°C for 10 min | |
| D1246Y R | 5′-TTAGGTTCTCTTAATAATGCT-3′ | ||||
ND, not done.
In a multiplex reaction with the pfcrt gene, 2 μl of amplified DNA from the 1F-1R primer pair was used as the template DNA for amplifying codons 72 to 76. In the pfmdr1 gene, 2 μl of amplified DNA from the 2F-2R primer pair was added to each of 2 PCR mixtures, using the primer pair N86Y F and N86Y R to amplify the fragments containing 86N/Y, and Y184F F and Y184F R were used to detect 184Y/F. DNA amplification from the 3F-3R primer pair was added to each of 3 PCR mixtures to amplify codons 1034, 1042, and 1246 of the pfmdr1 gene.
The single nucleotide polymorphisms of the pfcrt and pfmdr1 genes at their specific codons were determined by enzymatic digestion of specific restriction enzymes, as described previously (14, 28). ApoI, AflIII, DraI, DdeI, AsnI, and EcoRV (New England BioLabs) were used to identify the pfcrt codon K76 and pfmdr1 codons 86Y, 184F, 1034C, 1042D, and 1246Y, respectively. CQ-sensitive 3D7 and CQ-resistant Dd2 and 7G8 strains served as controls.
Sequencing of pfcrt and pfmdr1 genes.
The amplicons were purified from the agarose gel using a gel extraction kit (Qiagen). From 50 to 200 ng of the gel-purified amplicon, sequencing reactions were carried out with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit, which was run on a model 3730xl genetic analyzer (Applied Biosystems) (29). The sequencing PCR was performed in a volume of 20 μl with 1 μl of Terminator ready reaction mix (TRR), 3.2 pmol of gene-specific primer, and 0.5× sequencing buffer. Sequencing was carried out at the Indian Institute of Technology, Kharagpur (IIT, KGP), the National Institute for Cholera and Dysentery (NICED), and Chromus Biotech Company (Bangalore). The sequences were translated using the translation tool available online at the Expert Protein Analysis System proteomics server (http://www.expasy.org). The amino acid sequences were compared with the wild-type amino acid sequences (AF030694 for pfcrt and KJ690043.1 for pfmdr1). The single nucleotide polymorphisms (SNP) were confirmed by reading both the forward and reverse strands.
Statistical analysis.
Data are expressed as the univariate medians. Fisher's exact tests and the Mann-Whitney U test were used to study the relationships between IC50s and genotypes. The relationship between the in vivo and/or in vitro phenotype and the molecular genotypes was studied by Fisher's exact test and regression analysis. All analyses were done using a statistical package, Origin 6.1, and the GraphPad InStat software 3.0.
Nucleotide sequence accession numbers.
The sequences described were submitted to GenBank under the accession numbers given in Table 2.
TABLE 2.
Sequencing data of different pfcrt haplotypes and some wild-type and mutant alleles of the pfmdr1 gene
| Haplotype | DNA sequencea | GenBank accession no. |
|---|---|---|
| CVMNK | GTATGTGTAATGAATAAAATT | KM188437 |
| CVMNT | GTATGTGTAATGAATACAATT | KM188436 |
| CVIEK | GTATGTGTAATTGAAAAAATT | KM188439 |
| CVIET | GTATGTGTAATTGAAACAATT | KM188435 |
| SVMNT | GTAAGTGTAATGAATACAATT | KM188438 |
| Wild-type N86 allele | ATGAATTTAGGTG | KM056978 |
| Mutant 86Y allele | TGTATTTAGGT | KM056980 |
| Wild-type D1246 allele | AAGAGATCTTAGAAAC | KM056977 |
| Mutant 1246Y allele | AAGATATCTTAGAAAC | KM056979 |
The underlined nucleotides represent the mutant codons.
RESULTS
Sensitivity of patients to chloroquine.
Clinical data were available for 171 suspected cases of P. falciparum infection. One hundred twenty-six isolates (73.68%) out of 171 patients (age group, 1 to 78 years) were enrolled in this study, as they carried P. falciparum monoinfection. Monoclonal P. falciparum infection was confirmed after allelic family-specific nested-PCR. Twenty-eight (16.34%) patients were excluded, as they contained Plasmodium vivax infection. Another 11 (6.43%) patients were eliminated due to coinfection with P. vivax. Six (3.51%) patients had not completed the drug schedule. Thirty-three patients were found to be under the age of 5, and 19 patients were pregnant women. CQ treatment produced ACPR in 67 patients (53.17%). Depending on the parasite clearance time (median parasite clearance, 2 days; range, 1 to 3 days), early CQ treatment failure was detected in 43 patients (34.13%), whereas 16 patients (12.69%) were identified as LTF (median parasite clearance of day 9, 4 to 14 days) cases.
Assessment of clonality of infection.
The multiplicity of infection was analyzed for a subset of 171 patients. The proportion of the monoclonal infections was very high. One hundred twenty-six isolates (73.68%) with a single allelic form were considered to have a monoclonal infection. These isolates were included in the study.
In vitro sensitivity to chloroquine.
An in vitro CQ sensitivity test was conducted in monoclonal P. falciparum isolates, which yielded interpretable results with all 126 isolates. Only 23 isolates (18.25%) were sensitive to CQ (median IC50, 54.25 nM; range, 8 to 90 nM). One hundred three isolates (81.75%) were highly resistant to CQ (median IC50, 196.50 nM; range, 110 to 310 nM) (Fig. 1 and Table 3).
FIG 1.
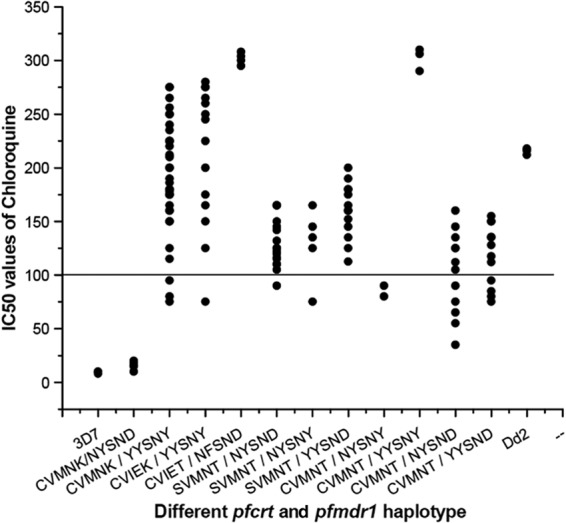
Relationship between in vitro IC50 of chloroquine and different pfcrt and pfmdr1 haplotypes. The solid line (corresponding to 100 nM, highly resistant) is hypothetical and shows the level of chloroquine resistance in vitro.
TABLE 3.
Distribution of different pfcrt and pfmdr1 haplotypes in relation to in vivo CQ treatment efficacy and in vitro CQ susceptibility
| No. of isolates | pfcrt haplotype (codons 72–76)a | pfmdr1 haplotype (codons 86, 184, 1034, 1042, and 1246) | CQ treatment efficacyb |
In vitro CQ responsea |
|||
|---|---|---|---|---|---|---|---|
| ACPR | ETF | LTF | S | R | |||
| 5 | CVMNK | NYSND | 5 | 5 | |||
| 37 | CVMNK | YYSNY | 5 | 26 | 6 | 4 | 33 |
| 11 | CVIEK | YYSNY | 1 | 8 | 2 | 1 | 10 |
| 4 | CVIET | NFSND | 2 | 1 | 1 | 4 | |
| 21 | SVMNT | NYSND | 17 | 4 | 1 | 20 | |
| 5 | SVMNT | NYSNY | 5 | 1 | 4 | ||
| 14 | SVMNT | YYSND | 9 | 3 | 2 | 14 | |
| 2 | CVMNT | NYSNY | 2 | 2 | |||
| 3 | CVMNT | YYSNY | 3 | 3 | |||
| 12 | CVMNT | NYSND | 10 | 1 | 1 | 5 | 7 |
| 12 | CVMNT | YYSND | 11 | 1 | 4 | 8 | |
The amino acids in bold type represent the mutant codons.
ACPR, adequate clinical and parasitological response; ETF, early treatment failure; LTF, late treatment failure.
S, susceptible (IC50, <100 nM); R, resistant (IC50, >100 nM).
pfcrt and pfmdr1 genotypes.
The pfcrt and pfmdr1 genes were amplified by PCR, followed by restriction digestion to detect each variant. We carefully examined the entire codons 72 to 76 of the pfcrt gene. The wild-type pfcrt (CVMNK) allele was found in 42 isolates (33.33%). The rare CVIEK haplotype was also found in 11 (8.73%) isolates. Mutations in 76T were found in 73 (54.76%) isolates. The SVMNT (31.75%) mutant haplotype was the most common pfcrt allele, followed by the CVMNT single mutant (23.02%) and CVIET triple mutant haplotype (Fig. 2). We checked codons 86, 184, 1034, 1042, and 1246 of the pfmdr1 gene. Of the isolates, 30.16% carried the wild-type pfmdr1 haplotype (NYSND). The rates of samples consisting of 86Y and 1246Y mutants were very high (61.11% and 46.03%, respectively). The YYSNY (40.48%) double mutant allele was most prevalent, followed by the YYSND (20.63%), NYSNY (5.55%), and NFSND (3.17%) single mutant haplotypes. No mutations were observed in the 1034C and 1042D alleles (Fig. 2).
FIG 2.
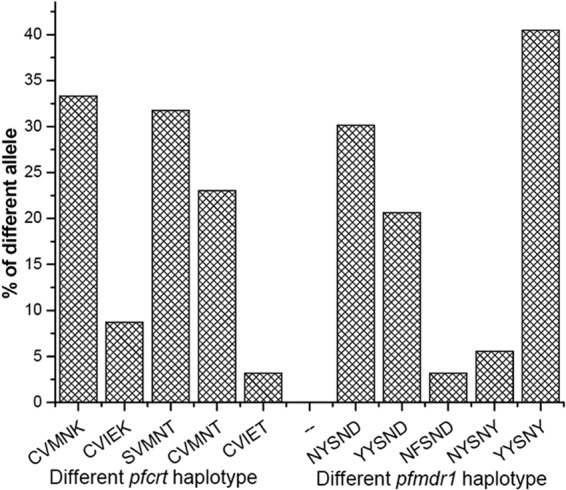
Frequency (%) of different pfcrt and pfmdr1 haplotypes in Jangalmahal.
Interestingly, 11 different combinations of parasite haplotypes (Table 3) were found. Only 5 isolates (3.97%) contained the wild-type pfcrt and pfmdr1 (CVMNK-NYSND) haplotypes. The prevalence of wild-type pfcrt and double pfmdr1 mutations (CVMNK-YYSNY) was observed. Thirty-seven isolates (29.36%) presented with this CVMNK-YYSNY allele. The CVIEK-YYSNY haplotype was found in 11 isolates (8.73%). The SVMNT-NYSND and SVMNT-YYSND haplotypes were observed in 21 (16.67%) and 14 (11.24%) isolates, respectively. The CVMNT-NYSND and CVMNT-YYSND alleles were detected in 12 patients. Four isolates represented the CVIET-NFSND allele.
Sequencing of pfcrt and pfmdr1 genes.
Sequencing of the pfcrt and pfmdr1 genes was carried out in all isolates, and it confirmed the restriction fragment length polymorphism (RFLP) results of the different codons of these two genes. Some isolates encoded a T mutation (ACA) instead of wild-type K (AAA) at codon 76 of the pfcrt gene, whereas 35 isolates encoded an S mutation (AGT) at codon 72. The mutations ATG to ATT and AAT to GAA were observed in codons 74 and 75, respectively, of the pfcrt gene, which ultimately encoded the I and E amino acid mutations, respectively. In the pfmdr1 gene, the prevalence of a Y mutation (TAT) was observed at codon 86 instead of the wild-type N (AAT). A large number of isolates also contained a Y mutation (TAT) at codon 1246 instead of the wild-type D (GAT) (Table 2). The GenBank accession numbers of 5 different haplotypes of the pfcrt gene and the 4 different wild-type and mutant alleles (N86Y and D1246Y) of the pfmdr1 gene are presented in Table 2. For checking true culture adaptation, we sequenced the parasite after culture adaptation; we found that the genotype of the culture-adapted line was identical to the genotype of the patient isolates.
pfcrt/pfmdr1 genotypes in relation to in vivo drug efficacy and in vitro susceptibility.
All 5 wild-type CVMNK-NYSND haplotypes produced ACPR after CQ treatment. It was clear that the phenotype of in vivo CQ treatment efficacy was related to the pfmdr1 genotypes at positions 86 and 1246 but not at 184, 1034, and 1042, as well as position 76 of the pfcrt gene (Fisher's test, CQ treatment failure [ETF], P < 0.01 for codons 86 and 1246; P was not significant at the level of 0.05 for codons 184, 1034, and 1042 of the pfmdr1 gene and codon 76 of the pfcrt gene) (Table 3). Regardless of the other mutations in the pfcrt and pfmdr1 genes, most interestingly, out of 43 ETF cases, 37 (86.05%) ETF patients were found with double mutations in pfmdr1 (YYSNY) (P < 0.01). On the other hand, out of 16 LTF cases, 8 LTF (50.00%) patients carried this double mutation in pfmdr1 (YYSNY) (P < 0.05). In a concise view, 26 ETF isolates (60.47%) out of 43 ETF cases were predominantly found to have the CVMNK-YYSNY haplotype, while 8 (18.60%) ETF isolates represented the CVIEK-YYSNY haplotype. Three patients who had the CVMNT-YYSNY allele were those in whom CQ treatment was ineffective. Three ETF cases were observed to have the SVMNT-YYSND haplotype. Each ETF case was observed to have the CVIET-NFSND and CVMNT-YYSND haplotypes (Table 3). Six (37.5%) LTF cases were found to have the CVMNK-YYSNY haplotype, whereas the CVIEK-YYSNY haplotype was found in 2 LTF cases. Eight (50.00%) LTF cases were associated with a 76T mutation in the pfcrt gene. It was found that the SVMNT-NYSND and SVMNT-YYSND alleles represented 4 LTF and 2 LTF cases, respectively.
The phenotype of in vitro CQ susceptibility was associated with both the pfmdr1 and pfcrt genotypes at positions 86 and 1246 for the pfmdr1 gene and codons 76 and 72 for the pfcrt gene, but not in codons 184, 1034, and 1042 of the pfmdr1 gene (CQ, P < 0.01 for pfmdr1 codons 86 and 1246, as well as pfcrt codons 76 and 72; P was not significant at the level of 0.05 for codons 184, 1034, and 1042) (Table 3). Isolates presenting the wild-type pfcrt and pfmdr1 (CVMNK-NYSND) alleles possessed low IC50s (median IC50, 16.00 nM) for CQ. Low IC50s were also found in the CVMNT-NYSND and CVMNT-NYSNY alleles. The CVMNK-YYSNY haplotype was associated with a very high IC50 (median IC50, 188.00 nM) for CQ and were resistant to CQ (P < 0.01). Thirty-three isolates (89.19%) out of 37 CVMNK-YYSNY haplotypes were highly in vitro CQ resistant. The CVIEK-YYSNY allele also showed high IC50s (median IC50, 235.00 nM) for CQ. Out of 11 isolates with CVIEK-YYSNY, 10 isolates produced in vitro CQ resistance. The SVMNT-NYSND double mutant and the SVMNT-YYSND and SVMNT-NYSNY triple mutant haplotypes were found to have moderate to high IC50s for CQ. The isolates presenting with the CVIET-NFSND (median IC50, 302.00 nM) and CVMNT-YYSNY (median IC50, 306.00 nM) alleles showed very high IC50s for CQ (Fig. 1 and 3). It is clear from Fig. 3 that the commonly found CVMNK-YYSNY haplotype possessed a lower median IC50 than did the control Dd2 strain, while the CVIEK-YYSNY allele had shown nearly identical IC50 to that of the Dd2 strain. The CVMNT-YYSNY triple mutant and CVIET-NFSND quadruple mutant alleles produced very high IC50 for CQ compared to that of the Dd2 strain. Molecular genotype and in vivo CQ treatment efficacy, as well as in vitro CQ resistance, were strongly correlated (r2 = 0.9991, P < 0.014).
FIG 3.
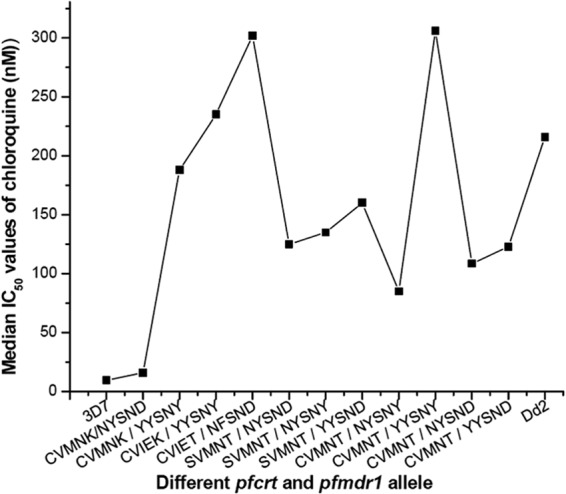
In vitro median IC50s of chloroquine in different pfcrt and pfmdr1 haplotype groups after 48 h of drug treatment.
DISCUSSION
The present work provides a comprehensive picture of antimalarial drug resistance in the Jangalmahal area of eastern India at a time that coincided with the implementation of a national drug policy. In 2009, the National Vector Borne Disease Control Program (NVBDCP) introduced ACT as a first-line option to treat all P. falciparum cases in India (30). This work involved in vivo CQ treatment efficacy, in vitro CQ susceptibility, and molecular genotyping of the pfcrt and pfmdr1 gene loci.
Forty-three (34.13%) ETF and 16 (12.69%) LTF cases were found after CQ treatment, whereas 103 isolates (81.75%) were found to have CQ resistance in vitro. These high rates of in vivo CQ treatment failure and in vitro CQ resistance indicated the enormous CQ drug pressure in this parasite population, as >45% of the cases resulted in CQ treatment failure in vivo. Previous studies stated that the higher number of pfcrt mutations produced higher levels of CQ resistance. It was reported from northern India that P. falciparum isolates with the CVIETS genotype showed higher levels of in vitro CQ resistance than did the isolates with the SVMNTS or CVMNTS allele (9, 31) Similarly, we observed that the CVIET isolates possessed higher in vitro IC50s for CQ than did isolates of the SVMNT or CVMNT haplotype. An increase in the number of these haplotypes was an alarming sign against malaria control.
The YYSNY double mutant (40.48%) allele was most common after the YYSND single mutant allele. Mutations at 86Y and 1246Y of the pfmdr1 gene were prevalent, showing very high IC50s for CQ. This type of finding was quite uncommon in India, but it was previously found in Madagascar, where polymorphisms at codons 86, 184, and 1246 of pfmdr1 were prevalent (12). The increase in pfmdr1 mutant alleles (86Y184Y1246D, 86Y184F1246D, and 86Y184F1246Y) and the low frequency of the pfcrt mutant genotype were related to CQ resistance in Madagascar. This might reflect the extensive use of antimalarial quinoline drugs, such as CQ and quinine (QU) (15).
It was confirmed that 76T mutations were associated with in vitro CQ resistance and also played a crucial role in LTF, but these mutations might not cause early CQ treatment failure. Forty isolates represented the SVMNT allele. Four LTF cases were associated with the SVMNT-NYSND haplotype, while another 2 LTF cases were observed to have the SVMNT-YYSND haplotype. Twenty-nine isolates (23.02%) contained the CVMNT haplotype, of which 12 isolates represented the CVMNT-NYSND haplotype, with low to moderate IC50s for CQ and resulting in only 1 ETF and 1 LTF case. When this CVMNT allele was associated with the YYSND or YYSNY allele, it resulted in very high IC50s for CQ. This finding implies that 76T mutations alone might not result in much selective pressure in CQ treatment efficacy in this population. One ETF case and 1 LTF case were detected among 4 CVIET-NFSND isolates. Our results also indicate that 76T might be the first mutation to occur in the pfcrt gene, followed by the 72S mutation. Moreover, mutations at codons 74 and 75 took place independently due to an increase in drug pressure (9). It was surely an alarming development that isolates with the SVMNT and CVIET genotypes started to increase in number.
Of the isolates, 29.37% contained the CVMNK-YYSNY haplotype. Of the isolates, 8.73% represented the CVIEK-YYSNY allele. We found that in vivo CQ treatment failure (ETF and LTF) and in vitro CQ resistance were strongly correlated with these CVMNK-YYSNY and CVIEK-YYSNY haplotypes. The presence of the wild-type CVMNK haplotype in the CQ nonresponder group was quite uncommon in India. A similar result was previously found in the nearby state Madhya Pradesh in 2005 (32).
The correlation between the pfmdr1 genotypes and CQ resistance often generated conflicting results, although it was suggested that the pfmdr1 86Y mutation might have correlated with increased CQ resistance in different parts of the world (10, 12). In India, field studies did not support these findings and the results of a P. falciparum genetic cross, which indicated that CQ resistance was not related to pfmdr1 gene mutations (19, 33). It was reported that in vitro CQ resistance and in vivo CQ treatment failure in India were solely related to pfcrt mutations (20). The substitution of lysine to threonine at codon 76 consistently was found in CQ-resistant P. falciparum isolates (34). Therefore, it was proposed that the detection of a 76T mutation in the pfcrt gene might allow information to be provided on the CQ resistance status of the parasite. As pfmdr1 mutations were strongly related to CQ resistance in Jangalmahal, it seemed that the detection of a mutation in pfcrt 76T alone is not enough to predict the drug resistance scenario in this part of India.
The impact of the pfcrt-pfmdr1 combination mutation depended on the genetic background of the strain, as demonstrated by studies with genetically manipulated lines or recombinant progeny of experimental crosses (35) and the history of the use of antimalarial drugs (CQ and quinine [QU] were the two main drugs used in India). Additional factors contributing to the uncommon drug resistance situation in this region might be its geographical position and broad range of malarial epidemiological strata in which the three Plasmodium species are present.
Our present findings imply that the CVMNK-YYSNY and CVIEK-YYSNY haplotypes were highly correlated (P < 0.01) with in vitro CQ resistance and in vivo CQ treatment failure (P < 0.01), while mutations in 76T and 72S increased the degree of resistance, i.e., resulted in very high IC50s. This might be a localized parasite population, since the presence of both 86Y and 1246Y mutations in our samples were largely dependent on their CQ response, indicating that CQ appeared to exert a selective pressure in this area, which seemed to be unusual compared to previous findings from India (20).
In conclusion, it seems that the clinical efficacy of CQ may rapidly wither away. Further studies are needed to obtain the full-length sequences of the pfcrt and pfmdr1 genes in this interesting parasite population.
ACKNOWLEDGMENTS
This work was not supported by any funding source.
We thank Vidyasagar University, Midnapore, India, for providing the facilities to execute these studies. We also thank the Council of Scientific and Industrial Research (CSIR), India, for fellowship to S.D. We also thank The Gautam Laboratories, Kolkata, India (NABL accredited laboratory, ISO 15189:2007-M-0423). Under their supervision, some work was done in collaboration with Vidyasagar University, Midnapore. Finally, we thank Purulia District Hospital, Bankura Medical College, and Midnapore Medical College for their assistance in completing the work.
We declare no conflicts of interest.
Footnotes
Published ahead of print 28 July 2014
REFERENCES
- 1.Trape JF, Pison G, Spiegel A, Enel C, Rogier C. 2002. Combating malaria in Africa. Trends Parasitol. 18:224–230. 10.1016/S1471-4922(02)02249-3 [DOI] [PubMed] [Google Scholar]
- 2.Aubouy A, Jafari S, Huart V, Migot-Nabias F, Mayombo J, Durand R, Bakary M, Le Bras J, Deloron P. 2003. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J. Antimicrob. Chemother. 52:43–49. 10.1093/jac/dkg294 [DOI] [PubMed] [Google Scholar]
- 3.Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K. 2010. Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 113:190–194. 10.1016/j.actatropica.2009.10.016 [DOI] [PubMed] [Google Scholar]
- 4.Laufer MK, Djimdé AA, Plowe CV. 2007. Monitoring and deterring drug-resistant malaria in the era of combination therapy. Am. J. Trop. Med. Hyg. 77(6 Suppl):160–169 [PubMed] [Google Scholar]
- 5.Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. 2009. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar. J. 8:89. 10.1186/1475-2875-8-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warhurst DC. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:299–302. 10.1056/NEJM200101253440411 [DOI] [PubMed] [Google Scholar]
- 7.Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257–263. 10.1056/NEJM200101253440403 [DOI] [PubMed] [Google Scholar]
- 8.Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic ZZ, Le Bras J. 2001. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 114:95–102. 10.1016/S0166-6851(01)00247-X [DOI] [PubMed] [Google Scholar]
- 9.Mittra P, Vinayak S, Chandawat H, Das MK, Singh N, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. 2006. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J. Infect. Dis. 193:1304–1312. 10.1086/502979 [DOI] [PubMed] [Google Scholar]
- 10.Duraisingh MT, Cowman AF. 2005. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 94:181–190. 10.1016/j.actatropica.2005.04.008 [DOI] [PubMed] [Google Scholar]
- 11.Das S, Chakraborty SP, Hati AK, Roy S. 2012. Association between prevalence of chloroquine resistance and unusual mutation in pfmdr1 and pfcrt genes in India. Am. J. Trop. Med. Hyg. 88:828–834. 10.4269/ajtmh.11-0795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Tichit M, Jahevitra M, Rabearimanana S, Raherinjafy R, Mercereau-Puijalon O, Durand R, Ménard D. 2010. Chloroquine clinical failures in P. falciparum malaria are associated with mutant pfmdr-1, not pfcrt in Madagascar. PLoS One 5:e13281. 10.1371/journal.pone.0013281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngo T, Duraising M, Reed M, Hipgrave D, Biggs B, Cowman AF. 2003. Analysis of pfcrt, pfmdr1, dhfr and dhps mutation and drug sensitivities in Plasmodium falciparum isolates from patients in Vietnam before and after treatment with artemisinin. Am. J. Trop. Med. Hyg. 68:350–356 [PubMed] [Google Scholar]
- 14.Duraisingh MT, Jones P, Sambou I, Pinder M, Warhurst DC. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13–23. 10.1016/S0166-6851(00)00201-2 [DOI] [PubMed] [Google Scholar]
- 15.Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Jahevitra M, Rabearimanana S, Radrianjafy R, Andrianaranjaka V, Randriantsoa T, Rason MA, Tichit M, Rabarijaona LP, Mercereau-Puijalon O, Durand R, Ménard D. 2009. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob. Agents Chemother. 53:4588–4597. 10.1128/AAC.00610-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valecha N, Srivastava P, Mohanty SS, Mittra P, Sharma SK, Tyagi PK, Pradhan K, Dev V, Singh R, Dash AP, Sharma YD. 2009. Therapeutic efficacy of artemether-lumefantrine in uncomplicated falciparum malaria in India. Malar. J. 8:107. 10.1186/1475-2875-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babiker HA, Pringale SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. 2001. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutation in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J. Infect. Dis. 183:1535–1538. 10.1086/320195 [DOI] [PubMed] [Google Scholar]
- 18.Sharma I, Aneja MK, Biswas S, Deb V, Ansari MA, Pasha ST, Sharma YD. 2001. Allelic variation in the cg2 gene does not correlate with chloroquine resistance among Indian Plasmodium falciparum isolates. Int. J. Parasitol. 31:1669–1672. 10.1016/S0020-7519(01)00286-7 [DOI] [PubMed] [Google Scholar]
- 19.Vathsala PG, Pramanik A, Dhanasekaran S, Devi CU, Pillai CR, Subarrao SK, Ghosh SK, Tiwari SN, Sathyanarayan TS, Deshpande PR, Mishra GC, Ranjit MR, Dash AP, Rangarajan PN, Padmanaban G. 2004. Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene haplotype SVMNT in P. falciparum malaria in India. Am. J. Trop. Med. Hyg. 70:256–259 [PubMed] [Google Scholar]
- 20.Valecha N, Joshi H, Mallick PK, Sharma SK, Kumar A, Tyagi PK, Shahi B, Das MK, Nagpal BN, Dash AP. 2009. Low efficacy of chloroquine: time to switch over to artemisinin-based combination therapy for falciparum malaria in India. Acta Trop. 111:21–28. 10.1016/j.actatropica.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 21.Directorate of Health Services, National Vector Borne Disease Control Program (NVBDCP). 2008. Status of drug resistance in India. National Vector Borne Disease Control Program, Delhi, India [Google Scholar]
- 22.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R Soc. Trop. Med. Hyg. 93:369–374. 10.1016/S0035-9203(99)90120-7 [DOI] [PubMed] [Google Scholar]
- 23.National Vector Borne Disease Control Program (NVBDCP). 2008. National drug policy on malaria. National Vector Borne Disease Control Program, Delhi, India [Google Scholar]
- 24.World Health Organization (WHO). 2003. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/hq/2003/WHO_HTM_RBM_2003.50.pdf?ua=1 [Google Scholar]
- 25.Das S, Chakraborty SP, Tripathy S, Hati AK, Roy S. 2012. Association between prevalence of pyrimethamine resistance and double mutation in pfdhfr gene in West Bengal, India. Asian Pac. J. Trop. Dis. 31–5. 10.1016/S2222-1808(12)60008-5 [DOI] [Google Scholar]
- 26.Basco KL, Ringwald P. 2000. Molecular epidemiology of malaria in Yaounde Cameroon. VI. Sequence variations in the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene and in vitro resistance to pyrimethamine and cycloguanil. Am. J. Trop. Med. Hyg. 62:271–276 [DOI] [PubMed] [Google Scholar]
- 27.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 28.Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil Pedro J, do Rosário VE, Cravo P. 2002. Molecular characterisation of drug-resistant Plasmodium falciparum from Thailand. Malar. J. 1:12. 10.1186/1475-2875-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Chakraborty SP, Hati AK, Roy S. 2013. Malaria treatment failure with novel mutation in Plasmodium falciparum dihydrofolate reductase (pfdhfr) gene in Kolkata, West Bengal, India. Int. J. Antimicrob. Agents 41:447–451. 10.1016/j.ijantimicag.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 30.Government of India, National Institute of Malaria Research, National Vector Borne Disease Control Programme. 2009. Guidelines for diagnosis and treatment of malaria in India. National Vector Borne Disease Control Programme, Delhi, India: http://nvbdcp.gov.in/Doc/Guidelines_for_Diagnosis___Treatment.pdf [Google Scholar]
- 31.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naudé B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861–871. 10.1016/S1097-2765(05)00077-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bharti PK, Alam MT, Boxer R, Shukla MM, Gautam SP, Sharma YD, Singh N. 2010. Therapeutic efficacy of chloroquine and sequence variation in pfcrt gene among patients with falciparum malaria in central India. Trop. Med. Int. Health 15:33–40. 10.1111/j.1365-3156.2009.02425.x [DOI] [PubMed] [Google Scholar]
- 33.Vinayak S, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. 2003. Prevalence of the K76T mutation in the pfcrt gene of Plasmodium falciparum among chloroquine responders in India. Acta Trop. 87:287–293. 10.1016/S0001-706X(03)00021-4 [DOI] [PubMed] [Google Scholar]
- 34.Sharma YD. 2012. Molecular surveillance of drug-resistant malaria in India. Curr. Sci. 102:696–703 [Google Scholar]
- 35.Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. U. S. A. 106:18883–18889. 10.1073/pnas.0911317106 [DOI] [PMC free article] [PubMed] [Google Scholar]


