Abstract
Kingella kingae is the major pathogen causing osteoarticular infections (OAI) in young children in numerous countries. Plasmid-borne TEM-1 penicillinase production has been sporadically detected in a few countries but not in continental Europe, despite a high prevalence of K. kingae infections. We describe here for the first time a K. kingae β-lactamase-producing strain in continental Europe and demonstrate the novel chromosomal location of the blaTEM-1 gene in K. kingae species.
TEXT
Kingella kingae is the major pathogen causing osteoarticular infections (OAI) in young children in numerous countries (1–4). Plasmid-borne TEM-1 penicillinase production has been sporadically detected in a few countries but not in continental Europe, despite a high prevalence of K. kingae infections (2–9).
We describe here for the first time a K. kingae β-lactamase-producing strain in continental Europe and demonstrate the novel chromosomal location of the blaTEM-1 gene in K. kingae species.
A healthy 2-year-old girl was admitted with a 1-day history of typical knee arthritis with moderate clinical and biologic features. A blood culture remained sterile. As the patient had spontaneous clinical improvement with neither pain nor fever at 48 h, she was discharged with no antibiotic treatment. However, since OAI was suspected, an oropharyngeal sample was obtained, for which a culture and specific PCR were positive for K. kingae (2, 10). This isolate was identified by the nitrocefin method (Oxoid, Ltd., Basingstoke, Hampshire, England) as a β-lactamase producer. The MICs of amoxicillin and amoxicillin-clavulanic acid, which were determined by the Etest method (bioMérieux, Marcy l'Etoile, France) (11), were 0.5 μg/ml and 0.094 μg/ml, respectively. Iterative throat samples from the patient's household contacts were obtained (with the written consent of her parents and with approval by the institutional review board [IRB00006477]) and cultured at 1, 2, 4, 6, 8, 10, 17, and 28 weeks after the arthritis episode. All the cultures for the patient and her 9-year-old brother were positive for the first 10 weeks (KWG and MAL strains, respectively), while PCR results were positive for the samples at up to 17 weeks for both of them. The parents' throat samples remained negative. Each isolate was a β-lactamase producer, and a multiplex PCR for β-lactamase genes identified a blaTEM-1 gene (12).
According to multilocus sequence typing, the strains harbored the new sequence type 41 (ST41) belonging to ST complex 14 (STc-14) (13, 14). Randomly chosen KWG and MAL isolates were indistinguishable (data not shown) by pulsed-field gel electrophoresis (PFGE) of EagI-digested DNA (New England BioLabs, Inc., Ipswich, MA) (7).
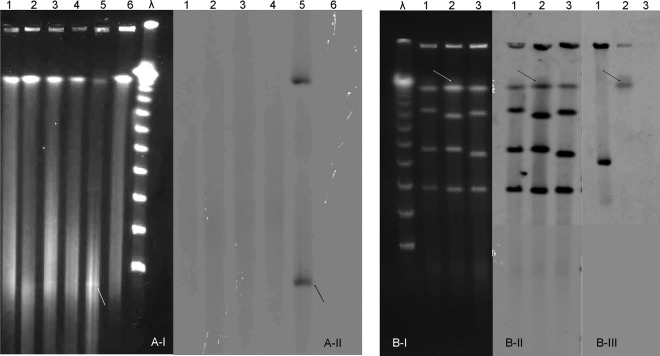
No plasmid bands were visualized for the KWG or MAL isolates by PFGE of S1 nuclease-digested DNA (Sigma-Aldrich, St. Louis, MO), even by Southern blot hybridization with a blaTEM-1 probe (PCR DIG probe synthesis kit; Roche Diagnostics, Indianapolis, IN) (15), in contrast to the results seen with the U.S. plasmid-borne TEM-1 producer C2005004457 (Fig. 1A) (9). Furthermore, we performed PFGE of I-CeuI-digested DNA (New England BioLabs, Inc.) by cutting a 26-bp restriction site in the 23S rRNA gene (16). The Southern blot experiments revealed that, for KWG1, one band hybridized with the blaTEM-1 probe and with a combined 16S/23S rRNA gene probe, while for the U.S. strain, the blaTEM-1 probe hybridized elsewhere outside of the chromosome (Fig. 1B). This result confirmed that blaTEM-1 is chromosomally located for the KWG1 strain.
FIG 1.
Pulsed-field gel electrophoresis (PFGE) and Southern blot analysis of Kingella kingae penicillinase-producing strains. (A) PFGE of S1 nuclease-digested DNA allowing visualization of plasmids (A-I) and Southern blot using a blaTEM-1 gene probe (A-II). Lane λ, molecular marker; lanes 1, KWG1, 1st isolate of the patient during arthritis episode at week 0; lanes 2, KWG4, 4th patient isolate at week 4; lanes 3, MAL1, 1st isolate of the patient's brother at week 1; lanes 4, MAL2, 2nd isolate of the patient's brother at week 4; lanes 5, C2005004457, U.S. plasmid-borne β-lactamase producer; lanes 6, ATCC 23330 type strain of K. kingae species, non-β-lactamase producer. Arrows show the plasmid band (A-I) hybridizing with the blaTEM-1 gene probe (A-II) for the U.S. strain. (B) PFGE of I-CeuI-digested DNA (B-I) and Southern blots using a 16S/23S rRNA gene probe (B-II) and a blaTEM-1 gene probe (B-III). Lane λ, molecular marker; lanes 1, C2005004457, U.S. plasmid-borne β-lactamase producer; lanes 2, KWG1, 1st patient isolate at week 0; lanes 3, ATCC 23330 type strain of K. kingae species, non-β-lactamase producer. Arrows show the band (B-I) which hybridized with the 16S/23S rRNA gene probe (B-II) and with the blaTEM-1 gene probe (B-III) for the patient isolate.
In conclusion, we describe here the first K. kingae strain in continental Europe with a chromosomal blaTEM-1 gene that belongs to the intercontinental and virulent STc-14 (13, 14), which did not previously include β-lactamase producers (8). We demonstrated the ability of the KWG strain to persist for a long time in the patient's oropharynx and to disseminate to her 9-year-old brother, although K. kingae is very rarely isolated in the oropharynx after the age of 4 years (17). These results emphasize the risk of emergence and spread of K. kingae β-lactam-resistant strains. Oropharyngeal sampling is more sensitive than osteoarticular sampling for isolating K. kingae (10) and could be an interesting approach to performing β-lactam susceptibility test.
ACKNOWLEDGMENTS
We acknowledge Céline Courroux and Julien Pansiot for their technical assistance.
We declare no conflicts of interest.
This work was supported in part by the Société Française de Pédiatrie. The funders had no role in the study design, data collection and analysis, publication decision, or preparation of the manuscript.
Footnotes
Published ahead of print 21 July 2014
REFERENCES
- 1.Yagupsky P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4:358–367. 10.1016/S1473-3099(04)01046-1 [DOI] [PubMed] [Google Scholar]
- 2.Ilharreborde B, Bidet P, Lorrot M, Even J, Mariani-Kurkdjian P, Liguori S, Vitoux C, Lefevre Y, Doit C, Fitoussi F, Pennecot G, Bingen E, Mazda K, Bonacorsi S. 2009. New real-time PCR-based method for Kingella kingae DNA detection: application to samples collected from 89 children with acute arthritis. J. Clin. Microbiol. 47:1837–1841. 10.1128/JCM.00144-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceroni D, Cherkaoui A, Ferey S, Kaelin A, Schrenzel J. 2010. Kingella kingae osteoarticular infections in young children: clinical features and contribution of a new specific real-time PCR assay to the diagnosis. J. Pediatr. Orthop. 30:301–304. 10.1097/BPO.0b013e3181d4732f [DOI] [PubMed] [Google Scholar]
- 4.Chometon S, Benito Y, Chaker M, Boisset S, Ploton C, Berard J, Vandenesch F, Freydiere AM. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26:377–381. 10.1097/01.inf.0000259954.88139.f4 [DOI] [PubMed] [Google Scholar]
- 5.Birgisson H, Steingrimsson O, Gudnason T. 1997. Kingella kingae infections in paediatric patients: 5 cases of septic arthritis, osteomyelitis and bacteraemia. Scand. J. Infect. Dis. 29:495–498. 10.3109/00365549709011861 [DOI] [PubMed] [Google Scholar]
- 6.Sordillo EM, Rendel M, Sood R, Belinfanti J, Murray O, Brook D. 1993. Septicemia due to beta-lactamase-positive Kingella kingae. Clin. Infect. Dis. 17:818–819. 10.1093/clinids/17.4.818 [DOI] [PubMed] [Google Scholar]
- 7.Yagupsky P, Slonim A, Amit U, Porat N, Dagan R. 2013. Beta-lactamase production by Kingella kingae in Israel is clonal and common in carriage organisms but rare among invasive strains. Eur. J. Clin. Microbiol. Infect. Dis. 32:1049–1053. 10.1007/s10096-013-1849-1 [DOI] [PubMed] [Google Scholar]
- 8.Basmaci R, Bonacorsi S, Bidet P, Balashova NV, Lau J, Munoz-Almagro C, Gene A, Yagupsky P. 26 April 2014. Genotyping, local prevalence, and international dissemination of beta-lactamase-producing Kingella kingae strains. Clin. Microbiol. Infect. 10.1111/1469-0691.12648 [DOI] [PubMed] [Google Scholar]
- 9.Banerjee A, Kaplan JB, Soherwardy A, Nudell Y, Mackenzie GA, Johnson S, Balashova NV. 2013. Characterization of TEM-1 beta-lactamase producing Kingella kingae clinical isolates. Antimicrob. Agents Chemother. 57:4300–4306. 10.1128/AAC.00318-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basmaci R, Ilharreborde B, Bidet P, Doit C, Lorrot M, Mazda K, Bingen E, Bonacorsi S. 2012. Isolation of Kingella kingae in the oropharynx during K. kingae arthritis in children. Clin. Microbiol. Infect. 18:E134–E136. 10.1111/j.1469-0691.2012.03799.x [DOI] [PubMed] [Google Scholar]
- 11.Yagupsky P. 2012. Antibiotic susceptibility of Kingella kingae isolates from children with skeletal system infections. Pediatr. Infect. Dis. J. 31:212. 10.1097/INF.0b013e31824041b8 [DOI] [PubMed] [Google Scholar]
- 12.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 13.Basmaci R, Yagupsky P, Ilharreborde B, Guyot K, Porat N, Chomton M, Thiberge JM, Mazda K, Bingen E, Bonacorsi S, Bidet P. 2012. Multilocus sequence typing and rtxa toxin gene sequencing analysis of Kingella kingae isolates demonstrates genetic diversity and international clones. PLoS One 7:e38078. 10.1371/journal.pone.0038078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amit U, Porat N, Basmaci R, Bidet P, Bonacorsi S, Dagan R, Yagupsky P. 2012. Genotyping of invasive Kingella kingae isolates reveals predominant clones and association with specific clinical syndromes. Clin. Infect. Dis. 55:1074–1079. 10.1093/cid/cis622 [DOI] [PubMed] [Google Scholar]
- 15.Lemaître C, Mahjoub-Messai F, Dupont D, Caro V, Diancourt L, Bingen E, Bidet P, Bonacorsi S. 2013. A conserved virulence plasmidic region contributes to the virulence of the multiresistant Escherichia coli meningitis strain S286 belonging to phylogenetic group C. PLoS One 8:e74423. 10.1371/journal.pone.0074423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878. 10.1073/pnas.90.14.6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagupsky P, Peled N, Katz O. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 40:4180–4184. 10.1128/JCM.40.11.4180-4184.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]