Abstract
The ability of different antibiotics to select for extended-spectrum β-lactamase (ESBL)-producing Escherichia coli remains a topic of discussion. In a mouse intestinal colonization model, we evaluated the selective abilities of nine common antimicrobials (cefotaxime, cefuroxime, dicloxacillin, clindamycin, penicillin, ampicillin, meropenem, ciprofloxacin, and amdinocillin) against a CTX-M-15-producing E. coli sequence type 131 (ST131) isolate with a fluoroquinolone resistance phenotype. Mice (8 per group) were orogastrically administered 0.25 ml saline with 108 CFU/ml E. coli ST131. On that same day, antibiotic treatment was initiated and given subcutaneously once a day for three consecutive days. CFU of E. coli ST131, Bacteroides, and Gram-positive aerobic bacteria in fecal samples were studied, with intervals, until day 8. Bacteroides was used as an indicator organism for impact on the Gram-negative anaerobic population. For three antibiotics, prolonged colonization was investigated with additional fecal CFU counts determined on days 10 and 14 (cefotaxime, dicloxacillin, and clindamycin). Three antibiotics (cefotaxime, dicloxacillin, and clindamycin) promoted overgrowth of E. coli ST131 (P < 0.05). Of these, only clindamycin suppressed Bacteroides, while the remaining two antibiotics had no negative impact on Bacteroides or Gram-positive organisms. Only clindamycin treatment resulted in prolonged colonization. The remaining six antibiotics, including ciprofloxacin, did not promote overgrowth of E. coli ST131 (P > 0.95), nor did they suppress Bacteroides or Gram-positive organisms. The results showed that antimicrobials both with and without an impact on Gram-negative anaerobes can select for ESBL-producing E. coli, indicating that not only Gram-negative anaerobes have a role in upholding colonization resistance. Other, so-far-unknown bacterial populations must be of importance for preventing colonization by incoming E. coli.
INTRODUCTION
Escherichia coli is a versatile and ubiquitous species that is regularly represented in the commensal flora of the gut. The species includes nonpathogenic, intestinal pathogenic, and extraintestinal pathogenic E. coli (ExPEC), all of which can be variably present in the human gut. ExPEC can cause a wide range of infections, from uncomplicated cystitis to life-threatening sepsis (1–3). Major sources for resistance in E. coli are plasmid-borne extended-spectrum β-lactamases (ESBL), which are enzymes capable of hydrolyzing, and thus conferring resistance toward, most β-lactam antibiotics, except for the cephamycins and carbapenems. In addition, ESBLs are inhibited by β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam (1, 4). Plasmids carrying ESBL genes often carry various other genes that cause resistance to other classes of antibiotics (e.g., aminoglycosides) (1, 4, 5). One of the most common types of ESBLs identified in the world is CTX-M-15, and the spread of its areas of endemicity seems to be associated with a few E. coli sequence types (ST), such as ST131. ST131 is most frequently linked to quinolone resistance and CTX-M-15, but it also harbors specific virulence genes coding for factors such as adhesins (fimH, papEF), toxins (sat), and capsules (kpsM II), contributing to its ability to colonize the human gut (2, 4–7). The possible human colonization is of great concern, since ST131 isolates often are resistant to several antibiotics and are known to cause urosepsis to a higher degree than non-ST131 isolates. In clinical settings, multiresistance, including production of ESBL, delays appropriate treatment, leading to extended hospital stays as well as increased mortality and morbidity (8–10). Finally, urinary tract infections are primarily caused by E. coli present in the patient's own microbiota, making knowledge on colonization by resistant E. coli of great importance (1, 4, 11).
Several case-control studies have identified recent antibiotic exposure, especially to cephalosporins and fluoroquinolones, and hospitalization as significant risk factors for acquiring an infection with an ESBL-producing Enterobacteriaceae (within 30 days) (10, 12–14). A systematic study of the selective ability of all antibiotic classes has been lacking. In this study, we wished to evaluate the ability of nine common antimicrobials, including antibiotics used for Gram-positive infections, to select for a CTX-M-15-producing E. coli ST131 isolate in a mouse intestinal colonization model.
(This work was presented as a poster at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 9 to 12 September 2012.)
MATERIALS AND METHODS
Strain.
For the colonizing pathogen, we used a clinical blood isolate of E. coli that belongs to the lineage B2-O25b-ST131. This isolate (65-Ec-09) carries some of the virulence factors previously seen in ST131 isolates found throughout Denmark in 2009, and it is similarly resistant toward many commonly used antibiotics (MICs when resistant: cefotaxime, >32 μg/ml; cefuroxime, >256 μg/ml; ceftazidime, 32 μg/ml; ampicillin, >256 μg/ml; aztreonam, 32 μg/ml; amoxicillin-clavulanate, >256 μg/ml; ciprofloxacin, >32 μg/ml; cloxacillin, >256 μg/ml; clindamycin, >256 μg/ml. MICs when susceptible are as follows: amdinocillin, 2 μg/ml; trimethoprim-sulfamethoxazole, ≤0.125 μg/ml; gentamicin, ≤0.5 μg/ml; piperacillin-tazobactam, 2 μg/ml; meropenem, ≤0.064 μg/ml; nitrofurantoin, 13 millimeters in zone diameter [mm]; trimethoprim, 27 mm) (15). Pheno- and genotypic characterizations were performed by using the MAST-test, PCR, and DNA sequence analysis according to methods used at the clinical laboratory of Department of Clinical Microbiology, Hvidovre Hospital, Denmark (HVH) or at Statens Serum Institut (SSI) as previously described (15). Sequence type verification was performed via full multilocus sequence typing (MLST, using the Achtmann scheme [http://mlst.warwick.ac.uk/mlst/dbs/Ecoli]) by SSI, who also found that the isolate harbored the virulence factors kpsM II and iutA (15–17).
Media.
To test bacterial growth in collected fecal samples from mice, we used selective agar plates, all from SSI Diagnostica, Hilleroed, Denmark. For the CTX-M-15-producing E. coli population, ID Flexicult agar containing cefotaxime at 32 mg/liter and vancomycin at 6 mg/liter were used. The Gram-positive aerobic population was selected on 5% blood agar plates containing gentamicin at 5 mg/liter, and the Gram-negative anaerobic population was selected on anaerobic plates containing gentamicin at 32 mg/liter and vancomycin at 16 mg/liter. Culturing of anaerobic species were performed under anaerobic conditions in GasPak EZ containers and an anaerobic atmosphere created by using AnaeroGen (Oxoid) (18).
MIC determinations.
Antibiotic susceptibility testing was performed as Etests when possible and as disc diffusion tests where no Etest was available (the MIC for dicloxacillin was found by using an Etest for cloxacillin, since neither Etest nor disks for dicloxacillin were available). The diffusion test methodology has been described elsewhere (15), and a similar Etest methodology was used according to guidelines from the Department of Clinical Microbiology, Hvidovre Hospital. All results were interpreted according to current recommendations from the European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/clinical_breakpoints/).
Antibiotics used for treatment.
Commonly used antibiotics were chosen for this study. These included different β-lactam antibiotics, ciprofloxacin, and clindamycin. All mouse dosages were calculated based on human doses (in mg per kg of body weight) from pharmacokinetic (PK) studies performed at SSI or from previously published mouse studies (Table 1) (19–26). Doses were chosen to mimic the serum antibiotic concentrations achieved in humans on standard doses. All antibiotics were administered subcutaneously once each day for 3 consecutive days. Concentrations of antibiotics in mouse feces had been measured in a previously published study for a β-lactam antibiotic, an expanded-spectrum cephalosporin, a carbapenem, clindamycin, and ciprofloxacin (19). The doses used in this study were similar or higher.
TABLE 1.
Antibiotics used as treatmentsa
| Antibiotic | Humanb |
Mouse |
|||
|---|---|---|---|---|---|
| Dose (g, i.v.) | Cmax (μg/ml) | Needed dose (mg/kg) | Cmax (μg/ml) | Dose given (mg/mouse/day) | |
| Cefuroxime | 1.5 | 65 | 120 | 50–60 | 4 |
| Cefotaxime | 1 | 40 | 60 | 100 | 2 |
| Ampicillin | 1 | 40 | 50 | 75 | 1.5 |
| Dicloxacillin | 1 | 30–40 | 60 | 90 | 2 |
| Benzylpenicillin | 2 mill. IEb | 60 | 70 | 60 | 2 |
| Amdinocillin | 0.4 | 30–40 | 60 | 30 | 2 |
| Meropenem | 0.5 | 26 | 50 | 50 | 1.5 |
| Clindamycin | 1.8 | 6 | 36 | 8 | 1.4 |
| Ciprofloxacin | 0.4 | 4 | 15 | 2 | 0.5 |
All doses were administered subcutaneously once a day, and the needed doses were calculated based on the expected average weight of the mice (weights given by provider), as described in previously published studies (18–25). All mouse dosages were calculated based on human doses from PK studies performed at SSI or from previously published mouse studies (19–26). i.v., intravenous.
Benzylpenicillin (1.2 grams = 2 millions units = 2 mill. IE) was administered intravenously.
Mouse intestinal colonization model. (i) Mice.
The animal experiment was approved by the Danish Centre for Animal Welfare and carried out at Statens Serum Institut in Copenhagen, Denmark. In all studies, 7- to 10-week-old female albino, outbred NMRI mice (Harlan, the Netherlands) weighing 26 to 30 g were used. The mice used in each study were all from the same litter and were brought simultaneously to the stable and housed in pairs of two per cage. At the end of the study, all mice were sacrificed to ensure that no mice were kept alive with no immediate purpose. Animals were housed, treated, and sacrificed according to current guidelines.
(ii) The mouse model.
The mouse intestinal colonization model was an experimental model where all mice were kept in pairs of two per cage. Two cages constituted one group and each group received one antibiotic. Thus, each antibiotic was given to a total of four mice in two different cages, and a total of 20 to 22 cages were included in the study. Treatment was given subcutaneously in the neck once a day for three consecutive days (day 1 to day 3). Inoculation of mice with the bacterial strain was done through a stainless steel orogastric feeding tube on day 1 prior to initiation of treatment. The intestinal flora was unaltered prior to the study, and no mice were anesthetized during the study. The experiment was conducted from day 1 to day 8, and cages were changed daily. At the end of day 8 all mice were sacrificed.
(iii) Experimental study.
We executed our full experimental study by using the described mouse intestinal colonization model. The study was conducted from day 1 to day 8 with feces collected prior to inoculation on day 1 and on days 2, 4, and 8. On day 1, mice were inoculated once with 0.25 ml of saline containing 108 CFU/ml of 65-Ec-09. Thus, each mouse was given an inoculum of 2.5 × 107 CFU.
The mice were left for 3 h before the first doses of antibiotics were administered subcutaneously. Each treatment group, consisting of four mice housed in two different cages, received cefotaxime, cefuroxime, ampicillin, dicloxacillin, amdinocillin, meropenem, clindamycin, ciprofloxacin, or benzylpenicillin (Table 1). A control group received 65-Ec-09 but no antibiotic treatment. The complete selection study of E. coli ST131 CTX-M-15 (65-Ec-09) in the mouse intestinal colonization model was performed twice, with the exception of CFU counts of the Gram-positive aerobic flora, which were only studied in the second run. The control group receiving 65-Ec-09 only was included in both runs. Furthermore, a group of mice that received treatment with cefotaxime without receiving the ESBL-producing strain was included in the first run only.
(iv) Prolonged presence of resistant pathogen after completed antibiotic treatment.
Additionally, the prolonged presence of the E. coli ST131 after completed antibiotic treatment was studied in the first run for three antibiotics (cefotaxime, dicloxacillin, and clindamycin). These three antibiotics were chosen based on their selective abilities found in the study. Treatment stopped, for all groups, on day 3. Cages were changed daily from days 1 to 14, and feces samples were collected on days 10 and 14. On day 14, the study was terminated and mice were killed.
Detection and quantification of bacteria in feces.
On specified days, 0.5 g of feces was collected from each cage. Feces were dissolved in 5 ml of saline and further diluted 10-fold in saline for a total of 6 times. Dilutions were plated on the different selective agar plates, and the log CFU per 0.5 g of stool for 65-Ec-09, the Gram-negative anaerobic flora, and the Gram-positive aerobic flora were calculated for each cage and two or three colonies from each day frozen. Each CFU count was performed on two agar plates and calculated as the average CFU count for the two plates. To ensure no presence of cefotaxime-resistant E. coli prior to inoculation, dilutions of feces from day 1, from each cage, were spotted on selective plates and this showed no growth of resistant E. coli. The lower detection limit was 10 CFU per 0.5 g of feces.
Molecular tests.
To ensure that the E. coli found in feces was identical to the isolate given through inoculation, we tested a total of 17 cefotaxime-resistant E. coli isolates found in feces during treatment and 4 E. coli isolates from day 1 for the presence of a CTX-M group 1 gene. Samples from both experimental runs were included (see below). Three to four isolates per frozen sample were used for DNA purification. As these tests were performed on isolates from both study runs, they were not performed until both runs had been completed. Thus, isolates tested were isolated from frozen samples. We were therefore likely to isolate the dominating E. coli strain and thereby determine if 65-Ec-09 had become the dominating E. coli of the microbiota (11, 27).
All of these 21 isolates were identified as E. coli by matrix-assisted laser desorption–time of flight analysis as described elsewhere (28). All resistant E. coli isolates contained a CTX-M group 1 gene, whereas none of the isolates from day 1 contained a CTX-M group 1 gene.
Isolates from the following groups were tested: cefotaxime (day 2 from both study runs and day 1 from the first run), cefuroxime (day 2 from both study runs and day 1 from the first run), ampicillin (day 2 from both study runs), dicloxacillin (days 1, 2, 4, and 8 from the first run and days 2 and 8 from the second run), clindamycin (days 1, 2, 4, and 10 from the first one and days 2 and 8 from the second run), and benzylpenicillin (day 2 from second run).
Additionally, five of the cefotaxime-resistant E. coli isolates and one isolate from day 1 were characterized by MLST (cefotaxime, cefuroxime [day 2], dicloxacillin [days 1 and 4], and clindamycin [days 4 and 10]). The five resistant E. coli were identified as ST131 and the one isolate from day 1 belonged to ST602.
Statistical analysis.
Data were analyzed with the use of SAS software, version 9.3 (SAS Institute). When just one group receiving one antibiotic was compared to the control group, this was done via a one-way analysis of variance (ANOVA). When multiple CFU counts were compared, it was done as a multiple variant analysis, adjusted for multiple comparisons with the Bonferroni correction. The conservative Bonferroni correction was used to avoid multiple comparisons. A P value of <0.05 was considered significant.
RESULTS
Results of fecal bacteriology were calculated as the log CFU/0.5 g of feces, and the mean CFU of four cages (two cages for Gram-positive aerobic flora) were used for statistical calculations. For graphic depictions, means and standard deviations (SD) were used. Data for the selective abilities of the different antibiotics on 65-Ec-09 are shown in Table 2 and Fig. 1 and 2 for E. coli, Gram-negative anaerobic flora, and Gram-positive flora, respectively.
TABLE 2.
Results of the CFU counts for E. coli ST131
| Antibiotic | Log CFU ± SD (P value) ona: |
||||
|---|---|---|---|---|---|
| Day 2 | Day 4 | Day 8 | Day 10 | Day 14 | |
| Cefotaxime | 7.75 ± 4.33 (<0.01) | 5.25 ± 6.54 (<0.01) | 3.61 ± 2.5 (0.011) | 1.5 ± 1.5 (0.98) | 0.5* ± 3.55 (1) |
| Clindamycin | 8.5 ± 1 (<0.01) | 7.75 ± 0.87 (<0.01) | 5.75 ± 0.87 (<0.01) | 2.5 ± 3.55 (0.44) | 1* ± 1.41 (1) |
| Dicloxacillin | 8.5 ± 1 (<0.01) | 9 ± 5.35 (<0.01) | 3 ± 3.74 (<0.01) | 0 (1) | 0 (1) |
| Cefuroxime | 7.5 ± 3 (<0.01) | 4.25 ± 5.89 (0.015) | 1.75 ± 4 (0.07) | ||
| Penicillin | 6.25 ± 3.57 (0.02) | 3.75 ± 6.76 (0.03) | 0 (1) | ||
| Amdinocillin | 2.25 ± 2.96 (0.3) | 1.25 ± 5.55 (0.5) | 0 (1) | ||
| Meropenem | 3.5 ± 2.96 (1) | 0 (1) | 0 (1) | ||
| Ciprofloxacin | 2.25 ± 2.6 (0.3) | 0 (1) | 0 (1) | ||
| Ampicillin | 3.25 ± 4.33 (0.8) | 2.5 ± 6.33 (0.14) | 0.75* ± 2.6 (0.4) | ||
| Control | 3.5 ± 1.73 | 0 | 0 | ||
P values for days 2 to 8 were determined via an ANOVA, while P values from days 10 and 14 were found by a multiple variant analysis with the Bonferroni correction.*, below the lower detection limit.
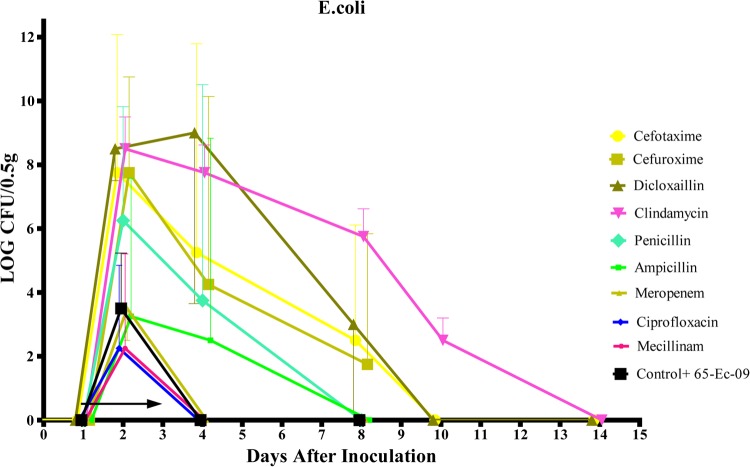
FIG 1.
The mean and SD of the log CFU/0.5 g of feces for 65-Ec-09 from day 1 to day 14. The arrow indicates treatment from day 1 to day 3.
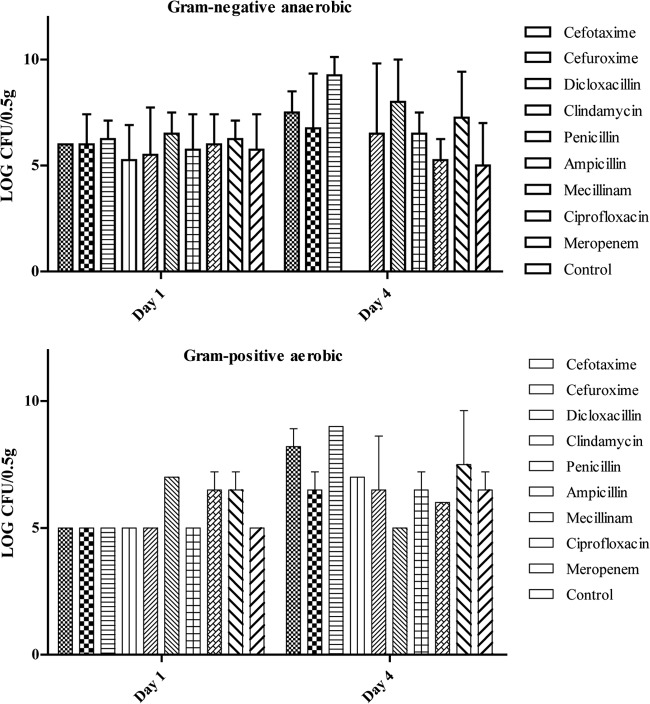
FIG 2.
The mean and SD of the log CFU/0.5 g of feces for Gram-negative anaerobic flora and Gram-positive aerobic flora, shown at day 1 and day 4 to compare the impact of 3 days of treatment. The SD was 0 for bars where the SD is not shown.
Effect of antibiotic treatment on establishment of resistant pathogens and on the indigenous microflora.
Data for the effects on colonization of 65-Ec-09 by the different antibiotics are shown in Fig. 1. Cefotaxime, dicloxacillin, and clindamycin promoted the colonization and overgrowth of 65-Ec-09 from day 2 through day 8 (P < 0.01 for dicloxacillin and clindamycin; P < 0.05 for cefotaxime). Benzylpenicillin and cefuroxime showed less overgrowth but selective abilities on days 2 and 4 (P < 0.05). Dicloxacillin and clindamycin showed the highest selective abilities (P < 0.01 for dicloxacillin on days 2 and 4 and P < 0.01 for clindamycin on all days). After treatment was completed on day 3, there was a decline in the colonization of 65-Ec-09 from day 4 to 8. In comparison, neither ampicillin, amdinocillin, meropenem, nor ciprofloxacin promoted overgrowth of 65-Ec-09 beyond day 2 (P > 0.05).
Data for the impact on the original microbiota as Gram-negative anaerobic flora, represented by Bacteroides, and Gram-positive flora, respectively, are shown in Fig. 2. None of the antibiotics used had an inhibiting or promoting impact on the Gram-positive flora (P > 0.05). Only clindamycin had an impact on the Gram-negative anaerobic flora, as it completely eliminated the Gram-negative anaerobic flora during treatment (P < 0.05). After treatment, the CFU counts for the Gram-negative anaerobic flora increased to counts equal to those before treatment.
Prolonged presence of 65-Ec-09 after completed antibiotic treatment.
Data for CFU of 65-Ec-09 on all sampling days are found in Fig. 1. After the initial increase, the CFU decreased for all three groups of antibiotics. For dicloxacillin and cefotaxime, the CFU counts dropped below the detection limit on day 10. For clindamycin, the CFU was measurable until day 14, the last day of the study. There were no significant differences in fecal CFU of 65-Ec-09 over time among the three antibiotics (P > 0.05).
DISCUSSION
To our knowledge, this is the first study to experimentally evaluate in vivo selection in the gut of a CTX-M-15-producing E. coli ST131 isolate by a range of commonly used antibiotics, including antibiotics with no activity against Gram-negative bacteria. With the mouse intestinal colonization model, we were able to illustrate the selective abilities of different antibiotics on the intestinal flora when a virulent isolate of CTX-M-15-producing E. coli ST131 was introduced. We found several interesting aspects of selection. First, we confirmed that antibiotics with activity against the colonizing strain did not promote proliferation, since neither amdinocillin nor meropenem resulted in colonization. Second, our findings confirmed that an antibiotic, clindamycin, that eliminates the Gram-negative anaerobic flora with no in vitro activity against Enterobacteriaceae promotes proliferation of ESBL-producing E. coli. It has been shown that antimicrobial impact on the total anaerobic population is proportional to the impact on the Bacteroides population (19). Third, of the β-lactams with no inhibiting effects on the ESBL-producing E. coli, cefotaxime showed the highest level of selection. Cefuroxime and benzylpenicillin did, however, show higher selection propensities than ampicillin, in agreement with what has previously been seen for high doses of penicillin (29). Furthermore, we discovered that dicloxacillin, with no obvious influence on either the Gram-negative anaerobic flora or other Gram-negative bacteria, promoted colonization with the ESBL-producing E. coli, while ciprofloxacin, with a limited in vitro effect on the Gram-negative anaerobic bacteria but an effect on Enterobacteriaceae, showed no abilities to select. These results could indicate that not only Gram-negative anaerobes have a key responsibility upholding colonization resistance. It appears that, as for penicillin (29), an antibiotic with limited impact on Gram-negative bacteria can select for a resistant E. coli isolate, suggesting that other bacterial populations, not measured in our study, such as certain anaerobic Gram positives, are of importance for preventing colonization from an incoming E. coli strain.
Finally, we found that the antibiotic with the highest impact on the anaerobic population, here represented by the Bacteroides population, seems to give room for prolonged colonization, i.e., clindamycin showed high to medium levels of 65-Ec-09 until day 14, compared to dicloxacillin and cefotaxime. This points to the possibility that even if unknown Gram-positive populations, anaerobic or aerobic, display colonization resistance, the Bacteroides population seems to play a role in preventing prolonged colonization.
Our study suggests that selection of E. coli ST131 is not derived alone by the antibiotic impact on the major population, such as anaerobes, as seen with dicloxacillin versus ampicillin and ciprofloxacin. This finding could potentially alter the perception of which antibiotics drive the spread of ESBL-producing E. coli, including ST131, even if clindamycin was the antibiotic that showed the highest level of selection and longest duration of colonization.
The model has some limitations, since it does not include clinical aspects of selection, such as treatment with multiple antimicrobials or long-term treatment, nor does it take into account reexposure to resistant pathogens or retreatment after exposure. Also, we did not investigate the antibiotics' effects on the total bacterial population of the intestines, including shifts in the dominating phyla. Such an investigation of changes in species and changes in the total Enterobacteriaceae population could potentially have described, in detail, factors influencing colonization. Future studies of the microbiota should be designed to fully describe antibiotic impacts on the different phyla. The lack of selective ability seen from ciprofloxacin on the CTX-M-15-producing E. coli ST131 isolate was not expected and was surprising. We have not tested the mice used in this study for the presence of ciprofloxacin-resistant strains, and such a presence could explain the lack of selection seen here. Furthermore, extrapolation of results from the model is limited by the physiological differences between mice and humans, even though studies have shown that subcutaneous treatment once a day in mice gives fecal antibiotic concentrations equivalent to those of humans (19). Finally, we studied only the fecal—and not the mucosal—microflora of mice. Yet, it has been postulated that, first, the mouse fecal flora is a mixture of mucosal and luminal flora and, second, the intestinal microflora of laboratory mice is comparable to the intestinal flora in humans (30, 31). We have no obvious explanation for the selective ability of dicloxacillin, since this drug had no impact on the Gram-negative anaerobic or the aerobic Gram-positive flora. More detailed evaluation of the mouse and human intestinal microbiomes may provide the reason for the change in intestinal flora imposed by dicloxacillin. A study of antibiotics' general impacts on the microflora of mice and men would further illustrate the elements involved in selection.
In summary, our study confirms that antibiotics with an impact on Gram-negative anaerobes support overgrowth and colonization of a CTX-M-15 producing E. coli ST131 isolate. Nonetheless, our study shows that selection could be driven by antibiotics with limited effect on anaerobes and no effect on competing Gram negatives, but not by other antimicrobials with a broader spectrum of activity. The results indicate a need for investigation of selective mechanisms of different drugs, to fully develop rules and guidelines for stewardship of antibiotics.
ACKNOWLEDGMENTS
This study was performed with financial support from PAR, an EU FP7-Health-2009-Single-Stage project. Additionally, the work was supported by The Danish Council for Strategic Research (DanCARD project 09-067075/DSF), Roskilde University, the Aase & Ejnar Danielsens Foundation, and the SSAC Foundation.
Furthermore, the work was performed in cooperation between Statens Serum Institut, Hvidovre University Hospital, Roskilde University, and Copenhagen University. We are grateful to and thank Anette M. Hammerum and Frank Hansen from SSI, who provided strain 65-Ec-09.
Footnotes
[This article was published on 23 September 2014 with the first author's surname incorrectly indexed as “Boetius Hertz” instead of “Hertz.” This has been corrected in the current version, posted on 14 August 2018.]
REFERENCES
- 1.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14. 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 2.Pitout JDD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 4.Pitout JDD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet 8:159–166. 10.1016/S1473-3099. [DOI] [PubMed] [Google Scholar]
- 5.Peirano G, Pitout JDD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321. 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS, Hansen G, Edelstein PH, Robicsek A, Cleary T, Adachi J, Paterson D, Quinn J, Hanson ND, Johnston BD, Clabots C, Kuskowski MA. 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob. Agents Chemother. 56:2364–2370. 10.1128/AAC.05824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahbi G, Mora A, López C, Alonso MP, Mamani R, Marzoa J, Coira A, García-Garrote F, Pita JM, Velasco D, Herrera A, Viso S, Blanco JJE, Blanco M. 2013. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 42:347–351. 10.1016/j.ijantimicag.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Tumbarello M, Spanu T, Di Bidino R, Marchetti M, Ruggeri M, Trecarichi EM, De Pascale G, Proli EM, Cauda R, Cicchetti A, Fadda G. 2010. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum beta-lactamase production and inadequate initial antibiotic therapy. Antimicrob. Agents Chemother. 54:4085–4091. 10.1128/AAC.00143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout JDD. 2010. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70:313–333. 10.2165/11533040-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Baño J, Alcala JC, Cisneros JM, Grill F, Oliver A, Horcajada JP, Tortola T, Mirelis B, Navarro G, Cuenca M, Peña C, Llanos AC, Canton R, Pascual A. 2008. Community infections caused by extended-spectrum beta-lactamase-producing. Arch. Intern. Med. 168:1897–1902. 10.1001/archinte.168.17.1897. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen KL, Dynesen P, Larsen P, Frimodt-Møller N. 2014. Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J. Med. Microbiol. 63:582–589. 10.1099/jmm.0.068783-0. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Baño J, Navarro MD, Romero L, Muniain MA, de Cueto M, Rios MJ, Hernandez JR, Pascual A. 2006. Bacteremia due to extended-spectrum beta-lactamases producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin. Infect. Dis 43:1407–1414. 10.1086/508877. [DOI] [PubMed] [Google Scholar]
- 13.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. 2001. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 32:1162–1171. 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- 14.Bellíssimo-Rodrigues F, Carolina A, Gomes F, Dinis A, Passos C, Achcar JA, Castro S, Martinez R, Gomes ACF, Passas ADC, Perdoná GDSC. 2006. Clinical outcome and risk factors related to extended-spectrum beta-lactamase-producing Klebsiella spp. infection among hospitalized patients. Mem. Inst. Oswaldo Cruz 101:415–421. 10.1590/S0074-02762006000600012. [DOI] [PubMed] [Google Scholar]
- 15.Hansen F, Olsen SS, Heltberg O, Justesen US, Fuglsang-Damgaard D, Knudsen JD, Hammerum AM. 2014. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb. Drug Resist. 20:316–3240. 10.1089/mdr.2013.0157. [DOI] [PubMed] [Google Scholar]
- 16.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277. 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen JB, Albayati A, Jørgensen RL, Hansen KH, Lundgren B, Schønning K. 2013. An abbreviated MLVA identifies Escherichia coli ST131 as the major extended-spectrum β-lactamase-producing lineage in the Copenhagen area. Eur. J. Clin. Microbiol. Infect. Dis. 32:431–436. 10.1007/s10096-012-1764-x. [DOI] [PubMed] [Google Scholar]
- 18.Imhof A. 1996. Continuous monitoring of oxygen concentrations in several systems for cultivation of anaerobic bacteria. J. Clin. Microbiol. 34:1646–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez F, Pultz MJ, Endimiani A, Bonomo RA, Donskey CJ. 2011. Effect of antibiotic treatment on establishment and elimination of intestinal colonization by KPC-producing Klebsiella pneumoniae in mice. Antimicrob. Agents Chemother. 55:2585–2589. 10.1128/AAC.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erlendsdottir H, Knudsen JD, Odenholt I, Cars O, Espersen F, Frimodt-møller N, Fuursted K, Kristinsson KG, Gudmundsson S. 2001. Penicillin pharmacodynamics in four experimental pneumococcal infection models. Antimicrob. Agents Chemother. 45:1078–1085. 10.1128/AAC.45.4.1078-1085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knudsen JD, Fuursted K, Frimodt-Møller N, Espersen F. 1997. Comparison of the effect of cefepime with four cephalosporins against pneumococci with various susceptibilities to penicillin, in vitro and in the mouse peritonitis model. J. Antimicrob. Chemother. 40:679–686. 10.1093/jac/40.5.679. [DOI] [PubMed] [Google Scholar]
- 22.Jakobsen L, Cattoir V, Jensen KS, Hammerum AM, Nordmann P, Frimodt-Møller N. 2012. Impact of low-level fluoroquinolone resistance genes qnrA1, qnrB19 and qnrS1 on ciprofloxacin treatment of isogenic Escherichia coli strains in a murine urinary tract infection model. J. Antimicrob. Chemother. 67:2438–2444. 10.1093/jac/dks224. [DOI] [PubMed] [Google Scholar]
- 23.Sandberg A, Jensen KS, Baudoux P, Van Bambeke F, Tulkens PM, Frimodt-Møller N. 2010. Intra- and extracellular activities of dicloxacillin against Staphylococcus aureus in vivo and in vitro. Antimicrob. Agents Chemother. 54:2391–2400. 10.1128/AAC.01400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerrn MB, Frimodt-Møller N, Espersen F. 2003. Effects of sulfamethizole and amdinocillin against Escherichia coli strains (with various susceptibilities) in an ascending urinary tract infection mouse model. Antimicrob. Agents Chemother. 47:1002–1009. 10.1128/AAC.47.3.1002-1009.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asahi Y, Miyazaki S, Yamaguchi K. 1995. In vitro and in vivo antibacterial activities of BO-2727, a new carbapenem. Antimicrob. Agents Chemother. 39:1030–1037. 10.1128/AAC.39.5.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frimodt-Møller N, Thomsen V. 1987. The pneumococcus and the mouse protection test: correlation of in vitro and in vivo activity for beta-lactam antibiotics, vancomycin, erythromycin and gentamicin. Acta Pathol. Microbiol. Immunol. Scand. 95:159–165. [DOI] [PubMed] [Google Scholar]
- 27.Achá SJ, Kühn I, Mbazima G, Colque-Navarro P, Möllby R. 2005. Changes of viability and composition of the Escherichia coli flora in faecal samples during long time storage. J. Microbiol. Methods 63:229–238. 10.1016/j.mimet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Haigh J, Degun A, Eydmann M, Millar M, Wilks M. 2011. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry system in the clinical microbiology laboratory. J. Clin. Microbiol. 49:3441. 10.1128/JCM.00576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frimodt-moller N, Ostri P, Pedersen IBK, Poulsen SR. 1980. Antibiotic prophylaxis in pulmonary surgery: a double-blind study of penicillin versus placebo. Ann. Surg. 195:444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson KH, Brown RS, Andersen GL, Tsang J, Sartor B. 2006. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe 12:249–253. 10.1016/j.anaerobe.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Krych L, Hansen CHF, Hansen AK, van den Berg FWJ, Nielsen DS. 2013. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS One 8:e62578. 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]