Abstract
A major obstacle in the process of discovery of drugs against Mycobacterium tuberculosis is its extremely slow growth rate and long generation time (∼20 to 24 h). Consequently, determination of MICs and minimum bactericidal concentrations (MBCs) of potential drug candidates using current methods requires 7 days (resazurin-based MIC assay [REMA]) and 1 month (CFU enumeration), respectively. We employed a synthetic luciferase operon optimized for expression in high-GC-content bacteria and adapted it for use in mycobacteria. Using luminescence-based readouts, we were able to determine the MICs and bactericidal activities of approved tuberculosis (TB) drugs, which correlated well with currently used methods. Although luminescence-based readouts have been used previously to determine the MICs and bactericidal activities of approved TB drugs, in this study we adapted this assay to carry out a pilot screen using a library of 1,114 compounds belonging to diverse chemical scaffolds. We found that MICs derived from a 3-day luminescence assay matched well with REMA-based MIC values. To determine the bactericidal potencies of compounds, a 1:10 dilution of the cultures from the MIC plate was carried out on day 7, and the bactericidal concentrations determined based on time to positivity in 2 weeks were found to be comparable with MBC values determined by the conventional CFU approach. Thus, the luminescent mycobacterium-based approach not only is very simple and inexpensive but also allowed us to generate the information in half the time required by conventional methods.
INTRODUCTION
Tuberculosis (TB) continues to be a major threat to public health worldwide, accounting for 1.3 million deaths annually, and is also a leading cause of death among people coinfected with HIV/AIDS. Despite the facts that the estimated number of people falling ill with tuberculosis each year is declining and that TB death rates dropped 45% over the last 2 decades, the rising incidence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB cases is alarming (1). Only half of MDR-TB cases are successfully treated, reflecting exceptionally high mortality rates. Moreover, 1/10 of MDR TB cases are redefined as XDR TB, as they are resistant to the majority of existing first- and second-line anti-TB drugs. The emergence of MDR and XDR TB strains compromises the existing treatment regimens and has prompted the search for new medicines. Therefore, an efficient, rapid, low-cost, high-throughput assay that would allow us to screen compound libraries and follow their effects on mycobacterial cells in real time would be of tremendous help.
The extremely slow growth rates of pathogenic mycobacteria and therefore the long incubation times needed to assay antimycobacterial activity represent major obstacles in drug discovery. Plating of bacterial cultures on specialized mycobacterial agar medium for CFU enumeration continues to be the “gold standard” for evaluating the effectiveness of antituberculosis compounds, despite being very labor-intensive and time-consuming (2). Moreover, CFU-based assays do not allow the detection of viable but nonculturable (VBNC) cells, involve long incubation periods (3 to 4 weeks), and are often prone to contamination. Monitoring viability and growth of mycobacterial cells in response to antibiotics in real time with high sensitivity would be an efficient approach and could replace conventional agar-based assays.
The early stage of antimycobacterial drug discovery involves routine assessment of the potency of the drug candidates against the bacterium under in vitro conditions. This is carried out usually by determining the MIC—the concentration of compound required to arrest bacterial growth (stasis) and the minimal bactericidal concentration (MBC)—the concentration of compound required to achieve a reduction in bacterial cell number (killing). Determination of MIC and MBC plays a critical role during the process of screening, prioritizing, and optimizing a chemical series during early drug discovery.
There are a number of in vitro assays currently available for monitoring viable mycobacteria and for high-throughput compound screening, including resazurin-based microtiter plate assay (REMA), microplate alamarBlue assay (MABA), and Bactec MGIT 960-based MIC readout (3–5). While Bactec MGIT 960 is expensive and lacks a high-throughput format, a higher sensitivity for resazurin and alamarBlue colorimetric assays would be desirable, along with a shorter detection time. The bacterial luciferase system serves as an alternative to these methods by providing a rapid, sensitive, real-time readout of bacterial cell response to antibiotics.
Bioluminescent bacteria are found in freshwater, marine, and terrestrial environments and include 4 genera: Photobacterium, Vibrio, Photorhabdus, and Alteromonas (Shewanellaceae family). The luminescent reaction in bacteria involves the oxidation of reduced flavin mononucleotide (FMNH2) using a long-chain fatty aldehyde substrate catalyzed by luciferase enzyme with the emission of blue-green light at 490 nm. Genes in the lux operon (luxCDABE) encode luciferase and all components required for the reaction. Luciferase is a heterodimeric enzyme composed of α and β subunits, encoded by luxA and luxB, respectively. The multienzyme complex responsible for catalyzing the fatty aldehyde substrate for luminescent reaction consists of a reductase, a transferase, and a synthetase encoded by luxC, luxD, and luxE, respectively (6). Bioluminescence-based approaches have been used previously with mycobacteria for evaluation of drug and vaccine efficacies in vitro and in vivo (7–17). Bacterial luciferase systems, therefore, represent an attractive way to readily screen various antimycobacterial compounds, since only viable cells will be luminescent, as the reaction requires the presence of FMNH2 and production of endogenous reaction substrate.
This study utilized a synthetic luxCDABE operon encoding the Photorhabdus luminescens Lux proteins, codon optimized for expression in high-GC bacteria (18), for reliable, rapid high-throughput screening of compounds targeting Mycobacterium tuberculosis. This system allows us to obtain MICs and MBCs for anti-TB compounds, is amenable for high-throughput screening, and has the advantage of significant reductions in material, time, and resources compared to traditional methods.
MATERIALS AND METHODS
Construction of bioluminescent plasmids and strains.
For constructing the luciferase plasmid, the 6.8-kb fragment containing the synthetic Lux genes luxCDABE was excised from pMU1* (generously provided by A. Craney and J. Nodwell, McMaster University) using NdeI and EcoRI (18). This fragment was cloned downstream of a strong mycobacterial promoter, pUV15 (kindly provided by Sabine Ehrt, Weill Cornell Medical College, New York, NY) (19), in the NheI site in the mycobacterial integrative vector pND239 (Hygr) after end filling of the overhangs using Klenow DNA polymerase (NEB). The resulting vector, pEG200, carries in addition to the Lux operon mycobacteriophage L5 attP-int sequences (20), a hygromycin resistance cassette, and an Escherichia coli origin of replication. Mycobacterial reporter strains were obtained by electroporation of pEG200 into Mycobacterium smegmatis mc2155 (Msm-lux) and M. tuberculosis H37Rv (Mtb-lux) and plating on 7H10 plates containing 50 μg/ml of hygromycin.
Bacterial strains and medium conditions.
M. smegmatis, Msm-lux, M. tuberculosis, and Mtb-lux were grown in Middlebrook 7H9 broth (Difco) containing 0.5% albumin, 0.085% NaCl, 0.2% glucose, 0.05% Tween 80, and 0.5% glycerol at 37°C to mid-log phase (optical density at 600 nm [OD600] = 0.5) and stored frozen as 0.5-ml aliquots in 10% glycerol in screw-cap cryovials (Corning, USA) at −80°C. For plating of mycobacteria, Middlebrook 7H10 agar (Difco) containing 10% oleic acid-albumin-dextrose-catalase (OADC; Difco) and 0.5% glycerol was used. For stress sensitivity assays, the Msm-lux strain was exposed to 1% SDS or 10 mM H2O2 for 3 h.
Luciferase assays with mycobacteria.
Luminescence measurements for M. smegmatis and M. tuberculosis were taken in 96-well or 384-well opaque white polystyrene plates (Corning 3917 or Corning 3570) using a Tecan Infinite M200 or M1000 microplate reader in luminescence mode with an integration time of 1 s. Alternatively, a sensitivity assay for M. smegmatis was performed using a Bio-Rad vacuum-driven Bio-dot system. One-hundred-microliter aliquots of 2-fold serial dilutions were loaded and subsequently transferred onto a nitrocellulose membrane, followed by immediate exposure to ECL chemiluminescence film (Amersham).
MIC assay.
MICs of antibiotics against M. tuberculosis H37Rv were determined in 7H9 broth by the standard microdilution method, with some modifications (4). Briefly, 1-μl quantities of serial 2-fold dilutions of test compound were added to a 384-well plate. Control wells included medium and culture controls. Forty microliters of culture (at 3 × 105 CFU/ml) was added to all the wells except the medium control wells. The plates were packed in gas-permeable polyethylene bags and incubated at 37°C for 6 days. Following this incubation period, MIC was determined using the resazurin-based microtiter plate assay (REMA); 8 μl of a freshly prepared 1:1 mixture of resazurin (0.02% in water) and 10% Tween 80 was added to all the wells. The plates were reincubated for an additional 24 h at 37°C, and the color conversion of all wells was recorded. A blue color in the well was interpreted as no growth, and a pink color was scored as growth. Absorbances at 575 nm and 610 nm were monitored and their ratios calculated. The untreated culture control values were considered 100% growth, and the lowest concentration of compound which yielded 80% inhibition of growth compared to the untreated control was defined as the MIC. Isoniazid (INH) was used as reference drug for this assay; it consistently gave an MIC of 0.22 to 0.45 μM.
For Lux-based MIC determination, 384-well opaque white polystyrene plates were used and the compound plates were prepared as described above. The assay plates were incubated at 37°C in a CO2 incubator for 7 days. Luminescence was measured on day 3 and day 7. The concentration that resulted in 80% or greater inhibition of relative light units (RLU) compared to that of untreated cells was defined as the MIC.
MBC assay.
Minimal bactericidal concentrations (MBCs) against M. tuberculosis H37Rv were determined in a 384-well plate format after 7 days of incubation with the compounds. For CFU-based enumeration, the culture was plated to enumerate the initial CFU before exposure to the drugs. Suitable dilutions of culture aliquots from wells containing strain H37Rv exposed to 3 to 5 concentrations of compounds greater than the MICs were plated on 7H10 agar plates. CFU were counted after incubation of the plates at 37°C for 21 to 28 days. The MBC was defined as the concentration which gave at least a 2-log10 reduction in CFU compared to the initial CFU.
For Lux-based determination of bactericidal activity, a 1:10 dilution of the cells from a Lux MIC plate was made on day 7 in a 384-well white plate using 7H9 broth and incubated at 37°C for 14 days. Luminescence was measured on day 14. The concentration at which there was 99% or greater inhibition of RLU compared to those of untreated cells was defined as the bactericidal concentration.
RESULTS
Construction of autoluminescent mycobacteria.
In the present study, we employed a synthetic luxCDABE operon that was designed previously for expression in high-GC bacteria, like Streptomyces coelicolor (18). This operon was cloned into a mycobacterial integration-proficient vector downstream of a strong promoter and a strong ribosome-binding site. We attempted to increase luciferase expression levels by cloning the expression cassette in an episomal plasmid but were unsuccessful, as in previous studies (14, 16). Using our construct, we were able to get stable and efficient expression of luciferase in mycobacteria that was easily detected by luminometry (14).
Evaluation of the luminescence reporter strains.
In order to test the sensitivity of the luciferase reporter, serial 2-fold dilutions of the Msm-lux strain were carried out and the luminescence was measured on a plate reader. Using this approach, the limit of detection was found to be in the range of 105 to 106 CFU/ml (see Fig. S1A in the supplemental material). This was further analyzed by spotting dilutions onto a nitrocellulose membrane and exposing it to ECL luminescence film. However, the sensitivity did not surpass that of a microplate reader, with 5.48 × 107 CFU/ml being the detection limit (see Fig. S1B).
In order to confirm that the luciferase expression was a rapid indicator of the metabolic state and viability of the cell, we exposed the Msm-lux strain to lethal concentrations of SDS or hydrogen peroxide. The luminescence signal dropped rapidly and was at background level within 1 h of stress exposure, which was consistent with CFU data (see Fig. S2 in the supplemental material).
The Mtb-lux strain was checked for antibiotic susceptibility pattern by determining the MICs of antimycobacterial drugs (isoniazid [INH], chloramphenicol, moxifloxacin, TMC207, ofloxacin, and PA824) by REMA. RLU were also measured daily and MICs were calculated every day for up to 7 days in order to determine if the same data could be derived earlier. We observed that the MIC values changed until day 3 and stabilized thereafter, suggesting that at least a 3-day incubation was required to obtain the expected MIC (Table 1).
TABLE 1.
Comparison of MICs for antimycobacterial drugs determined using Mtb-lux- and resazurin-based methods
| Standard drug | Lux-based MIC (μM) |
Expected REMA MIC range (μM) | ||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | ||
| Moxifloxacin | 0.5 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125–0.25 |
| Chloramphenicol | >384 | 48 | 12 | 12 | 12 | 12 | 12 | 12–24 |
| Ofloxacin | 5.2 | 1.4 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7–1.4 |
| PA824 | 2.4 | 1.2 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3–0.6 |
| TMC207 | 4.5 | 0.9 | 0.45 | 0.45 | 0.45 | 0.45 | 0.45 | 0.22–0.45 |
| Isoniazid | >7.25 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22–0.45 |
Determination of LOQ and assay robustness using Mtb-lux.
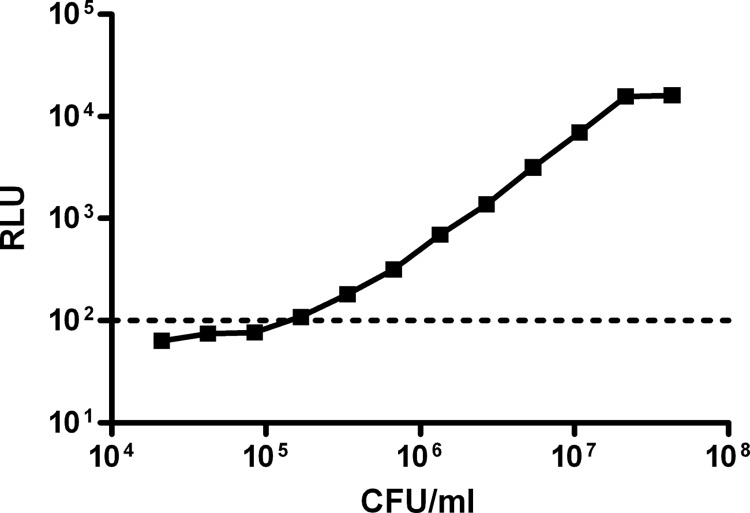
In order to determine the correlation of CFU with RLU for the Mtb-lux strain, an actively growing culture of Mtb-lux was diluted serially and luminescence was measured. Appropriate dilutions of the original culture were plated on 7H10 agar to determine the cell number. The RLU obtained were plotted against the cell numbers (estimated based on the dilutions of the original culture) to obtain the correlation between the CFU and RLU (Fig. 1). The media control values were in the range of 20 to 100 RLU. Therefore, we decided to work at a range about 10-fold higher than the media control (1,000 RLU), which corresponded to ∼106 CFU/ml and was therefore considered the limit of quantification (LOQ) for this method.
FIG 1.
Limit of quantification of M. tuberculosis using luminescence. An exponentially growing culture of Mtb-lux was plated to determine CFU. Two-fold serial dilutions were prepared, and luminescence was measured using a microplate reader. The dashed line indicates the background RLU for media.
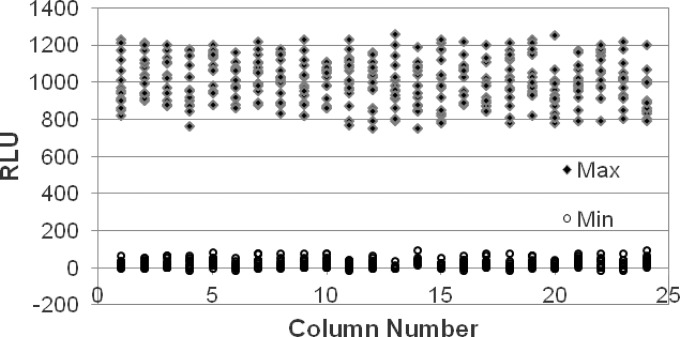
For routine screening in the drug discovery process, a 384-well microtiter plate format is preferred. Hence, the robustness of the assay was evaluated using luminescence-based reporter assays in this format. To determine if any variability in the readouts occurred that could be attributed to positional effects caused by drying or malfunctioning of the dispensing instrument, a Min-Max plate was prepared as follows: sterile medium (minimum [min]) and culture control (maximum [max]) were added to alternate rows of each column, and the RLU were recorded after 3 days of incubation, which was used to calculate the Z′ as follows: Z′ = 1 − [(3 × SDmax) + (3 × SDmin)]/(Avmax − Avmin), where SD is standard deviation and Av is average. A Z′ value of 0.5 and above was considered to be indicative of a robust and reproducible assay. We derived a Z′ value of 0.56 for the Lux assay using the Mtb-lux strain in the 384-well format (Fig. 2).
FIG 2.
Determining assay robustness using Mtb-lux in 384-well microplate with 3 days of incubation. Min, sterile media; Max, Mtb-lux culture. Z′ = 0.56.
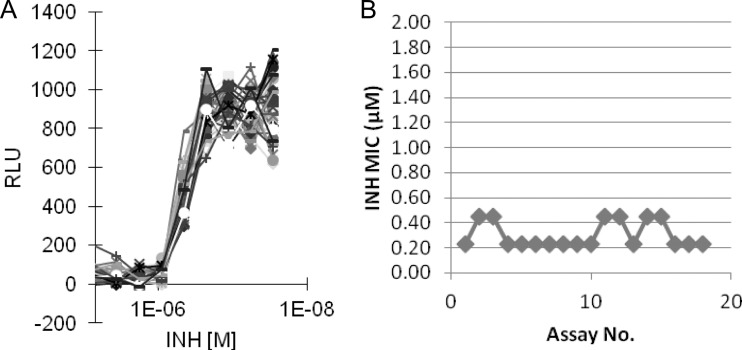
To further evaluate the possible influence of positional effects in determining the MIC of a given compound, INH was dispensed as a 9-point concentration response (CR) with a 2-fold dilution and a starting concentration of 7.2 μM. Thirty-two replicates of INH as a CR, along with a single minimum (sterile media), maximum (no-drug control), and reference drug (0.5 μM INH), were dispensed in a 384-well plate in horizontal and vertical orientations to obtain a random but well-distributed representation of various positions within the plate to which Mtb-lux was added as previously described. The RLU were recorded after a 3-day incubation period and plotted against the concentration of INH. The profiles overlapped well, and a consistent MIC value of 0.45 μM was determined for this drug, indicating that no positional effect was detected for this assay in the 384-well microtiter plate format (Fig. 3A). A Manhattan plot for INH MIC derived from multiple independent assays also showed consistency (∼2-fold variation) across the various repeats (Fig. 3B).
FIG 3.
(A) Determination of assay robustness for Mtb-lux-based MIC in 384-well format. INH MIC = 0.45 μM. (B) Manhattan plot for Mtb-lux-based INH MICs derived from multiple independent assays.
Comparison of MICs derived from Mtb-lux and REMA methods for a diverse set of compounds.
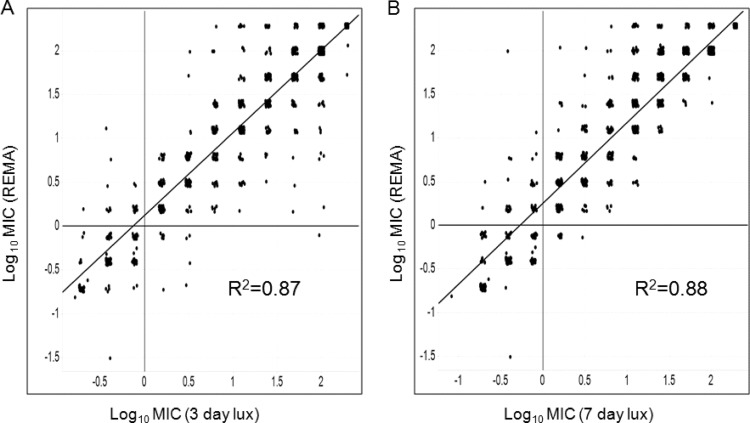
A set of 1,114 compounds that were known to act on different targets in M. tuberculosis was selected from the AstraZeneca corporate library. This set included a range of bactericidal, bacteriostatic, and inactive compounds. To compare the Mtb-lux assay with the REMA assay, MICs were determined by both these methods in parallel for these compounds. RLU were measured and MICs determined after both 3 days and 7 days of exposure to the compounds and compared to the 7-day MIC determined by REMA (Fig. 4). For comparison purposes, a 4-fold variation in MIC was considered to be in the acceptable range, and any variation that was 8-fold or higher between the two methods was recorded as a mismatch. A 3-day incubation was sufficient to produce the same MIC by Mtb-lux assay as observed after 7 days of exposure to drugs in the REMA method for the majority (>90%) of the compounds. Some compounds were found to be inactive on day 3 but had potent MICs on day 7, suggesting that their activity was exposure dependent. Overall, comparable MICs were obtained for ∼94% of the compounds by the two methods (Table 2).
FIG 4.
Scatter plot to show correlation between REMA MIC values versus 3-day Lux MIC values (A) or 7-day Lux MIC values (B) for compounds belonging to diverse chemical scaffolds.
TABLE 2.
Comparison of MICs between REMA- and Lux-based methods for a diverse set of 1,114 compounds
| Analysis parameter | REMA- vs D3 Luxa | REMA- vs D7 Luxa | D3 Lux vs D7 Lux exposure |
|---|---|---|---|
| No. of compounds with ≤4-fold shift in MIC | 1,072 | 1,064 | 1,053 |
| No. of compounds with ≥8-fold shift in MIC | 42 | 50 | 61 |
| % mismatch | 3.7 | 4.4 | 5.4 |
Lux-based assay with 3-day (D3) or 7-day (D7) exposures.
Use of TTP curve to determine bactericidal activity with Mtb-lux.
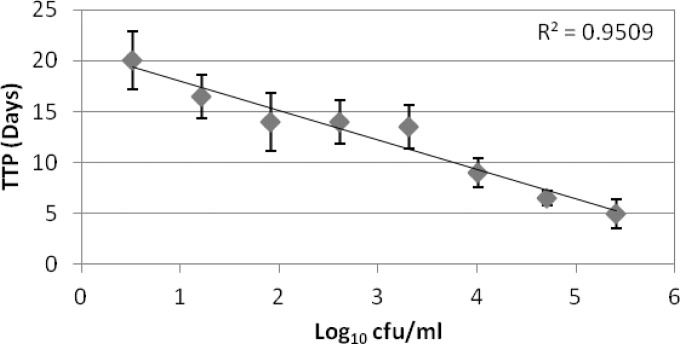
Since the LOQ for Mtb-lux was ∼106 CFU/ml, we perceived that determination of the MBC by directly correlating the RLU to CFU would be misleading. However, if the cells surviving at the end of MIC determination period could be diluted and allowed to grow back to reach a threshold RLU which could be consistently correlated to a specific CFU, then an indirect method of assessing the cell number in the original sample could be designed based on the time taken to achieve this threshold RLU. We therefore derived a standard time-to-positivity (TTP) curve for the Mtb-lux strain. Briefly, 2-fold dilutions of Mtb-lux cells designed to get a range of cell numbers (determined by plating and enumerating CFU/ml) were grown at 37°C and the RLU were recorded every day. Since the LOQ of 106 CFU/ml corresponded to 1,000 RLU, the time taken for a given cell number to reach a value above 1,000 RLU was considered the TTP for that cell number. A standard curve was obtained by plotting the TTP against the various starting cell concentrations (Fig. 5).
FIG 5.
Standard TTP curve for the Mtb-lux strain.
The MBC determined by CFU assay is considered the concentration at which there is a 2-log or greater reduction in bacterial cell number compared to the initial CFU. Bactericidal concentrations for various compounds were determined based on this standard TTP curve. The initial CFU in MIC experiments was 3 × 105 to 5 × 105 CFU/ml, as specified by the CLSI guidelines (21). Therefore, typically after a 7-day exposure to a bactericidal compound, the cell number would be ∼103 CFU/ml or lower. In order to determine the TTP, and also to reduce the concentration of the compound, the cells were diluted 10-fold, thereby achieving ≤102 CFU/ml of Mtb-lux in case of MBC wells. Based on the TTP standard curve (Fig. 5), this cell number is expected to take >14 days to reach the threshold RLU. For compounds that are either bacteriostatic or inactive on M. tuberculosis, the TTP is expected to be much less than 14 days, and therefore, such compounds can be easily differentiated from the bactericidal compounds (Table 3).
TABLE 3.
TTP-based classification of inhibitors using Mtb-lux
| Factor used to classify inhibitors | Value for indicated inhibitor type |
||
|---|---|---|---|
| No inhibition | Bacteriostatic compound | Bactericidal compound | |
| Undiluteda | 107 CFU/ml | 105 CFU/ml | ≤103 CFU/ml |
| 1:10 dilutionb | 106 CFU/ml | 104 CFU/ml | ≤102 CFU/ml |
| TTPc | <6 days | 7–11 days | >14 days |
Expected bacterial number after 7 days of exposure in MIC assay.
Expected bacterial number after 1:10 dilution of the MIC plate.
As derived from Fig. 5.
The above hypothesis was tested using standard drugs that were known to be bactericidal or bacteriostatic against M. tuberculosis. CFU enumeration was also carried out in parallel to verify the results derived from the TTP assay for Mtb-lux. There was a maximum of 2-fold variation in the bactericidal concentration determined by the two methods used, except in the case of chloramphenicol, which was determined to be a static drug by the CFU-based method, while the Lux-based assay determined 50 μM (16 μg/ml) as the bactericidal concentration (Table 4). Detailed analysis of the actual killing by chloramphenicol at these concentrations revealed a 1-log reduction in cell number at 50 μM and higher concentrations (Table 5). Since the definition for CFU-based MBC was a 2-log reduction in cell number, these concentrations were not considered bactericidal by this method. We also noted that the Lux-based method could not differentiate between compounds that showed a 1-log or 2-log killing, and we therefore refer to the Lux-based determination of bactericidal activity as bactericidal concentration, in order to differentiate from MBC, which is defined as a 2-log killing. Therefore, the Lux-based method potentially cannot be used to quantify the extent of killing but can be used qualitatively to identify bactericidal compounds faster (2 weeks) than the CFU-based method, which requires a longer turnaround time (4 weeks).
TABLE 4.
Bactericidal concentrations of antimycobacterial drugs as determined by CFU-based and Lux-based methods
| Drug | CFU-based MBC (μM) | Lux-based bactericidal activity (μM) |
|---|---|---|
| Ciprofloxacin | 1.5 | 3 |
| Moxifloxacin | 0.3 | 0.6 |
| Chloramphenicol | >198 | 50 |
| Rifampin | 0.009 | 0.018 |
| PA824 | 0.7 | 1.4 |
| Ofloxacin | 2.7 | 2.7 |
| Penicillin | >382 | >382 |
TABLE 5.
Cell numbers determined at various concentrations of chloramphenicola
| Chloramphenicol concn (μM) | Log10 CFU/ml |
|---|---|
| 198 | 4.3 |
| 99 | 4.3 |
| 50 | 4.7 |
| 25 | 5.7 |
| 12 | 5.8 |
The initial cell number was 5.74 log10 CFU/ml.
Evaluation of Mtb-lux-based determination of MBC.
To further evaluate the TTP-based determination of bactericidal activity, 119 compounds exhibiting a broad range of MICs, as described above, were shortlisted. We ensured that these compounds spanned a variety of targets and chemical space, to represent a diverse set. CFU-based MBC data were available for all of these compounds, which were used to include a mix of bacteriostatic and bactericidal compounds within this set. The Lux-based bactericidal activity assay was set up in parallel with plating for CFU enumeration for the compound set described above. The minimum concentration required to restrict the luminescence values below 1,000 RLU on day 14 was considered to be the bactericidal concentration for the given compound. These data were compared with MBCs derived from the CFU-based method. Out of the 119 compounds tested, 68 compounds were determined to be bactericidal at similar concentrations (within 4-fold variation) by both methods. In addition, 22 compounds that were determined to be bactericidal by the Lux method were found to induce 1- to 1.5-log killing of M. tuberculosis as determined by CFU enumeration. Twenty-nine compounds were determined to be nonbactericidal by both methods.
DISCUSSION
In the present study, we demonstrate the expression of an entirely synthetic 5.6-kb luxCDABE operon, optimized for expression in high-GC bacteria, in mycobacteria. Efficient expression of luciferase genes in our reporter strains was achieved by the use of a strong transcriptional promoter and an optimal Shine-Dalgarno sequence. Due to the sensitivity of detection, the lux-containing expression cassette is useful for monitoring viability of mycobacterial cells in in vitro cultures. The biochemical requirements for the production of light by luxCDABE products, such as the presence of FMNH2 cofactor and oxygen, are available in the cytoplasm of aerobically growing mycobacteria. Therefore, viable mycobacterial cells that express the luxCDABE operon emit light spontaneously at 490 nm.
The natural luxCDABE operon from Photorhabdus luminescens is relatively rich in codons having either A or T in the wobble position (69%), suggesting that these genes would not be efficiently expressed in mycobacteria with a high GC content (>65%). There have been several reports describing use of luciferase in mycobacteria (7–17). Most of these studies are based on light production by firefly luciferase in the presence of ATP and rely on an exogenously added substrate. In contrast, reporter strains employing bacterial luxCDABE are autoluminescent, since the operon encodes the enzyme as well as the aldehyde substrate, eliminating the need for addition of expensive substrates. Moreover, the system is very amenable to high-throughput screening, and samples can be assayed multiple times. Recently, a few studies have succeeded in expressing the whole luxCDABE operon from Photorhabdus luminescens, on an integrating vector under the control of strong promoters in M. smegmatis and M. tuberculosis, for in vivo imaging and evaluation of drug and vaccine efficacy (14, 16).
Several attempts have been made toward maximizing the luminescence signal output, for example, by cloning of additional promoters within the Lux operon (14) or by increasing the substrate concentrations. n-Decyl aldehyde (decanal), a substrate of bacterial luciferase, is characterized by rapid uptake and was supplied exogenously to E. coli expressing bacterial luciferase (22). Decanal was also used as a substrate for luciferase expressed in M. smegmatis cells (11). However, aldehyde levels exceeding 100 μM were shown to inhibit the in vitro bioluminescence reaction (23). We also tried a broad range of decanal concentrations, from 0.001% to 1%, added to Msm-lux cells but failed to detect any further increase in luminescence (data not shown).
The experimental results shown here indicate that the luxCDABE luciferase system is strongly indicative of the viability and metabolic state of mycobacterial cells, probably due to its strong dependence on FMNH2 cofactor. Bacterial luciferase from Vibrio harveyi was shown to be relatively destabilized in M. tuberculosis H37Ra, with half-life of ∼19 h (24). Therefore, the luminescence signal observed probably reflects active synthesis and metabolism rather than accumulated products and thus could serve as a dynamic readout of the state of the cell. Interestingly, the LOQ for Msm-lux was 105 to 106 CFU/ml, while that for Mtb-lux was 106 CFU/ml. This could be due to the differential efficiencies of the encoded transcriptional signals in these two mycobacterial species.
As mentioned before, an obvious advantage of this luminescence assay is the possibility to take measurements frequently, thus obtaining very detailed information about kinetics of killing of mycobacteria. Moreover, the luciferase construct proved to be an inexpensive, versatile tool allowing us to monitor cell viability in real time as a whole-cell sensor without addition of any exogenous substrate and to obtain MICs in only 3 days. In contrast, the conventional agar proportion method requires 3 to 4 weeks to visualize colonies (25), the Bactec MGIT 960 system requires 13.3 days, the Bactec 460TB system requires 10.6 days, the microdilution resazurin assay requires 7 days, and MABA requires 5 days (4, 26, 27).
Early drug discovery for TB is heavily dependent on the evaluation of the potency of compounds as determined by MIC and MBC. Resazurin-based MIC determination is widely used as the standard method in TB drug discovery and requires 1 week to derive the results. We have explored the option of using a luminescence-based readout that can be detected in half the time. Our evaluation shows that MIC can be determined for most compounds as early as 3 days of incubation. We have observed that some compounds (<10% of the compounds tested), depending on their chemical nature and mode of action, need to have a longer exposure to exhibit their activity on M. tuberculosis cells. MICs for such compounds are more potent on day 7 than on day 3. We did not encounter any compound that was more potent on day 3 than on day 7. Andreu et al. describe the determination of standard drugs using a Lux reporter strain and report that MICs for all the drugs tested can be determined by day 3 (15). We find that this is true for the standard drugs, but some early compounds that are yet to be optimized for their killing properties may need longer exposure time. We recommend that the day 3 MIC be used to improve the DMTA (design-make-test-analyze) cycle in drug discovery but also that the day 7 MIC be considered to identify compounds that require longer exposure time. Since medium control values were often in the range of 20 to 100 RLU, we set a 10-fold-higher value, 1,000 RLU (corresponding to 106 CFU/ml), as the minimum cutoff for the determination of bactericidal activity based on time to positivity. After a 7-day exposure of Mtb-lux to compounds during the MIC evaluation in a 384-well plate format, all the cells were diluted 10-fold into fresh media. This dilution served dual purposes: (i) to facilitate growth, thereby allowing for differentiation between dead, damaged, and live cells, and (ii) to dilute the compound levels so that any inhibitory effect of the compound could be minimized. The diluted cells were then incubated to allow for growth and the resulting luminescence to cross a threshold value (cutoff) equal to the LOQ. In principle, a sample with a higher cell number will cross the cutoff value much faster than a sample with a lower cell number. The standard TTP curve confirmed the above hypothesis and indicated that the cells in a sample with a bactericidal compound would require 14 days or more to achieve the cutoff RLU. This was used as the minimum time to positivity required for compounds to be considered bactericidal. In order to determine if there is a problem due to compound carryover in plates for the Lux-based bactericidal-activity assay, we evaluated 119 compounds by making a 1:10 as well as a 1:100 dilution of the MIC plate into fresh media. There was no significant difference in the bactericidal concentrations derived from either dilution plates compared to the data from CFU plates. However, we did observe that variability was higher in the 1:100 dilution plates, probably due to the additional dilution step. We therefore used the data generated from the 1:10 dilution plates for our analysis. We also attempted incubating the cells for an additional week to see if we could correlate the TTP to CFU/ml more accurately, but as indicated by the slope of the standard TTP curve (Fig. 5), there is often overlap in the range of TTP values for a 1-log difference in cell number. Therefore, an accurate quantification of the CFU/ml was not possible. Andreu et al. describe luminescence-based determination of the MBC in 6 days (3 days for the MIC and 3 days for the MBC) (15). Zhang et al. describe a similar effort of determining bacterial counts under in vitro and in vivo conditions for standard TB drugs (16). Although this method works well with standard drugs, we observed that for early compounds in discovery which have suboptimal killing properties, a clear distinction between bactericidal and bacteriostatic properties could be ascertained only upon extended incubation time. We therefore arrived at the TTP-based determination of bactericidal activity.
Evaluation of bactericidal activity using luminescence resulted in comparable data between Lux- and CFU-based methods for all the standard drugs tested, with the exception of INH. We observed that the RLU increased after 7 days of incubation in the bactericidal-activity assay plate. This could be attributed to the INH-resistant subpopulation, which resulted in regrowth of bacteria after an initial kill, as previously reported (28, 29). As mentioned above, a total of 119 compounds across various scaffolds were tested in parallel; 29 compounds were determined to be nonbactericidal by both methods, while 68 compounds which gave 2-log killing as determined by CFU enumeration were also considered bactericidal based on the Lux method. Interestingly, an additional 22 compounds that induced 1- to 1.5-log killing (determined by CFU enumeration) were also picked up as bactericidal by the Lux method. This observation suggests that while the Lux method cannot differentiate between 2-log or 1-log killing, it definitely can identify bactericidal compounds. We also noticed that compounds showing ≤0.5-log killing were not picked up by the Lux method, as the luminescence values had crossed 1,000 RLU by day 14 for such compounds. Therefore, Lux-based methods did not pick up any false positives or false negatives in our validation set. Although the extent of killing may not be quantifiable by this method, this assay represents a major advantage in terms of turnaround time needed to determine the bactericidal property of an early compound, which will be of significant impact in any drug discovery program for tuberculosis.
Overall, we demonstrated that the Lux system is readily available for easy, rapid, reliable real-time high-throughput screening of antibacterial compounds. In addition, there is considerable savings in terms of plate pouring efforts, plating efforts, plates, and medium requirements. Therefore, we recommend the Lux-based determination of bactericidal activity as a faster and easier alternative to the REMA- and CFU-based methods used routinely in TB drug discovery, on the basis of its significant benefits in terms of time, material, and resource savings for the TB research community.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Shaw for his guidance with statistical analysis. We are grateful to Arryn Craney and Justin Nodwell for providing the pMU1* plasmid. We are grateful to Sunita DeSousa and Shridhar Narayanan for their efforts to make this endeavor possible and acknowledge John McKinney for his support.
Footnotes
Published ahead of print 21 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03205-14.
REFERENCES
- 1.WHO. 2014. Tuberculosis fact sheet. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs104/en/ Accessed 28 April 2014 [Google Scholar]
- 2.Sirgel FA, Wiid IJ, van Helden PD. 2009. Measuring minimum inhibitory concentrations in mycobacteria. Methods Mol. Biol. 465:173–186. 10.1007/978-1-59745-207-6_11 [DOI] [PubMed] [Google Scholar]
- 3.Taneja NK, Tyagi JS. 2007. Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J. Antimicrob. Chemother. 60:288–293. 10.1093/jac/dkm207 [DOI] [PubMed] [Google Scholar]
- 4.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kontos F, Maniati M, Costopoulos C, Gitti Z, Nicolaou S, Petinaki E, Anagnostou S, Tselentis I, Maniatis AN. 2004. Evaluation of the fully automated Bactec MGIT 960 system for the susceptibility testing of Mycobacterium tuberculosis to first-line drugs: a multicenter study. J. Microbiol. Methods 56:291–294. 10.1016/j.mimet.2003.10.015 [DOI] [PubMed] [Google Scholar]
- 6.Meighen EA. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrew PW, Roberts IS. 1993. Construction of a bioluminescent mycobacterium and its use for assay of antimycobacterial agents. J. Clin. Microbiol. 31:2251–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooksey RC, Crawford JT, Jacobs WR, Jr, Shinnick TM. 1993. A rapid method for screening antimicrobial agents for activities against a strain of Mycobacterium tuberculosis expressing firefly luciferase. Antimicrob. Agents Chemother. 37:1348–1352. 10.1128/AAC.37.6.1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arain TM, Resconi AE, Hickey MJ, Stover CK. 1996. Bioluminescence screening in vitro (Bio-Siv) assays for high-volume antimycobacterial drug discovery. Antimicrob. Agents Chemother. 40:1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hickey MJ, Arain TM, Shawar RM, Humble DJ, Langhorne MH, Morgenroth JN, Stover CK. 1996. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob. Agents Chemother. 40:400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snewin VA, Gares MP, Gaora PO, Hasan Z, Brown IN, Young DB. 1999. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 67:4586–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deb DK, Srivastava KK, Srivastava R, Srivastava BS. 2000. Bioluminescent Mycobacterium aurum expressing firefly luciferase for rapid and high throughput screening of antimycobacterial drugs in vitro and in infected macrophages. Biochem. Biophys. Res. Commun. 279:457–461. 10.1006/bbrc.2000.3957 [DOI] [PubMed] [Google Scholar]
- 13.Heuts F, Carow B, Wigzell H, Rottenberg ME. 2009. Use of non-invasive bioluminescent imaging to assess mycobacterial dissemination in mice, treatment with bactericidal drugs and protective immunity. Microbes Infect. 11:1114–1121. 10.1016/j.micinf.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 14.Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. 2010. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777. 10.1371/journal.pone.0010777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreu N, Fletcher T, Krishnan N, Wiles S, Robertson BD. 2012. Rapid measurement of antituberculosis drug activity in vitro and in macrophages using bioluminescence. J. Antimicrob. Chemother. 67:404–414. 10.1093/jac/dkr472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Li SY, Nuermberger EL. 2012. Autoluminescent Mycobacterium tuberculosis for rapid, real-time, non-invasive assessment of drug and vaccine efficacy. PLoS One 7:e29774. 10.1371/journal.pone.0029774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreu N, Zelmer A, Sampson SL, Ikeh M, Bancroft GJ, Schaible UE, Wiles S, Robertson BD. 2013. Rapid in vivo assessment of drug efficacy against Mycobacterium tuberculosis using an improved firefly luciferase. J. Antimicrob. Chemother. 68:2118–2127. 10.1093/jac/dkt155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craney A, Hohenauer T, Xu Y, Navani NK, Li Y, Nodwell J. 2007. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 35:e46. 10.1093/nar/gkm086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. 10.1093/nar/gni013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and Bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115. 10.1073/pnas.88.8.3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2008. Laboratory detection and identification of mycobacteria; approved guideline. CLSI document M48-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 22.Schultz DW, Yarus M. 1990. A simple and sensitive in vivo luciferase assay for tRNA-mediated nonsense suppression. J. Bacteriol. 172:595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holzman TF, Baldwin TO. 1982. Isolation of bacterial luciferases by affinity chromatography on 2,2-diphenylpropylamine-Sepharose: phosphate-mediated binding to an immobilized substrate analogue. Biochemistry 21:6194–6201. 10.1021/bi00267a026 [DOI] [PubMed] [Google Scholar]
- 24.Roberts EA, Clark A, Friedman RL. 2005. Bacterial luciferase is naturally destabilized in Mycobacterium tuberculosis and can be used to monitor changes in gene expression. FEMS Microbiol. Lett. 243:243–249. 10.1016/j.femsle.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 25.Rastogi N, Goh KS, David HL. 1989. Drug susceptibility testing in tuberculosis: a comparison of the proportion methods using Lowenstein-Jensen, Middlebrook 7H10 and 7H11 agar media and a radiometric method. Res. Microbiol. 140:405–417. 10.1016/0923-2508(89)90016-8 [DOI] [PubMed] [Google Scholar]
- 26.Garrigó M, Aragon LM, Alcaide F, Borrell S, Cardenosa E, Galan JJ, Gonzalez-Martin J, Martin-Casabona N, Moreno C, Salvado M, Coll P. 2007. Multicenter laboratory evaluation of the MB/BacT Mycobacterium detection system and the BACTEC MGIT 960 system in comparison with the BACTEC 460TB system for susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 45:1766–1770. 10.1128/JCM.02162-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M, Sala C, Hartkoorn RC, Dhar N, Mendoza-Losana A, Cole ST. 2012. Streptomycin-starved Mycobacterium tuberculosis 18b, a drug discovery tool for latent tuberculosis. Antimicrob. Agents Chemother. 56:5782–5789. 10.1128/AAC.01125-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Steenwinkel JE, de Knegt GJ, ten Kate MT, van Belkum A, Verbrugh HA, Kremer K, van Soolingen D, Bakker-Woudenberg IA. 2010. Time-kill kinetics of anti-tuberculosis drugs, and emergence of resistance, in relation to metabolic activity of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65:2582–2589. 10.1093/jac/dkq374 [DOI] [PubMed] [Google Scholar]
- 29.Gumbo T, Louie A, Liu W, Ambrose PG, Bhavnani SM, Brown D, Drusano GL. 2007. Isoniazid's bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J. Infect. Dis. 195:194–201. 10.1086/510247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.