A new crystal structure of the human nNOS heme domain and a better diffracting crystal form of human eNOS heme domain are described.
Keywords: nitric oxide synthase
Abstract
Mammals produce three isoforms of nitric oxide synthase (NOS): neuronal NOS (nNOS), inducible NOS (iNOS) and endothelial NOS (eNOS). The overproduction of NO by nNOS is associated with a number of neurodegenerative disorders; therefore, a desirable therapeutic goal is the design of drugs that target nNOS but not the other isoforms. Crystallography, coupled with computational approaches and medicinal chemistry, has played a critical role in developing highly selective nNOS inhibitors that exhibit exceptional neuroprotective properties. For historic reasons, crystallography has focused on rat nNOS and bovine eNOS because these were available in high quality; thus, their structures have been used in structure–activity–relationship studies. Although these constitutive NOSs share more than 90% sequence identity across mammalian species for each NOS isoform, inhibitor-binding studies revealed that subtle differences near the heme active site in the same NOS isoform across species still impact enzyme–inhibitor interactions. Therefore, structures of the human constitutive NOSs are indispensible. Here, the first structure of human neuronal NOS at 2.03 Å resolution is reported and a different crystal form of human endothelial NOS is reported at 1.73 Å resolution.
1. Introduction
Nitric oxide (NO) is an important signaling molecule involved in many fundamental physiological functions in the nervous, immune and cardiovascular systems (Moncada & Higgs, 1991 ▶). In humans and other mammals the primary source of NO is nitric oxide synthase (NOS), three different isoforms of which are localized and expressed in various tissues or cell types: neuronal NOS (nNOS), inducible NOS (iNOS) and endothelial NOS (eNOS). All NOS isoforms catalyze the same reaction that first oxidizes l-Arg to N-hydroxy-l-Arg followed by a further oxidation to citrulline and NO (Stuehr & Griffith, 1992 ▶). The mammalian NOSs share a similar domain architecture, with an N-terminal catalytic domain containing a heme active site, a nearby cofactor site for tetrahydrobiopterin (H4B) and a C-terminal reductase domain consisting of FMN, FAD and NADPH binding sites. NADPH provides the electrons needed for catalysis, which are first passed on to FAD and FMN and then to the heme (Stuehr & Griffith, 1992 ▶). The electron flow within NOS is monitored by binding of CaM/Ca2+ in the linker region between the two major domains. For iNOS, binding of CaM occurs even at a basal cellular level of Ca2+; therefore, CaM functions like a subunit of iNOS (Cho et al., 1992 ▶). The expression of iNOS is induced by certain cytokines. In contrast, both nNOS and eNOS are expressed constitutively, but the activities of both isoforms are regulated by CaM binding, which itself requires a much higher influx of cellular Ca2+ (Bredt et al., 1992 ▶; Mayer et al., 1992 ▶).
The overproduction or underproduction of NO is linked to a number of pathophysiological conditions (Moncada & Higgs, 1991 ▶). Therefore, the development of NOS inhibitors may have a potential therapeutic benefit in treating NOS-related diseases (Hobbs et al., 1999 ▶). We have been focused on nNOS selective inhibitors that do not interfere with the vital function of eNOS in the cardiovascular system (Silverman, 2009 ▶). Soon after the initial combinatorial chemistry screening for active dipeptide NOS inhibitors, we adopted a structure-based approach in our NOS inhibitor studies (Poulos & Li, 2013 ▶). At the time, crystal structures of the NOS heme domain were available for murine iNOS (Crane et al., 1998 ▶), bovine eNOS (Raman et al., 1998 ▶), human iNOS (Li et al., 1999 ▶; Fischmann et al., 1999 ▶), human eNOS (Fischmann et al., 1999 ▶) and rat nNOS (Li et al., 2002 ▶). Inhibitor-binding studies require the structures of the two constitutive NOSs at favorable resolution (around 2.0 Å), and only rat nNOS and bovine eNOS met the required standards. Considering the highly conserved primary sequence (>90%) of the same NOS isoform from different mammalian species, inhibitor binding to rat nNOS and bovine eNOS was thought to closely reflect the binding behaviors to human nNOS and eNOS, respectively. This is an acceptable assumption because the immediate vicinity of the NOS active site for each isoform is essentially identical across mammalian species. We found a few differences, such as Asp597/Met336 in rat nNOS versus Asn368/Val106 in bovine eNOS, that influence inhibitor-binding behavior, and these differences have served as a structural basis for the design and synthesis of highly selective nNOS inhibitors (Flinspach et al., 2004 ▶). Fortunately, the amino-acid composition of Asp/Met in rat nNOS as well as Asn/Val in bovine eNOS are conserved in human nNOS and eNOS, respectively. For small inhibitors the structures of rat nNOS and bovine eNOS provide good models for human nNOS and eNOS. However, we also noticed that some bulkier inhibitors can reach farther away from the heme active site into a hydrophobic pocket (Xue et al., 2010 ▶) where the sequence diversity between the human isoforms and their rat and bovine counterparts is high, as shown in Table 1 ▶. In these cases, subtle variations in this pocket indeed affect inhibitor binding affinity. For instance, for a series of aminoquinoline compounds we found that the inhibitory potency had a fivefold to sixfold difference between rat and human nNOS (Cinelli et al., 2014 ▶). We therefore decided to shift our efforts to human NOSs both in the inhibitory assays and in structural characterizations. Here, we report the first human nNOS structure at 2.03 Å resolution in the monoclinic space group C2 and also a new human eNOS crystal form that exhibits much better diffraction power to 1.73 Å resolution in the orthorhombic space group P212121.
Table 1. Sequence diversity in a hydrophobic pocket outside the heme active site.
| Rat nNOS | Met336 | Leu337 | Tyr706 | Trp306(B) |
| Human nNOS | Met341 | His342 | Tyr711 | Trp311(B) |
| Bovine eNOS | Val106 | Leu107 | Tyr477 | Trp76(B) |
| Human eNOS | Val104 | Phe105 | Tyr475 | Trp74(B) |
| Murine iNOS | Met114 | Asn115 | Tyr485 | Trp84(B) |
| Human iNOS | Met120 | Thr121 | Tyr491 | Trp90(B) |
2. Methods
2.1. Protein cloning, expression and purification
The production of human eNOS and nNOS heme-domain proteins requires the expression and partial purification of full-length eNOS and nNOS followed by limited trypsinolysis that generates the heme domain. Further purification followed the procedures used for bovine eNOS (Flinspach et al., 2004 ▶) and rat nNOS (Li et al., 2002 ▶). The trypsin digestion of human eNOS produced a 49 kDa heme-domain protein identical to the size of the bovine eNOS heme domain (Raman et al., 1998 ▶) because the trypsin sites in the N-terminal end (Arg39) and in the CaM-binding motif are conserved across the species. However, the trypsin digestion of human nNOS showed a different pattern from that observed for rat nNOS. A comparison of the primary sequence indicated that the major N-terminal cut site of rat nNOS at Arg296 is replaced by Lys301 in human nNOS. To improve the efficiency in the trypsin digest, a human nNOS mutant, K301R, was engineered. A secondary trypsin site known for rat nNOS at Arg349 is also conserved in human nNOS as Arg354. Therefore, a double mutant of human nNOS, K301R/R354A, was produced to avoid the side product generated from the secondary trypsin site.
The original wild-type human nNOS plasmid was kindly provided by Dr Linda Roman in Dr Bettie Sue Masters’ laboratory (University of Texas at San Antonio). The nNOS coding region was then subcloned via the NdeI and XbaI sites into another pCWori vector that contains a six-His tag right before the NdeI site; this was the vector that we used for rat nNOS expression. The K301R/R354A mutations were introduced into this His-tagged version of the pCWori construct using the QuikChange site-directed mutagenesis kit (Stratagene). The mutations and the fidelity of the cloning process were confirmed by sequencing the entire coding region of human nNOS.
The E. coli expression host strain BL21(DE3) was first transformed with the plasmid of human CaM, pACYChCaM, and plated onto LB–agar medium containing 35 µg ml−1 chloramphenicol. Colonies obtained by overnight incubation at 37°C were used to prepare competent cells according to the Clontech procedure. Competent cells were then co-transformed by the plasmid of human eNOS or nNOS and spread onto LB–agar medium containing 100 µg ml−1 ampicillin and 35 µg ml−1 chloramphenicol. The agar plates were incubated at 37°C overnight. The colonies obtained were used to inoculate the small starter LB culture. The overnight-grown starter was then used to inoculate large-scale cell growth in TB culture containing 0.5 mM CaCl2, 100 µg ml−1 ampicillin and 35 µg ml−1 chloramphenicol. The cell cultures were placed in a 37°C shaker with 220 rev min−1 agitation until 1.5 < OD600 nm < 2.0 and were then induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), 0.4 mM 5-aminolevulinic acid and 3 µM riboflavin. Antibiotics were also replenished at induction to maintain the selection. The post-induction incubation was continued for 40 h at 25°C and 100 rev min−1. Cells were harvested by centrifugation and stored at −80°C.
The first column for human nNOS purification was an Ni–NTA column. The same protocol (Li et al., 2002 ▶) for cell lysis and Ni column chromatography as used for rat nNOS was adopted, with the only difference being that the Tris–HCl buffer was replaced by sodium phosphate buffer to improve the protein binding to the Ni resin. An adenosine 2′,5′-diphosphate (2′,5′-ADP) affinity column (25 ml) was the second step. The running buffer was 50 mM Tris–HCl pH 7.8, 10% glycerol, 5 mM βME, 1 mM l-Arg, 10 µM H4B, 200 mM NaCl, 0.5 mM PMSF. Pooled fractions from the Ni column were loaded onto the column pre-equilibrated with Tris buffer. After loading, the column was washed with 250 ml Tris buffer before elution with 50 ml 10 mM NADP+. For human eNOS the binding affinity to the Ni column was poor, making the column-chromatography step less effective. Therefore, the ADP column was used as the first step for human eNOS with the same buffer and procedure as described for human nNOS.
The partially purified full-length human eNOS or nNOS was subjected to limited trypsin digestion at room temperature for 1 h at a protein:trypsin weight ratio of 100:1. The digested sample contained the red heme domain and the yellow FAD/NADPH domain, which could be separated by a Superdex 200 gel-filtration column, as described for rat nNOS (Li et al., 2002 ▶). The Tris buffer used for this last column step was the same as that for the ADP column but was often free of l-Arg to avoid potential competition with the inhibitor binding to the crystals.
2.2. Crystal preparation
The purified human eNOS heme domain did not produce crystals under the conditions reported in the literature (Fischmann et al., 1999 ▶), nor were any hits obtained with various commercial screening kits (Hampton Research and Molecular Dimensions). We decided to try the cross-seeding technique by setting up crystallization under the conditions for bovine eNOS with PEG 3350, cacodylate buffer pH 6.0 and magnesium acetate at 4°C (Raman et al., 1998 ▶). To expand the pH range, bis-tris buffer pH 7.0 was also used. Some red quasi-crystalline spheres of human eNOS (at 10 mg ml−1) appeared after cross-seeding with bovine eNOS crystals in sitting drops containing bis-tris buffer. The reservoir conditions were then used to set up a 96-well additive screen with the Hampton Research Solubility & Stability Screen. Tiny crystals were found with GdCl3 as an additive. Further fine-tuning of the conditions with streak-seeding led to diffraction-quality crystals with a well solution consisting of 10–12% PEG 3350, 0.1 M bis-tris pH 7.5, 0.3 M magnesium acetate, 0.1 M GdCl3.xH2O, 5 mM tris(2-carboxyethyl)phosphine (TCEP). The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 62, b = 110, c = 154 Å, which closely resemble those of bovine eNOS crystals (a = 58, b = 106, c = 158 Å; Raman et al., 1998 ▶).
The human nNOS sample at 10 mg ml−1 was used for crystal screening with commercial screening kits (Hampton Research and Molecular Dimensions). Fine-tuning of the hit conditions from the Hampton Research PEG/Ion kit led to the following conditions, which produced crystals that diffracted to 2.5 Å resolution at a synchrotron source. Crystals were grown at 4°C by the sitting-drop vapor-diffusion method. The well solution consisted of 16% PEG 3350, 150–175 mM sodium acetate, 5% Tacsimate, 0.5% 2-propanol, 5 mM TCEP.
The crystals belonged to the monoclinic space group P21, with unit-cell parameters that were very similar to those of rat nNOS: a = 51.7, b = 164.5, c = 108.8 Å, β = 91.2°, with two independent dimers per asymmetric unit. A close comparison of the crystal packing between the human and rat nNOS structures revealed that a tighter packing between the two NCS-related human nNOS dimers interrupted the higher orthorhombic symmetry observed in the rat nNOS structure. The closer dimer-to-dimer packing interaction (a hydrogen bond) in human nNOS is from the main-chain amide N atom of Gly357 in one dimer to the main-chain carbonyl O atom of Pro518 in the other, whereas in rat nNOS the equivalent residue is Asp352, which uses its side chain to make a hydrogen bond to the carbonyl O atom of Arg514 in the second dimer.
To closely mimic the rat nNOS crystal that diffracted to higher resolution, we decided to further mutate Gly357 to Asp. The resulting triple K301R/R354A/G357D mutant of the human nNOS protein was cloned, expressed and purified in the same way as for the double mutant described above. The new sample at 10 mg ml−1 was used for new rounds of crystal screening. Optimizing from one of the hit conditions, Hampton Research PEG/Ion 2 condition No. 41, led to diffraction-quality crystals. The sitting-drop set up at 4°C had a well solution consisting of 11–13% PEG 3350, 50 mM citric acid, 50 mM bis-tris propane pH 5.0, 10% glycerol, 5 mM TCEP. Crystals of large size were obtained by touch-seeding into drops that had been pre-equilibrated for a few hours. The crystals belonged to the monoclinic space group C2, with unit-cell parameters a = 174.9, b = 84.7, c = 166.7 Å, β = 90.9°.
2.3. N-terminal human nNOS triple-mutant construct
Although the heme domain generated from the triple-mutant full-length human nNOS produced diffraction-quality crystals, the expression yield of this protein was marginal compared with the double mutant or the wild type. To overcome this problem, we made another triple-mutant construct that contained only the N-terminal half of nNOS: residues 1–753, a 83.8 kDa protein. Following the same protocol as outlined above, the expression of this N-terminal half of human nNOS showed an improved yield, which provided sufficient protein for the needs of crystallization. The new construct contained both sites for the trypsinolysis that produces the same heme-domain protein (49.7 kDa) as that generated from the digest of full-length human nNOS. More importantly, the heme-domain protein produced from either full-length or N-terminal human nNOS resulted in the same crystal form as described above.
2.4. Diffraction data collection, processing and structure refinement
Cryogenic (100 K) X-ray diffraction data were collected remotely at the Stanford Synchrotron Radiation Lightsource (SSRL) or at the Advanced Light Source (ALS) using the Blu-Ice data-collection control software (McPhillips et al., 2002 ▶) and a crystal-mounting robot. Raw ADSC Q315r CCD data frames were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). Both human nNOS and eNOS structures were solved by molecular replacement with Phaser (McCoy et al., 2007 ▶) using the rat nNOS (PDB entry 1om4; Li et al., 2002 ▶) and bovine eNOS (PDB entry 1nse; Raman et al., 1998 ▶) structures as the search model, respectively. The resulting models were further refined with REFMAC to generate the initial electron-density maps (Murshudov et al., 2011 ▶). The sequence matching (mutations) and model building were performed in Coot (Emsley & Cowtan, 2004 ▶) and refinement was performed using REFMAC. For the human nNOS structure, water molecules were added in REFMAC and checked using Coot. The TLS (Winn et al., 2001 ▶) protocol was implemented in the final stage of refinement with each subunit as one TLS group. The two human eNOS structures were initially refined in REFMAC and then continued in PHENIX (Adams et al., 2010 ▶) to handle the surface Gd site with the ReadySet! feature in the package. The water updates and TLS refinements (each subunit as a TLS group) were both performed in PHENIX. For the two Gd3+ ions, anisotropic B factors and occupancies were also refined. The OMIT F o − F c density maps were calculated by repeating the last round of TLS refinement in REFMAC or running one round of simulated annealing at 2000 K in PHENIX with the inhibitor coordinates removed from the input PDB file to generate the map coefficients DELFWT and PHDELWT. The coordinates of the refined structures were deposited in the Protein Data Bank. The crystallographic data-collection and structure-refinement statistics are summarized in Table 2 ▶. The Arg608 residues in all four chains of human nNOS and its equivalent Arg372 in two chains of the human eNOS structure are located after a tight turn and are listed as outliers in the Ramachandran plot, but their backbone geometries (ϕ ≃ −115° and ψ ≃ −132°) are well supported by the electron density.
Table 2. Crystallographic data-collection and refinement statistics.
| Data set | Human nNOS–Arg | Human eNOS–Arg | Human eNOS–inhibitor |
|---|---|---|---|
| Data collection | |||
| PDB code | 4d1n | 4d1o | 4d1p |
| Space group | C2 | P212121 | P212121 |
| Unit-cell parameters | |||
| a (Å) | 174.9 | 62.4 | 62.2 |
| b (Å) | 84.7 | 110.1 | 110.4 |
| c (Å) | 166.7 | 153.3 | 153.7 |
| β (°) | 90.4 | ||
| Resolution (Å) | 2.03 (2.07–2.03) | 1.82 (1.85–1.82) | 1.73 (1.78–1.73) |
| R merge | 0.066 (0.721) | 0.099 (>1.000) | 0.074 (>1.000) |
| R p.i.m. | 0.041 (0.446) | 0.049 (0.594) | 0.037 (0.500) |
| CC1/2 | n/a (0.666) | n/a (0.533) | n/a (0.671) |
| 〈I/σ(I)〉 | 24.0 (1.8) | 19.9 (1.0) | 22.8 (1.1) |
| No. of unique reflections | 149403 | 95342 | 109589 |
| Completeness (%) | 94.9 (92.0) | 99.8 (98.2) | 99.9 (99.6) |
| Multiplicity | 3.5 (3.5) | 4.9 (4.0) | 4.9 (4.6) |
| Refinement | |||
| Resolution (Å) | 2.03 | 1.82 | 1.73 |
| No. of reflections used | 141877 | 95245 | 109492 |
| R work/R free | 0.177/0.209 | 0.155/0.191 | 0.156/0.185 |
| No. of atoms | |||
| Protein | 13559 | 6447 | 6442 |
| Ligand/ion | 332 | 224 | 217 |
| Water | 856 | 769 | 777 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.011 | 0.009 | 0.006 |
| Bond angles (°) | 1.39 | 1.16 | 1.14 |
| Ramachandran plot | |||
| Preferred regions | 1602 [97.27%] | 765 [97.58%] | 767 [97.71%] |
| Allowed regions | 40 [2.43%] | 16 [2.04%] | 16 [2.04%] |
| Outliers | 5 [0.30%] | 3 [0.38%] | 2 [0.25%] |
3. Results and discussion
3.1. Structure comparison of human and bovine eNOS
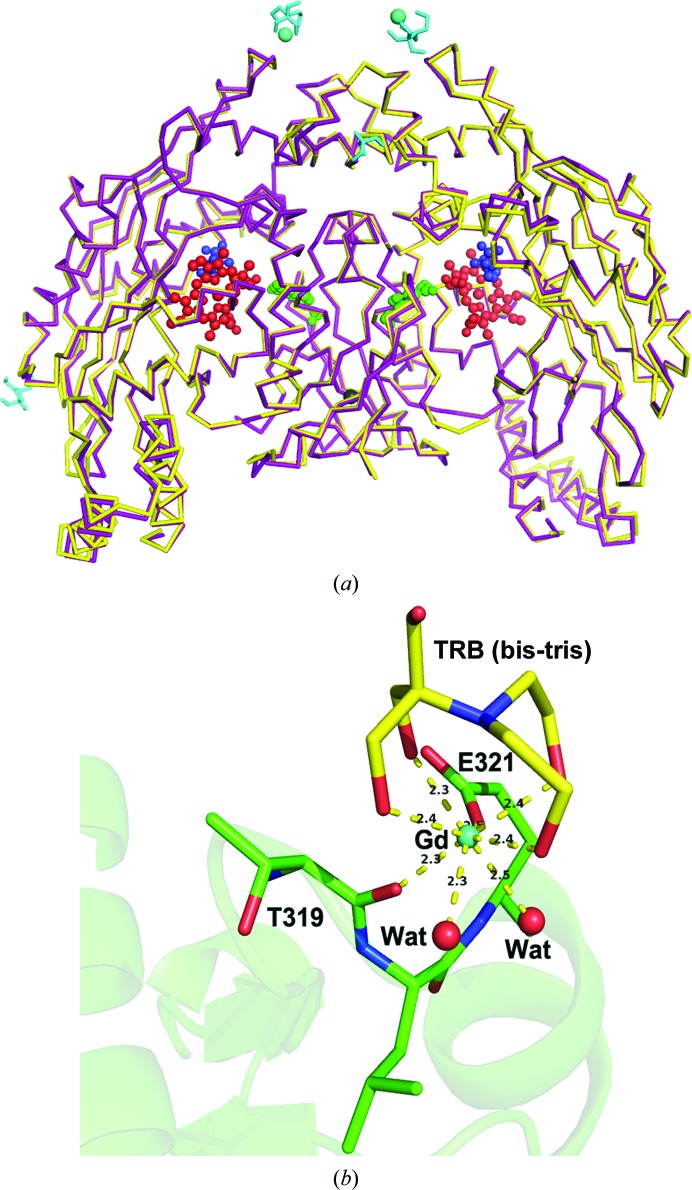
As expected, the structure of human eNOS highly resembles that of bovine eNOS; it crystallizes in the same space group with one dimer per asymmetric unit. Residues Lys67–Trp480 are visible in the human eNOS structure, corresponding to residues Lys69–Trp482 observed in the bovine eNOS structure. The often-disordered loop region (residues Arg109–Pro120) in bovine eNOS is also invisible in human eNOS (residues Arg107–Pro118). The overall r.m.s.d. between the two dimeric structures is 0.65 Å when 803 Cα atoms are superimposed, as shown in Fig. 1 ▶(a).
Figure 1.
(a) Superimposition of bovine eNOS (purple) and human eNOS (yellow) structures. Heme (red), tetrahydrobiopterin (green) and l-Arg (blue) are shown as ball-and-stick representations. A few surface Gd and bis-tris clusters are highlighted in cyan. The overall r.m.s.d. for dimeric eNOS is 0.65 Å with 803 residues compared. (b) A surface Gd3+ site next to Glu321 in chain B stabilized by bis-tris. The 2F o − F c density level for Gd3+ is higher than 40σ. The ligation bond distances from eight oxygen donors are marked in Å.
Although the protein structure of human eNOS is not greatly different from that of bovine eNOS, quite a few clusters of positive density in human eNOS showed up next to some acidic residues on the protein surface that apparently provided additional crystal contacts. The sites next to Asp384 in chain A and Glu321 in chain B are identified as Gd3+ ions owing to the extremely high electron-density level. The sites next to Glu377 in chain A and Glu298 in chain B are bulky clusters but without high density; thus, no heavy metal is involved. These surface clusters turned out to be very difficult to model. The crystallization buffer components Tris, bis-tris, glycerol and Mg ion were considered as potential models for the density. However, the only well ordered site is the Gd3+ site next to Glu321 in chain B, as shown in Fig. 1 ▶(b). The Gd3+ ion is chelated by four hydroxyl groups from bis-tris and is coordinated by the Glu321 side chain, the carbonyl of Thr319 and two water molecules, forming an all-oxygen eight-coordination sphere. The two water ligands were also modeled as chloride ions but resulted in negative difference density. In addition to serving as an additive to promote crystallization, GdCl3 is a good reagent for use in heavy-atom phasing. From what we have observed, it is likely that Gd3+ binding will be supported by the use of bis-tris buffer or some similar chelating reagent that promotes Gd3+ binding to surface acidic residues.
3.2. Structure comparison of human and rat nNOS
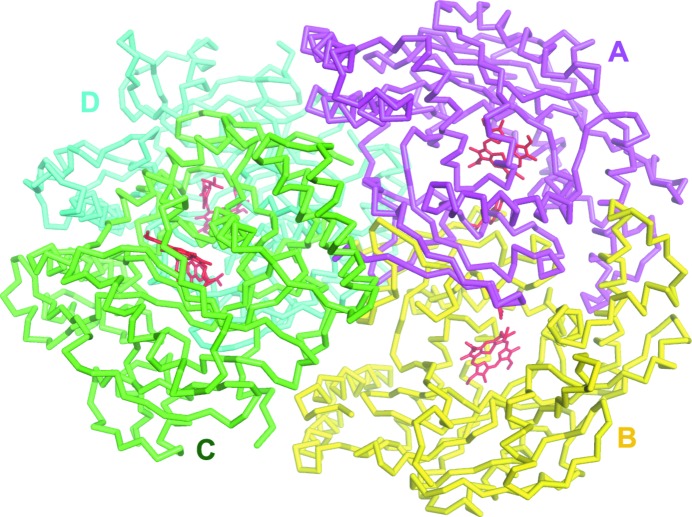
The structure of the human nNOS heme domain contains residues Cys302–Trp721. In contrast to a slightly disordered N-terminus (lacking Cys297 and Pro298) in the rat nNOS structure, the N-terminus of human nNOS is intact, including all residues after the trypsin cleavage site at Arg301 (Arg was introduced into human nNOS by the K301R mutation). The fully ordered N-termini in the human nNOS structure are likely to be owing to crystal-packing interactions. There are two dimers per asymmetric unit in the monoclinic C2 space group, which are tightly packed together in a handshake manner (Fig. 2 ▶). The Cys302 side chains in subunits B and D are almost joined to each other (but without making a disulfide bond), whereas Cys302 in subunits A or C packs closely against the backbone of His346 in subunits C or A, respectively.
Figure 2.
Two dimers in the asymmetric unit of human nNOS. Dimer AB packs tightly against dimer CD in a handshake manner.
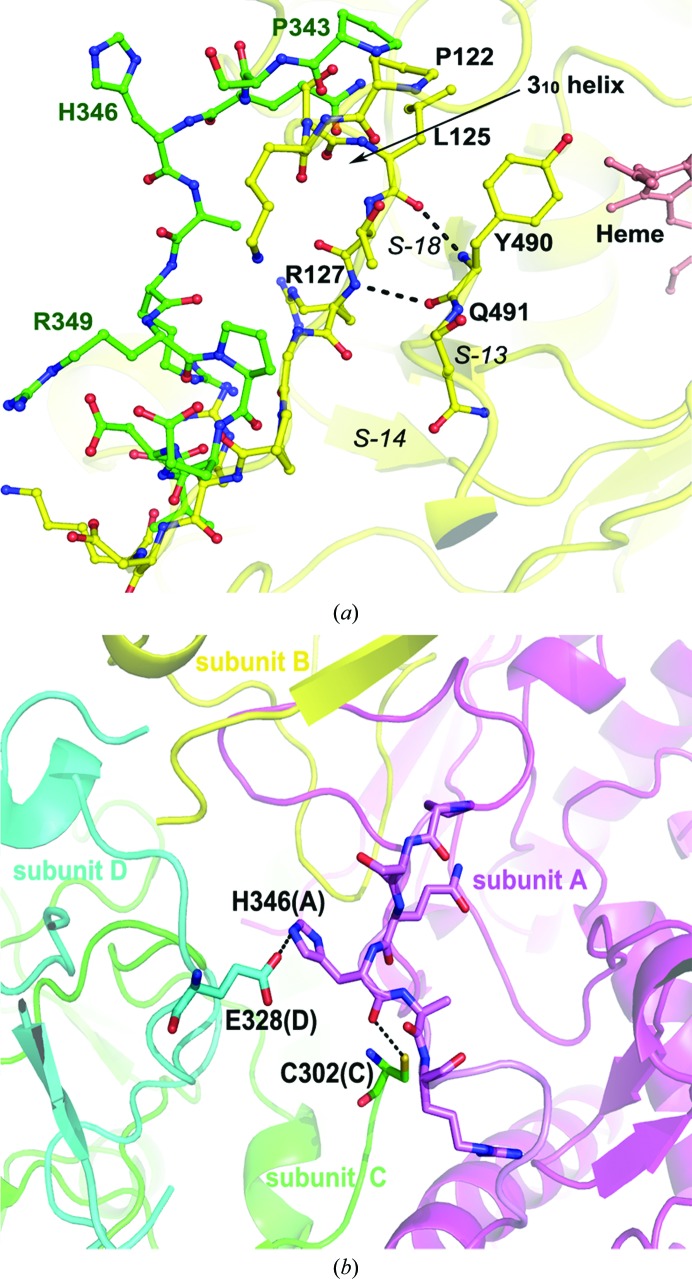
Consistent with the sequence identity of as high as 94%, the structures of human and rat nNOS are almost identical, with an overall r.m.s.d. of 0.338 Å with 816 Cα atoms superimposed. The only exception is a flexible surface loop downstream from the Zn2+-binding site. This loop is usually disordered in the rat nNOS (residues Ser339–Asp347) or bovine eNOS (residues Arg109–Pro120) structures. In the human nNOS structure this loop becomes fully ordered in one subunit of each dimer, but residues Ser344–Asp352 are still missing in the other subunit. The same loop region is always fully ordered in human iNOS (residues Lys123–Asp131; Fig. 3 ▶ a). The loop consists of a small 310-helix (residues Pro122–Leu125) followed by a short β-strand (residues Leu125–Arg127) that forms main-chain to main-chain hydrogen bonds with another β-strand (residues Tyr490 and Gln491 near the C-terminus of the heme domain), which itself is an extension from a three-stranded (S13, S14 and S18) β-sheet (Fischmann et al., 1999 ▶). In contrast, the loop observed in the human nNOS structure lacks any secondary structure, being merely random coil from Pro343 to Arg349. The most likely reason that this loop becomes more fully ordered is owing to crystal-packing interactions. The backbone of His346 in subunit A makes close van der Waals contacts with the N-terminal Cys302 of subunit C from the second dimer. In addition, the His346 side chain can make a hydrogen bond to Glu328 of subunit D (Fig. 3 ▶ b). The ordered loop seen in the human nNOS structure reflects its flexible nature since it can easily adapt to the local environment, which in this case involves crystal packing. Without such packing, this region remains disordered in all mammalian nNOS and eNOS isoforms. The fact that this loop is disordered in constitutive NOSs but not in iNOS might have implications for its potential function. The iNOS isoform is known to bind CaM more tightly and lacks a regulatory insert in the FMN-binding subdomain. Therefore, the NOS–CaM interactions in iNOS are expected to be unique compared with those in the constitutive nNOS or eNOS. A recent hydrogen–deuterium exchange mass-spectrometric study (Smith et al., 2013 ▶) with a few constructs of iNOS mapped out interaction surfaces between the heme domain, the FMN subdomain and CaM. Most importantly, the contact surface between the heme domain and CaM was identified for the first time. The disordered loop found in nNOS or eNOS is in the vicinity of this contact surface with CaM and thus is likely to be involved in the interactions with CaM, especially when the FMN subdomain is in position to transfer electrons from FMN to heme.
Figure 3.
(a) Comparison of the surface loop in human nNOS (green) with that in human iNOS (yellow). The secondary structure of iNOS is also shown as a cartoon with a three-stranded β-sheet labeled (S-13, S-14, S-18). The backbone–backbone hydrogen bonds from the loop to an extension (residues 490 and 491) from the β-sheet are shown by dashed lines. In contract to the 310-helix (residues 122–125) observed in iNOS, the loop in nNOS is a random coil from Pro343 to Arg349. (b) The flexible loop in human nNOS is stabilized by some intersubunit packing contacts, which are shown as dashed lines.
3.3. The active site of human eNOS
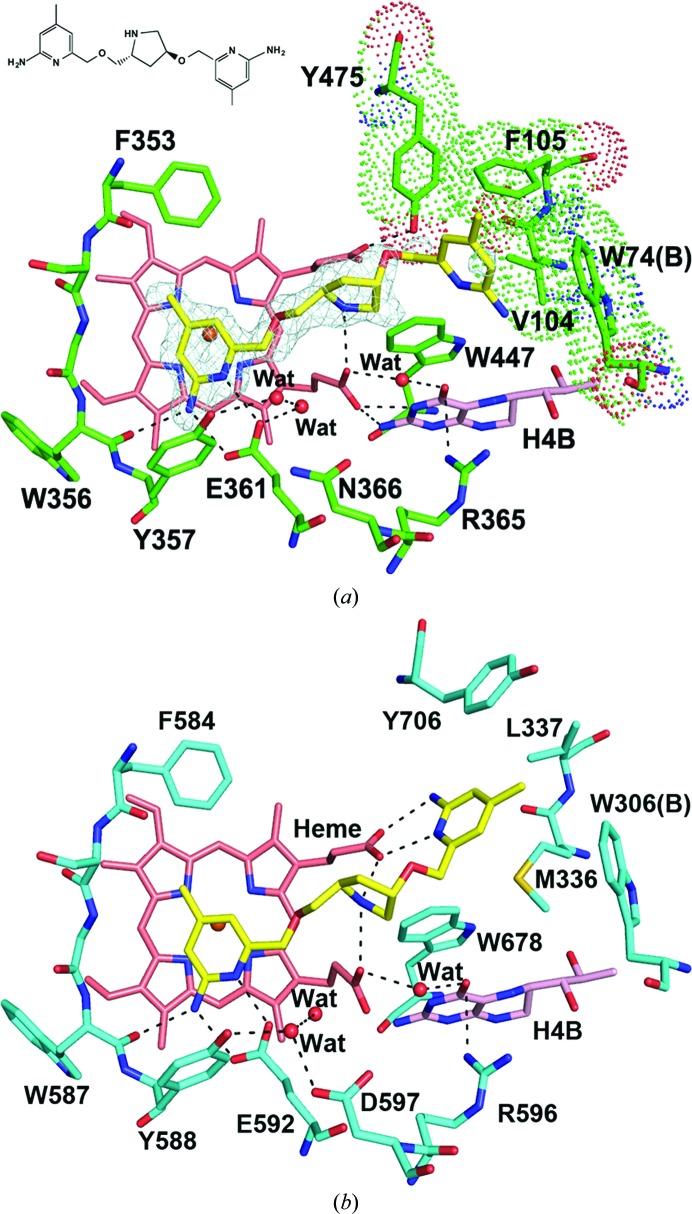
We have determined two human eNOS structures in complex with l-Arg and with an aminopyridine inhibitor. The active site of human eNOS is identical to that of bovine eNOS. The Asn366 and Val104 residues in human eNOS are conserved from those (Asn368 and Val106) in bovine eNOS, which have been shown to be responsible for the eNOS versus nNOS inhibitor selectivity (Flinspach et al., 2004 ▶). There is one residue difference that can potentially impact the inhibitor binding: the bulkier Phe105 in human versus Leu107 in bovine eNOS. This variation is in a hydrophobic pocket that is on the outside of the heme active site but is accessible by bulkier inhibitors. The human eNOS aminopyridine inhibitor complex structure shown in Fig. 4 ▶(a) illustrates the situation. The inhibitor uses one of its two aminopyridines to make hydrogen bonds to Glu361 and its pyrrolidine ring N atom to anchor it between the two heme propionate groups. While the second aminopyridine ring is poorly defined, it is clear that this aminopyridine cannot access heme propionate D because the Tyr475 side chain remains in its in-rotamer conformation and hydrogen-bonds to the heme propionate (Fig. 4 ▶ a). This also is the situation that is observed in the bovine eNOS structure in complex with the same inhibitor (Jing et al., 2014 ▶). In comparison, as shown in Fig. 4 ▶(b), the same inhibitor bound to rat nNOS can establish a double-headed binding mode with the second aminopyridine directly hydrogen-bonded to heme propionate D (Jing et al., 2014 ▶). To achieve this double-headed binding, the Tyr706 side chain (equivalent to Tyr475 in human eNOS or Tyr477 in bovine eNOS) in rat nNOS can adopt an out-rotamer position, leaving room for the inhibitor to interact with the heme. The side-chain mobility of this conserved Tyr residue directly influences the inhibitor-binding modes. In human eNOS, an additional potential obstacle to double-headed binding is the bulky Phe105 side chain, which might make unfavorably close contacts with the second aminopyridine of the inhibitor if it were hydrogen-bonded to heme propionate D (Li et al., 2014 ▶).
Figure 4.
(a) The active site of human eNOS with an aminopyridine inhibitor bound. The OMIT F o − F c density for the inhibitor is contoured at 3.0σ. Major hydrogen bonds are depicted as dashed lines. The hydrophobic pocket on the outside of the active site is lined with Val104, Phe105, Tyr475 and Trp74 from chain B and is highlighted with a surface dot representation. (b) The active site of rat nNOS with the same inhibitor bound (PDB entry 4c39; Jing et al., 2014 ▶) but in a double-headed binding mode with both aminopyridine rings involved in hydrogen bonds to protein and heme.
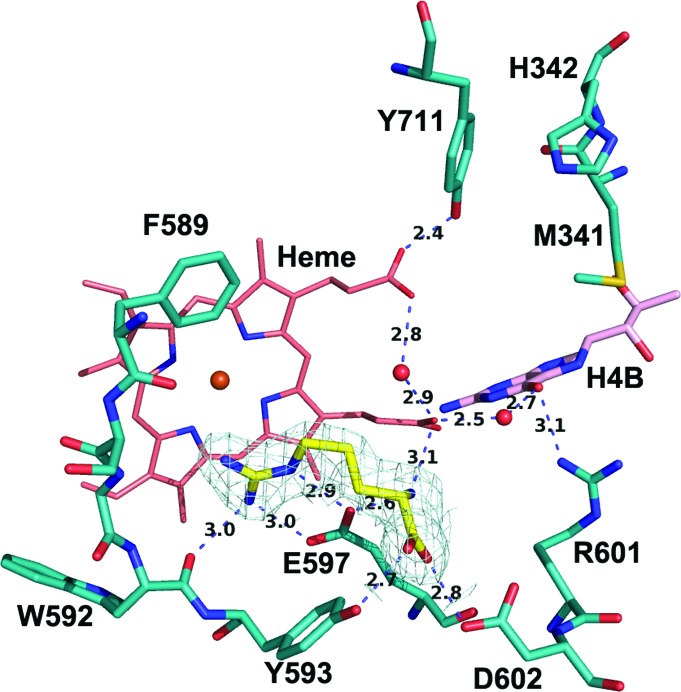
3.4. The active site of human nNOS
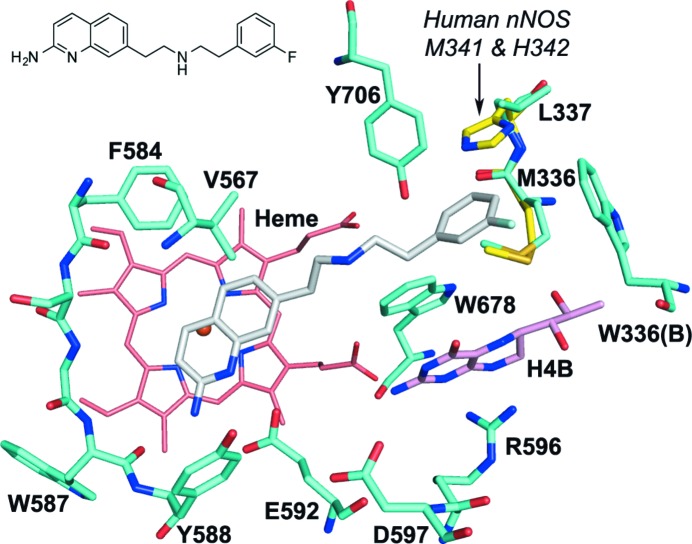
The two isoform-specific residues Met341 and Asp602 in human nNOS and Met336 and Asp597 in rat nNOS are conserved. Therefore, it is no surprise that the active site of human nNOS is identical to that of rat nNOS (Fig. 5 ▶). However, there is an amino-acid difference, His342 in human versus Leu337 in rat, located in the aforementioned hydrophobic pocket, which is potentially important in inhibitor binding. We have found that some of the bulky rat nNOS inhibitors exhibited a fivefold to sixfold weaker affinity for human nNOS (Cinelli et al., 2014 ▶), which might be influenced by the polarity and bulkiness of this His342 residue. Fig. 6 ▶ shows one of the aminoquinoline inhibitors bound to the rat nNOS active site with the human nNOS residues Met341 and His342 overlaid. Apparently, the fluorophenyl tail of the inhibitor may clash with the His342 side chain, which may force the inhibitor to adjust its binding conformation. In the past, without a crystal structure of human nNOS, we used the rat nNOS L337H mutant to explore the binding affinity of inhibitors to the human form (Fang et al., 2009 ▶). In the future, the inhibitory assay and structural characterization will be performed with human nNOS samples.
Figure 5.
The active site of human nNOS with l-Arg bound. The 2F o − F c electron density for l-Arg is contoured at 1.0σ. The extensive hydrogen-bonding networks involving protein, heme, H4B and l-Arg are depicted as dashed lines with distances marked in Å.
Figure 6.
An aminoquinoline inhibitor bound to the rat nNOS active site (PDB entry 4cdt; Cinelli et al., 2014 ▶). For comparison, two residues from the human nNOS structure, Met341 and His342 (yellow), are overlaid. The potential clash between the fluorophenyl ring and the His342 side chain is obvious.
Our efforts in the development of NOS inhibitors have been focused on the two constitutive NOSs, inhibiting nNOS but sparing eNOS. With diffraction-quality crystals now available for both human nNOS and eNOS, we can further our studies with human protein samples, which represents a step forward towards our ultimate goal of searching for compounds that can serve as therapeutic drugs for NOS-related human diseases.
Supplementary Material
PDB reference: human nNOS–Arg, 4d1n
PDB reference: human eNOS–Arg, 4d1o
PDB reference: human eNOS–inhibitor, 4d1p
Acknowledgments
This work was supported by NIH grants GM57353 (TLP) and GM049725 (RBS). We thank the SSRL and ALS beamline staff for their support during remote data collection.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bredt, D. S., Ferris, C. D. & Snyder, S. H. (1992). J. Biol. Chem. 267, 10976–10981. [PubMed]
- Cho, H. J., Xie, Q.-W., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D. & Nathan, C. (1992). J. Exp. Med. 176, 599–604. [DOI] [PMC free article] [PubMed]
- Cinelli, M. A., Li, H., Chreifi, G., Martásek, P., Roman, L. J., Poulos, T. L. & Silverman, R. B. (2014). J. Med. Chem. 57, 1513–1530. [DOI] [PMC free article] [PubMed]
- Crane, B. R., Arvai, A. S., Ghosh, D. K., Wu, C., Getzoff, E. D., Stuehr, D. J. & Tainer, J. A. (1998). Science, 279, 2121–2126. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fang, J., Ji, H., Lawton, G. R., Xue, F., Roman, L. J. & Silverman, R. B. (2009). J. Med. Chem. 52, 4533–4537. [DOI] [PMC free article] [PubMed]
- Fischmann, T. O., Hruza, A., Niu, X. D., Fossetta, J. D., Lunn, C. A., Dolphin, E., Prongay, A. J., Reichert, P., Lundell, D. J., Narula, S. K. & Weber, P. C. (1999). Nature Struct. Biol. 6, 233–242. [DOI] [PubMed]
- Flinspach, M. L., Li, H., Jamal, J., Yang, W., Huang, H., Hah, J.-M., Gómez-Vidal, J. A., Litzinger, E. A., Silverman, R. B. & Poulos, T. L. (2004). Nature Struct. Mol. Biol. 11, 54–59. [DOI] [PubMed]
- Hobbs, A. J., Higgs, A. & Moncada, S. (1999). Annu. Rev. Pharmacol. Toxicol. 39, 191–220. [DOI] [PubMed]
- Jing, Q., Li, H., Roman, L. J., Martasek, P., Poulos, T. L. & Silverman, R. B. (2014). ACS Med. Chem. Lett. 5, 56–60. [DOI] [PMC free article] [PubMed]
- Li, H., Jamal, J., Delker, S., Plaza, C., Ji, H., Jing, Q., Huang, H., Kang, S., Silverman, R. B. & Poulos, T. L. (2014). Biochemistry, 53, 5272–5279. [DOI] [PMC free article] [PubMed]
- Li, H., Raman, C. S., Glaser, C. B., Blasko, E., Young, T. A., Parkinson, J. F., Whitlow, M. & Poulos, T. L. (1999). J. Biol. Chem. 274, 21276–21284. [DOI] [PubMed]
- Li, H., Shimizu, H., Flinspach, M., Jamal, J., Yang, W., Xian, M., Cai, T., Wen, E. Z., Jia, Q., Wang, P. G. & Poulos, T. L. (2002). Biochemistry, 41, 13868–13875. [DOI] [PubMed]
- Mayer, B., Klatt, P., Böhme, E. & Schmidt, K. (1992). J. Neurochem. 59, 2024–2029. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Moncada, S. & Higgs, E. A. (1991). Eur. J. Clin. Invest. 21, 361–374. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Poulos, T. L. & Li, H. (2013). Acc. Chem. Res. 46, 390–398. [DOI] [PMC free article] [PubMed]
- Raman, C. S., Li, H., Martásek, P., Král, V., Masters, B. S. & Poulos, T. L. (1998). Cell, 95, 939–950. [DOI] [PubMed]
- Silverman, R. B. (2009). Acc. Chem. Res. 42, 439–451. [DOI] [PMC free article] [PubMed]
- Smith, B. C., Underbakke, E. S., Kulp, D. W., Schief, W. R. & Marletta, M. A. (2013). Proc. Natl Acad. Sci. USA, 110, 3577–E3586. [DOI] [PMC free article] [PubMed]
- Stuehr, D. J. & Griffith, O. W. (1992). Adv. Enzymol. Relat. Areas Mol. Biol. 65, 287–346. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Xue, F., Li, H., Fang, J., Roman, L. J., Martásek, P., Poulos, T. L. & Silverman, R. B. (2010). Bioorg. Med. Chem. Lett. 20, 6258–6261. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human nNOS–Arg, 4d1n
PDB reference: human eNOS–Arg, 4d1o
PDB reference: human eNOS–inhibitor, 4d1p