Crystallization experiments of the BRPF1 bromodomain in complex with its H4K12ac and H2AK5ac histone ligands yielded crystals that are suitable for high-resolution X-ray diffraction analysis. These structures will be solved in order to elucidate the molecular mechanism of histone recognition.
Keywords: BRPF1 bromodomain, H4K12ac, H2AK5ac
Abstract
The bromodomain-PHD finger protein 1 (BRPF1) is an essential subunit of the monocytic leukemia zinc (MOZ) histone acetyltransferase (HAT) complex and is required for complex formation and enzymatic activation. BRPF1 contains a structurally conserved bromodomain, which recognizes specific acetyllysine residues on histone proteins. The MOZ HAT plays a direct role in hematopoiesis, and deregulation of its activity is linked to the development of acute myeloid leukemia. However, the molecular mechanism of histone-ligand recognition by the BRPF1 bromodomain is currently unknown. The 117-amino-acid BRPF1 bromodomain was overexpressed in Escherichia coli and purified to homogeneity. Crystallization experiments of the BRPF1 bromodomain in complex with its H4K12ac and H2AK5ac histone ligands yielded crystals that were suitable for high-resolution X-ray diffraction analysis. The BRPF1 bromodomain–H4K12ac crystals belonged to the tetragonal space group P43212, with unit-cell parameters a = 75.1, b = 75.1, c = 86.3 Å, and diffracted to a resolution of 1.94 Å. The BRPF1 bromodomain–H2AK5ac crystals grew in the monoclinic space group P21, with unit-cell parameters a = 60.9, b = 55.6, c = 82.1 Å, β = 93.6°, and diffracted to a resolution of 1.80 Å. Complete data sets were collected from both crystal forms using synchrotron radiation on beamline X29 at Brookhaven National Laboratory (BNL).
1. Introduction
Covalent modification of histone proteins plays a key role in determining the outcome of many nuclear processes, including transcription, DNA repair, recombination and replication (Iizuka et al., 2006 ▶; Peña et al., 2008 ▶; Matthews et al., 2007 ▶; Sterner & Berger, 2000 ▶). They are central to cell homeostasis, as alterations in chromatin structure contribute to the development of cancer and other human diseases (Delmore et al., 2011 ▶; Guetg et al., 2010 ▶). The monocytic leukemic zinc-finger/MOZ-related factor (MOZ/MORF) histone acetyltransferase (HAT) is involved in chromosomal translocations found in a subtype of acute myeloid leukemia (AML) associated with a poor prognosis and a median survival of only six months (Brown et al., 2012 ▶; Borrow et al., 1996 ▶; Panagopoulos et al., 2001 ▶). MOZ functions as a multi-subunit HAT complex and acetylates free histones H3, H4, H2A and H2B in vitro (Ullah et al., 2008 ▶; Champagne et al., 2001 ▶). Acetylation of histones located near gene promoters is associated with up-regulation of gene transcription, and the acetylation activity of MOZ has been shown to control the expression of homeobox (HOX) genes (Camós et al., 2006 ▶). The MOZ HAT also plays a direct role in hematopoiesis and is essential for the development and maintenance of hematopoietic stem cells (HSCs; Katsumoto et al., 2006 ▶). As such, MOZ has also been shown to be a strong co-activator of runt-related transcription factor 1 (RUNX1), the master regulator of hematopoiesis (Perez-Campo et al., 2009 ▶; Kitabayashi et al., 2001 ▶). Furthermore, disruption of the MOZ HAT activity has been linked to the development of disease, particularly leukemias (Carapeti et al., 1998 ▶; Crowley et al., 2005 ▶; Esteyries et al., 2008 ▶). MOZ was first identified as a fusion partner with the CREB binding protein (CBP) HAT in a t(8;16)(p11;p13) translocation found in AML, and disruption of the normal acetylation activity of MOZ leads to leukemogenic transformations and oncogenesis (Borrow et al., 1996 ▶). The MOZ HAT functions as a quaternary complex with the hEaf6, BRPF1 and ING5 subunits (Ullah et al., 2008 ▶). The BRPF1 subunit has been shown to mediate complex formation of MOZ/MORF with the ING5 and hEAF6 proteins (Ullah et al., 2008 ▶). Deletion-mapping studies have also revealed that the acetyltransferase domain of MOZ/MORF is sufficient for BRPF1 interaction (Ullah et al., 2008 ▶). Therefore, BRPF1 is thought to play a key role in assembling and activating MOZ/MORF HAT complexes. Human BRPF1 is a large multi-domain protein that contains multiple effector domains with known functions in gene transcription and chromatin binding and remodeling (Laue et al., 2008 ▶). These include a unique double PHD finger and zinc knuckle domain (PZP), a bromodomain, a chromo/Tudor-related Pro-Trp-Trp-Pro (PWWP) domain and a leucine-zipper motif. Interestingly, the noncatalytic subunits of the MOZ HAT complex stimulate its acetylation activity, and chromatin reader domains within the complex help to recruit MOZ/MORF to distinct sites of active chromatin (Vezzoli et al., 2010 ▶; Champagne et al., 2008 ▶; Qin et al., 2011 ▶; Qiu et al., 2012 ▶; Carlson & Glass, 2014 ▶). Bromodomains are evolutionary conserved structural motifs generally known to bind histones acetylated at specific lysine residues (Dhalluin et al., 1999 ▶). All bromodomains share a conserved fold that comprises a left-handed bundle of four α-helices (αZ, αA, αB and αC) linked by loop regions of variable length (ZA and BC loops), which line the acetylated lysine (Kac) binding pocket and determine the binding specificity (Mujtaba et al., 2007 ▶). To date, there is only limited structural information available on the BRPF1 bromodomain in complex with its histone ligands. An unpublished NMR structure of the BRPF1 bromodomain complexed with an H4K5ac histone ligand has been deposited in the PDB (PDB entry 2rs9; X. Qin, F. Hayashi & S. Yokoyama, unpublished work); however, we recently found that the BRPF1 bromodomain preferentially selects for the H2AK5ac, H4K12ac and H3K14ac acetylation modifications (Poplawski et al., 2014 ▶). Here, we report the crystallization of the BRPF1 bromodomain in complex with H2AK5ac and H4K12ac histone peptides, which is the first step towards solving the structures of these complexes.
2. Materials and methods
2.1. Plasmid construction
Human BRPF1 (UniProt code P55201) cDNA was provided by Dr Xiang-Jiao Yang, McGill University Department of Medicine. The bromodomain region (residues 629–742) was PCR-amplified and cloned into the pDEST15 vector encoding an N-terminal GST tag using the Gateway Cloning technology (Invitrogen) as described previously (Poplawski et al., 2014 ▶). The DNA sequence was verified and the plasmid was transformed into Escherichia coli Rosetta 2(DE3)pLysS competent cells (Novagen), which supply tRNA for rare codons.
2.2. Protein expression and purification
E. coli cells containing the GST-tagged BRPF1 bromodomain were grown in 4 l Terrific broth (TB) at 37°C. The culture was induced by the addition of 0.25 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the OD600 reached 1.3, and the cells were harvested by pelleting after 16 h of incubation at 16°C with shaking at 225 rev min−1. The bacterial pellet was resuspended in 100 ml lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% Nonidet P-40, 1 mM DTT) containing 1 ml lysozyme and the cells were disrupted by sonication. The cell lysate was cleared by centrifugation (10 000 rev min−1 for 20 min). The supernatant was added to 12.5 ml glutathione agarose resin (Thermo Scientific) and incubated on ice (4°C) while agitating. After incubation, the suspension was centrifuged for 5 min at 500g to collect the beads. The collected beads were poured into a 25 ml Econo-Column Chromatography Column (Bio-Rad) and washed with four column volumes of wash buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM DTT). The GST tag was cleaved overnight at 4°C by the addition of PreScission Protease (GE Healthcare) and the eluted BRPF1 bromodomain protein was concentrated to a total volume of approximately 3 ml. The sample was further purified using gel-filtration chromatography on a HiPrep 16/60 Sephacryl S-100 High Resolution column (GE Healthcare) equilibrated with wash buffer. Eluted fractions corresponding to the BRPF1 bromodomain were pooled and concentrated to 69.9 mg ml−1 at 4°C. The protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific) and was calculated from the absorption at 562 nm and the BRPF1 bromodomain extinction coefficient of 7450 M −1 cm−1. The purity of the BRPF1 bromodomain was verified by SDS–PAGE gels stained with GelCode Blue Safe protein stain (Thermo Scientific).
2.3. Crystallization
The purified 117-residue BRPF1 bromodomain protein (114 residues from the bromodomain and –GPL from the N-terminal GST tag) was premixed with either the H2AK5ac or H4K12ac histone peptides. The histone H2A (residues 1–12) and histone H4 (residues 4–17) peptides were synthesized at the University of Colorado, Denver with amino-acid sequences SGRGKacQGGKARA and GKGGKGLGKacGGAKR, respectively, where Kac represents an acetylated lysine. The purified BRPF1 bromodomain protein at 2.3 mM was mixed with 5 mM of either the H2AK5ac or H4K12ac modified peptide in a 1.5 ml microcentrifuge tube. Crystallization screens were set up using the sitting-drop method in 96-well VDX plates (Hampton Research), with drops consisting of 1 µl protein–peptide mixture plus 1 µl reservoir solution and a reservoir volume of 100 µl. The initial screens tested were Crystal Screen and Crystal Screen 2 (Hampton Research). Crystals were observed under several conditions (Table 1 ▶).
Table 1. Initial crystallization conditions of the BRPF1 bromodomain in complex with H4K12ac or H2AK5ac.
Structures were solved from the conditions shown in bold.
| Crystal | Commercial crystallization screen | Condition No. |
|---|---|---|
| BRPF1 bromoH4K12ac | Crystal Screen | 6 |
| Crystal Screen 2 | 22, 30, 37, 38 | |
| BRPF1 bromoH2AK5ac | Crystal Screen | 16 |
| Crystal Screen 2 | 26, 32, 42 |
The H4K12ac ligand was able to co-crystallize with the BRPF1 bromodomain in five different conditions at 4°C: Crystal Screen condition No. 6 [0.2 M magnesium chloride hexahydrate, 0.1 M Tris–HCl pH 8.5, 30%(w/v) polyethylene glycol 4000], Crystal Screen 2 condition No. 22 [0.1 M MES monohydrate pH 6.5, 12%(w/v) polyethylene glycol 20 000], Crystal Screen 2 condition No. 30 [0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol 6000, 5%(v/v) (±)-2-methyl-2,4-pentanediol], Crystal Screen 2 condition No. 37 [0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol 8000, 8%(v/v) ethylene glycol] and Crystal Screen 2 condition No. 38 [0.1 M HEPES pH 7.5, 20%(w/v) polyethylene glycol 10 000]. The best single, well ordered and large crystals formed in condition Nos. 30 (Fig. 1 ▶ a), 37 and 38 of Crystal Screen 2 (Table 1 ▶). The crystals were mounted in cryoloops and flash-cooled in liquid nitrogen at 100 K as described by Garman (1999 ▶).The crystals from Crystal Screen 2 condition No. 30 diffracted to 2.5 Å resolution but had a strong ice ring. The crystals from Crystal Screen 2 condition No. 37 diffracted to 2.0 Å resolution with no ice, while the crystals from Crystal Screen 2 condition No. 38 diffracted well with no ice but were mosaic. The crystals from Crystal Screen 2 condition No. 30 [0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol 6000, 5%(v/v) (±)-2-methyl-2,4-pentanediol] were reproduced in hanging drops (1 µl protein solution plus 1 µl mother liquor) using 24-well VDX plates (Hampton Research) containing 500 µl mother liquor in the reservoir. They were cryoprotected by serially transferring them into a reconstituted drop solution (50% wash buffer:50% mother liquor) with 10% MPD added followed by transfer into another reconstituted drop solution containing 15% MPD before flash-cooling in liquid nitrogen. The cryoprotected crystals were tested at the University of Vermont (UVM) Center for X-ray crystallography on a Rigaku RUH3R generator equipped with an MAR345 image-plate detector and diffracted to 2.0 Å resolution with no ice. These crystals were then saved, along with the crystals from Crystal Screen 2 condition No. 37 [0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol 8000, 8%(v/v) ethylene glycol], for further data collection at the synchrotron light source.
Figure 1.
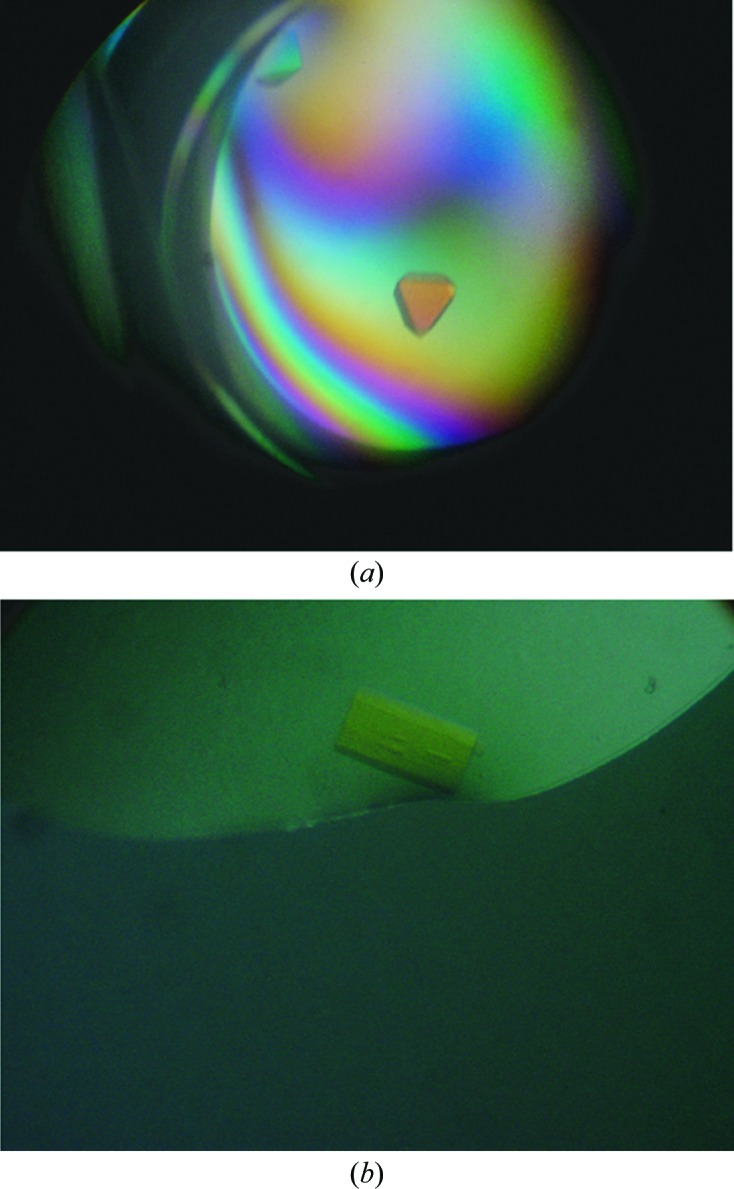
Crystals of the BRPF1 bromodomain in complex with its histone ligands. (a) Crystals of the BRPF1 bromodomain bound to the H4K12ac peptide from Crystal Screen 2 condition No. 30. (b) Crystals of BRPF1 bromodomain bound to the H2AK5ac peptide after optimization of Crystal Screen 2 condition No. 26.
The H2AK5ac ligand co-crystallized with the BRPF1 bromodomain in four different conditions: Crystal Screen condition No. 16 (0.1 M HEPES sodium pH 7.5, 1.5 M lithium sulfate monohydrate), Crystal Screen 2 condition No. 26 [0.2 M ammonium sulfate, 0.1 M MES monohydrate pH 6.5, 30%(w/v) polyethylene glycol monomethyl ether 5000], Crystal Screen 2 condition No. 32 (0.1 M sodium chloride, 0.1 M HEPES pH 7.5, 1.6 M ammonium sulfate) and Crystal Screen 2 condition No. 42 [1.5 M ammonium sulfate, 0.1 M Tris–HCl pH 8.5, 12%(v/v) glycerol] (Table 1 ▶). Crystals of the BRPF1 bromodomain in complex with the H2AK5ac ligand grown from condition No. 26 of Crystal Screen 2 [0.2 M ammonium sulfate, 0.1 M MES monohydrate pH 6.5, 30%(w/v) polyethylene glycol monomethyl ether 5000] were selected for screening and optimization based on size and their well ordered and single appearance (Fig. 1 ▶ b). Since this condition provided suitable cryoprotection, the crystals were mounted directly into cryoloops and flash-cooled in liquid nitrogen at 100 K. Crystal diffraction was tested at the UVM Center for X-ray Crystallography. The initial crystals diffracted to 2.0 Å resolution but were highly mosaic with a smeared lattice that could not be indexed. We optimized the BRPF1 bromodomain–H2AK5ac crystals by reproducing them in hanging drops (1 µl protein solution plus 1 µl mother liquor) using 24-well VDX plates (Hampton Research) containing 500 µl mother liquor in the reservoir. New crystals were grown by seeding the new drops with a 1/10 dilution mixture containing a crushed, previously grown crystal (of the same kind) in a reconstituted drop solution, as described by Bergfors (2003 ▶). After several cycles of optimization including seeding, adjusting the pH, the PEG and the salt concentrations, new crystals were obtained that diffracted to better than 2 Å resolultion but without much improvement in the quality of the diffraction lattice or the mosaicity. Additives, including 1% propylene glycol, 1% dioxane, 1% propanediol and 1% xylitol, were then added to the crystallization condition to try to improve the crystal quality further. The addition of 1% propylene glycol produced crystals with reduced mosaicity that diffracted to 2.0 Å resolution, but determination of the correct space group was still difficult owing to overlapping spots in the crystal lattice. The highest quality crystals were saved for further analysis at the synchrotron light source.
2.4. Data collection
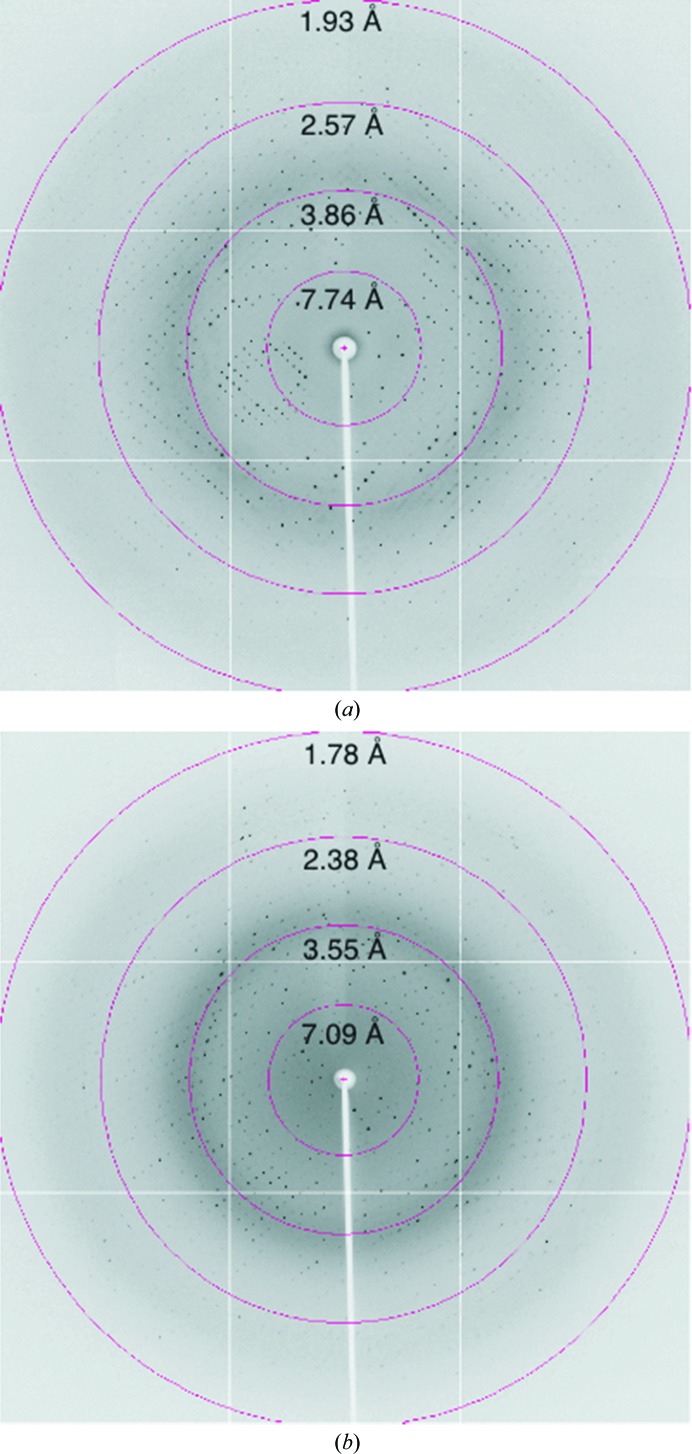
As mentioned above, crystals of BRPF1 bromodomain in complex with either the H4K12ac or H2AK5ac histone peptides were initially tested at the UVM Center for X-ray Crystallography using a Rigaku RUH3R generator and diffracted to around 2.0 Å resolution. Complete data sets for both crystal types were collected during the RapiData Collection and Structure Solving course at Brookhaven National Laboratory (BNL) using synchrotron radiation on beamline X29 equipped with an ADSC Quantum 315r CCD detector (Figs. 2 ▶ a and 2 ▶ b). The data were processed with the HKL-2000 software package (Otwinowski & Minor, 1997 ▶) and Table 2 ▶ summarizes the data-collection statistics.
Figure 2.
Diffraction patterns of the BRPF1 bromodomain crystals using synchrotron radiation on beamline X29 at Brookhaven National Laboratory. (a) BRPF1 bromodomain in complex with H4K12ac. (b) BRPF1 bromodomain in complex with H2AK5ac.
Table 2. Data-collection statistics for the BRPF1 bromodomain in complex with the H2AK5ac and H4K12ac histone ligands.
Values in parentheses are for the outermost resolution shell.
| BromodomainH4K12ac | BromodomainH2AK5ac | |
|---|---|---|
| No. of crystals | 1 | 1 |
| Beamline | X29, BNL | X29, BNL |
| Wavelength () | 1.075 | 1.075 |
| Detector | ADSC Quantum 315r CCD | ADSC Quantum 315r CCD |
| Crystal-to-detector distance (mm) | 250 | 225 |
| Rotation range per image () | 1 | 0.5 |
| Total rotation range () | 360 | 284 |
| Exposure time per image (s) | 12 | 12 |
| Resolution range () | 34.431.94 (2.011.94) | 41.021.80 (1.831.80) |
| Space group | P43212 | P21 |
| Unit-cell parameters | ||
| a, b, c () | 75.1, 75.1, 86.3 | 60.9, 55.6, 82.1 |
| , , () | 90.0, 90.0, 90.0 | 90.0, 93.6, 90.0 |
| Total No. of measured intensities | 521674 | 204355 |
| Unique reflections | 18780 | 48421 |
| Multiplicity | 27.7 (27.2) | 4.0 (3.9) |
| Mean I/(I) | 25.84 (14.66) | 21.04 (3.0) |
| Completeness (%) | 99.6 (96.9) | 99.9 (100) |
| R merge † (%) | 13.9 (27.9) | 8.1 (38.6) |
| Wilson B factor (2) | 26.5 | 21.0 |
R
merge = 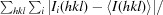
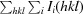 .
.
3. Results and discussion
Previously, we used nuclear magnetic resonance (NMR) titration and isothermal titration calorimetry (ITC) experiments to identify the acetylated histone ligands of the BRPF1 bromodomain (Poplawski et al., 2014 ▶). The H2AK5ac, H4K12ac and H3K14ac histone ligands showed the highest affinities for the BRPF1 bromodomain. However, to elucidate the molecular mechanism of acetyllysine recognition by the BRPF1 bromodomain, atomic resolution structural information is needed. We set up co-crystallization trials of the 117-amino-acid bromodomain of BRPF1 with its histone ligands using Crystal Screen and Crystal Screen 2 and were able to obtain crystals of the BRPF1 bromodomain in complex with the H2AK5ac and H4K12ac ligands. Crystallization attempts of the BRPF1 bromodomain in complex with the H3K14ac ligand were unsuccessful. As shown in Table 1 ▶, the H4K12ac ligand was able to co-crystallize with the BRPF1 bromodomain in five different conditions at 4°C, while the H2AK5ac ligand co-crystallized with the BRPF1 bromodomain in four different conditions.
The diffraction quality of the crystals was examined at the Center for X-ray Crystallography at the University of Vermont using a Rigaku RUH3R generator equipped with a MAR345 image-plate detector. Although many of the crystals tested diffracted to a resolution better than 3 Å, the BRPF1 bromodomain–H2AK5ac crystals showed a significant level of mosaicity (Crystal Screen 2 condition No. 26) and better cryoprotection of the BRPF1 bromodomain–H4K12ac crystals was needed (Crystal Screen 2 condition No. 30). Once the crystallization growth and cryoprotection conditions had been optimized, high-resolution data sets for both crystal forms were collected during the RapiData Collection and Structure Solving course at Brookhaven National laboratory (BNL) using synchrotron radiation on beamline X29 equipped with an ADSC Quantum 315r CCD detector.
The BRPF1 bromodomain–H4K12ac crystals (from Crystal Screen 2 condition No. 30) diffracted to a maximum resolution of 1.94 Å (Fig. 2 ▶ a). The data-collection statistics are summarized in Table 2 ▶. The BRPF1 bromodomain–H4K12ac crystal belonged to the primitive tetragonal space group P43212, with unit-cell parameters a = b = 75.1, c = 86.3 Å. The self-rotation function (data not shown) implies the presence of only one monomer per asymmetric unit, with a calculated Matthews coefficient V M of 4.44 Å3 Da−1 and a solvent content of 72.3% (Matthews, 1968 ▶). The collected diffraction data were 99.6% complete, with an overall 〈I/σ(I)〉 of 25.84 (Table 2 ▶). The BRPF1 bromodomain–H2AK5ac crystals (from Crystal Screen 2 condition No. 26) diffracted to a resolution of 1.8 Å (Fig. 2 ▶ b). This crystal belonged to the monoclinic space group P21, with unit-cell parameters a = 60.9, b = 55.6, c = 82.1 Å, β = 93.6°. The Matthews coefficient (V M = 2.53 Å3 Da−1) and the self-rotation function (data not shown) imply the presence of four monomers per asymmetric unit and a solvent content of 51.4% (Matthews, 1968 ▶). The collected diffraction data were 99.9% complete, with an overall 〈I/σ(I)〉 of 21.0 (Table 2 ▶). We are currently solving the X-ray crystal structures of the bromodomain in complex with the H2AK5ac and H4K12ac acetylated histone ligand(s) using molecular-replacement techniques with the solution structure of the bromodomain of peregrin (PDB entry 2d9e; RIKEN Structural Genomics/Proteomics Initiative, unpublished work) as a search model. The study proposed here will increase our understanding of the molecular mechanism utilized by structurally diverse bromodomains to recognize their histone ligands. Furthermore, structural and functional characterization of the BRPF1 bromodomain–histone interaction will be important for unraveling its role in modulating the genomic binding targets of the MOZ HAT and its potential as a therapeutic target in MOZ-related leukemias.
Acknowledgments
We thank Dr Brian E. Eckenroth for his help optimizing the crystal-growth and cryoprotection conditions, as well as for his assistance in screening crystals for diffraction at the Center for X-ray Crystallography at the University of Vermont. We are grateful to Dr Robert Sweet, Dr Rebecca Page, Dr William Grath and all of the staff members providing support during the 2013 RapiData course at the Brookhaven National Laboratory for their help with crystal data collection and processing. We are equally thankful to Dr X.-J. Yang at McGill University Department of Medicine for providing us with the BRPF1 cDNA. This study was supported by a National Institutes of Health grant, NIGMS 1R15GM104865, and an ACPHS internal research award to KCG. DNA sequencing was performed in the Vermont Cancer Center DNA Analysis Facility. Crystal growth, screening and initial data collection was carried out at the Center for X-ray Crystallography at the University of Vermont, which is supported by the National Institutes of Health grants P01CA098993 and R01CA52040 awarded by the National Cancer Center.
References
- Bergfors, T. (2003). J. Struct. Biol. 142, 66–76. [DOI] [PubMed]
- Borrow, J., Stanton, V. P. Jr, Andresen, J. M., Becher, R., Behm, F. G., Chaganti, R. S., Civin, C. I., Disteche, C., Dubé, I., Frischauf, A. M., Horsman, D., Mitelman, F., Volinia, S., Watmore, A. E. & Housman, D. E. (1996). Nature Genet. 14, 33–41. [DOI] [PubMed]
- Brown, T., Swansbury, J. & Taj, M. M. (2012). Leuk. Lymphoma, 53, 338–341. [DOI] [PubMed]
- Camós, M., Esteve, J., Jares, P., Colomer, D., Rozman, M., Villamor, N., Costa, D., Carrió, A., Nomdedéu, J., Montserrat, E. & Campo, E. (2006). Cancer Res. 66, 6947–6954. [DOI] [PubMed]
- Carapeti, M., Aguiar, R. C., Goldman, J. M. & Cross, N. C. (1998). Blood, 91, 3127–3133. [PubMed]
- Carlson, S. & Glass, K. C. (2014). J. Cell. Physiol. 229, 1571–1574 [DOI] [PMC free article] [PubMed]
- Champagne, N., Pelletier, N. & Yang, X.-J. (2001). Oncogene, 20, 404–409. [DOI] [PubMed]
- Champagne, K. S., Saksouk, N., Peña, P. V., Johnson, K., Ullah, M., Yang, X.-J., Côté, J. & Kutateladze, T. G. (2008). Proteins, 72, 1371–1376. [DOI] [PMC free article] [PubMed]
- Crowley, J. A., Wang, Y., Rapoport, A. P. & Ning, Y. (2005). Leukemia, 19, 2344–2345. [DOI] [PubMed]
- Delmore, J. E. et al. (2011). Cell, 146, 904–917. [DOI] [PMC free article] [PubMed]
- Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M.-M. (1999). Nature (London), 399, 491–496. [DOI] [PubMed]
- Esteyries, S., Perot, C., Adelaide, J., Imbert, M., Lagarde, A., Pautas, C., Olschwang, S., Birnbaum, D., Chaffanet, M. & Mozziconacci, M. J. (2008). Leukemia, 22, 663–665. [DOI] [PubMed]
- Garman, E. (1999). Acta Cryst. D55, 1641–1653. [DOI] [PubMed]
- Guetg, C., Lienemann, P., Sirri, V., Grummt, I., Hernandez-Verdun, D., Hottiger, M. O., Fussenegger, M. & Santoro, R. (2010). EMBO J. 29, 2135–2146. [DOI] [PMC free article] [PubMed]
- Iizuka, M., Matsui, T., Takisawa, H. & Smith, M. M. (2006). Mol. Cell. Biol. 26, 1098–1108. [DOI] [PMC free article] [PubMed]
- Katsumoto, T., Aikawa, Y., Iwama, A., Ueda, S., Ichikawa, H., Ochiya, T. & Kitabayashi, I. (2006). Genes Dev. 20, 1321–1330. [DOI] [PMC free article] [PubMed]
- Kitabayashi, I., Aikawa, Y., Nguyen, L. A., Yokoyama, A. & Ohki, M. (2001). EMBO J. 20, 7184–7196. [DOI] [PMC free article] [PubMed]
- Laue, K., Daujat, S., Crump, J. G., Plaster, N., Roehl, H. H., Kimmel, C. B., Schneider, R. & Hammerschmidt, M. (2008). Development, 135, 1935–1946.
- Matthews, A. G. et al. (2007). Nature (London), 450, 1106–1110.
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mujtaba, S., Zeng, L. & Zhou, M.-M. (2007). Oncogene, 26, 5521–5527. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Panagopoulos, I., Fioretos, T., Isaksson, M., Samuelsson, U., Billström, R., Strömbeck, B., Mitelman, F. & Johansson, B. (2001). Hum. Mol. Genet. 10, 395–404. [DOI] [PubMed]
- Peña, P. V., Hom, R. A., Hung, T., Lin, H., Kuo, A. J., Wong, R. P., Subach, O. M., Champagne, K. S., Zhao, R., Verkhusha, V. V., Li, G., Gozani, O. & Kutateladze, T. G. (2008). J. Mol. Biol. 380, 303–312. [DOI] [PMC free article] [PubMed]
- Perez-Campo, F. M., Borrow, J., Kouskoff, V. & Lacaud, G. (2009). Blood, 113, 4866–4874. [DOI] [PMC free article] [PubMed]
- Poplawski, A., Hu, K., Lee, W., Natesan, S., Peng, D., Carlson, S., Shi, X., Balaz, S., Markley, J. L. & Glass, K. C. (2014). J. Mol. Biol. 426, 1661–1676. [DOI] [PMC free article] [PubMed]
- Qin, S., Jin, L., Zhang, J., Liu, L., Ji, P., Wu, M., Wu, J. & Shi, Y. (2011). J. Biol. Chem. 286, 36944–36955. [DOI] [PMC free article] [PubMed]
- Qiu, Y., Liu, L., Zhao, C., Han, C., Li, F., Zhang, J., Wang, Y., Li, G., Mei, Y., Wu, M., Wu, J. & Shi, Y. (2012). Genes Dev. 26, 1376–1391. [DOI] [PMC free article] [PubMed]
- Sterner, D. E. & Berger, S. L. (2000). Microbiol. Mol. Biol. Rev. 64, 435–459. [DOI] [PMC free article] [PubMed]
- Ullah, M., Pelletier, N., Xiao, L., Zhao, S. P., Wang, K., Degerny, C., Tahmasebi, S., Cayrou, C., Doyon, Y., Goh, S.-L., Champagne, N., Côté, J. & Yang, X.-J. (2008). Mol. Cell. Biol. 28, 6828–6843. [DOI] [PMC free article] [PubMed]
- Vezzoli, A., Bonadies, N., Allen, M. D., Freund, S. M., Santiveri, C. M., Kvinlaug, B. T., Huntly, B. J., Göttgens, B. & Bycroft, M. (2010). Nature Struct. Mol. Biol. 17, 617–619. [DOI] [PubMed]