McbB, a multifunctional enzyme responsible for catalysing Pictet–Spengler cyclization, decarboxylation and oxidation reactions in the biosynthesis of β-carboline, was expressed and crystallized. The crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 66.06, b = 85.48, c = 106.19 Å, α = 90.00, β = 106.77, γ = 90.00°.
Keywords: β-carboline alkaloids, Pictet–Spengler cyclization, McbB, Marinactinospora thermotolerans
Abstract
β-Carboline alkaloids (βCs), with tricyclic pyrido[3,4-b]indole rings, have important pharmacological and therapeutic value. In the biosynthesis of βCs, the Pictet–Spengler (PS) cyclization reaction is responsible for the formation of ring structures. McbB is one of a few enzymes that are known to catalyse PS cyclization. It can also catalyse decarboxylation and oxidation. Here, the expression, crystallization and preliminary data analysis of McbB are reported. The crystals diffracted to 2.10 Å resolution and belonged to the monoclinic space group P21, with unit-cell parameters a = 66.06, b = 85.48, c = 106.19 Å, α = 90.00, β = 106.77, γ = 90.00°. These results provide a basis for solving the crystal structure and elucidating the catalytic mechanism for McbB.
1. Introduction
Indole alkaloids are a group of compounds that are widely distributed in plants, bacteria, fungi and marine microorganisms (Cao et al., 2007 ▶). They have attracted great attention owing to their important pharmacological and therapeutic value (Ma et al., 2004 ▶). One important subclass of indole alkaloids, β-carbolines, possess a tricyclic pyrido[3,4-b]indole ring system (Zhang & Larock, 2001 ▶). The characteristic tricyclic structure of β-carbolines endows them with wide-spectrum biological activities: some β-carbolines and derivatives have been discovered to be inhibitors of CDKs (cyclin-dependent kinases) and IkB kinase (Ritzeler et al., 2000 ▶; Song et al., 2002 ▶, 2004 ▶); some possess antiparasitic effects; and some (norharmane, harmane and harmine) possess anti-Toxoplasma gondii effects and exhibit no cytotoxic effects on the host cell line (Alomar et al., 2013 ▶). Similarly, β-carbolines isolated from Marinactinospora thermotolerans demonstrate antiplasmodial activity (Huang et al., 2011 ▶). Additionally, several β-carboline indole alkaloids from the genus Tabernaemontana have proved to have an effect on regulation of multidrug resistance in mouse lymphoma cell lines (Mansoor et al., 2009 ▶).
Recently, there has been great interest in the biosynthesis of alkaloids because of their diverse arrays of scaffolds and complicated biosynthetic pathways (Ma et al., 2004 ▶). Alkaloid biosynthetic pathways involve multiple steps, including Pictet–Spengler (PS) cyclization, oxidation and decarboxylation etc. Several enzymes are known to catalyse PS cyclization. One good example is strictosidine synthase (STR1), which catalyses the condensation of secologanin and tryptamine to form 3α-(S)-strictosidine (Stöckigt et al., 2011 ▶), which is the precursor of more than 2000 alkaloids (Maresh et al., 2008 ▶; Bernhardt et al., 2007 ▶; Ma et al., 2006 ▶). It can also catalyse the reaction inducing the formation of 12-aza-strictosidine, which is an important intermediate for cytotoxic alkaloids (Zou et al., 2010 ▶). In addition, an oxidase is often found in alkaloid biosynthetic pathways. Secologanin synthase, an NADPH-dependent P450 oxidase, is used to catalyse the oxidative cleavage of loganin to generate secologanin (Irmler et al., 2000 ▶). Similar to secologanin synthase, it has been discovered that cytochrome P450 monooxygenases are involved in the biosynthesis of secologanin, cornoside and shikonin (Inoue, 2005 ▶). Another reaction frequently detected in the biosynthesis of alkaloids is decarboxylation. Lys decarboxylase (LDC) was firstly discovered to synthesize the quinolizidine alkaloids via cyclization of the cadaverine unit (Bunsupa, Yamazaki et al., 2012 ▶). Several LDCs from other species have been isolated, cloned and expressed (Bunsupa, Katayama et al., 2012 ▶). Usually, different enzymes are involved in PS cyclization, oxidation and decarboxylation reactions. Multifunctional enzymes, which can catalyse PS cyclization, decarboxylation and oxidation reactions, have rarely been reported.
Recently, marinacarbolines A–D (MCB1–MCB4) and 1-acetyl-β-carboline have been isolated from the deep South China Sea derived M. thermotolerans SCSIO 00652 (Huang et al., 2011 ▶). Three genes, mcbABC, are responsible for MCB scaffold biosynthesis. Interestingly, McbB alone can mediate β-carboline core construction, which involves PS cyclization, decarboxylation and oxidation reactions (Chen et al., 2013 ▶). McbB exhibits very low identities to other PS cyclases, oxidases and decarboxylases. It exhibits 10% identity to strictosidine synthase (GenBank ACL06003.1) from Desulfatibacillum alkenivorans AK-01, 4% identity to lysine decarboxylase (YP_004502371.1) and 6% identity to secologanin synthase (ENH66901.1). Although McbB has low homology with these three groups of enzymes, it possesses the activities of all of them. The McbB sequence was aligned with the sequences of several PS cyclases, and some conserved residues were found in the alignment. However, none of these conserved residues are located in the active site. It was shown that Glu97 in McbB is vital for its activities (Chen et al., 2013 ▶). A conserved glutamic acid is also found in the sequence of strictosidine synthase STR1 (Ma et al., 2006 ▶). This implies that McbB may possess a similar catalytic mechanism to that of STR1 for PS cyclization. In this work, protein expression, crystallization and diffraction data collection of McbB are presented. These results will provide a foundation for solving the crystal structure and elucidating the catalytic mechanism.
2. Materials and methods
2.1. Macromolecule production
The gene of mcbB was cloned into the NdeI and HindIII sites of the pET-28a(+) vector (Chen et al., 2013 ▶). It was transformed into Escherichia coli BL21 (DE3) grown in LB broth containing 34 µg ml−1 kanamycin sulfate overnight at 310 K. A single colony was picked, inoculated into 5 ml LB medium overnight and cultured in 1 l LB medium. When the OD600 was around 0.6, the cells were induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 289 K. The cells were harvested by centrifugation (Thermo Scientific ST 16R) at 5000g for 20 min at 277 K and then suspended in 30 ml lysis buffer consisting of 20 mM Tris, 0.5 M KCl pH 8.3. The cells were cooled by immersion in liquid nitrogen and stored at 193 K until further use.
To purify recombinant McbB, the cells were lysed by crushing via a high-pressure homogenizer (Constant Systems TS) at 277 K and the cell lysate was removed by centrifugation at 12 000g for 1 h at 277 K. The recombinant McbB was purified by chromatography with 5 ml Ni–NTA resin (GE Healthcare). It was washed with 15 ml lysis buffer (20 mM Tris, 500 mM KCl pH 8.3), 25 ml wash buffer 1 (30 mM imidazole, 20 mM Tris, 500 mM KCl pH 8.3) and 15 ml wash buffer 2 (60 mM imidazole, 20 mM Tris, 500 mM KCl pH 8.3) and finally eluted with 15 ml elution buffer (200 mM imidazole, 20 mM Tris, 0.5 M KCl pH 8.3). The purified protein was concentrated to 2 ml by ultrafiltration using an Amicon Ultra concentrator (Millipore) with a 10 kDa cutoff filter at 277 K. The protein was then further purified by size-exclusion chromatography on a Superdex 75 column (16/60, GE Healthcare) equilibrated with 500 mM KCl, 20 mM Tris pH 8.3. The peak corresponding to McbB was collected, concentrated, flash-cooled in liquid nitrogen and stored at 193 K. Macromolecule-production information is summarized in Table 1 ▶.
Table 1. Macromolecule-production information.
| Source organism | M. thermotolerans SCSIO 00652 |
| DNA source | SCSIO 00652 marinacarbolines gene cluster, complete sequence (GenBank KC541560.1) |
| Forward primer | CATATGAGGCAGATCGAAATCGAA |
| Reverse primer | AAGCTTTCATGCGTCCAGATAGCGGTA |
| Cloning vector | pET-28a(+) |
| Expression vector | pET-28a(+) |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHSSGLVPRGSHVRQIEIEWVQPGITVTADLSWERNPELAELLWTGLLPYNSLQNHALVSGNHLYHLIADPRLVYTEARYKEDRTKSPDGTVFLSQLQHLAVKYGPLTEYLPAAPVGSVVPEDIDALREAGRACWKAAWETKQPIEVRVRRKGEAVTDFALPRTPPVDHPGVQKLVEEIQDETERVWITPPAEIVDMHQGRIASRAGSYDQYFSTLVFLNGEVRPLGYCALNGLLKICRTTDLTLNDLKRITPTFIKTPAEFLGYTGLDTLWRFTQQVLTLLPDVETREQYFALVNALALYANMLNTWNLHFFPWQHGTDYRY |
2.2. Crystallization
The McbB protein was concentrated to 20 mg ml−1. The crystallization trials were performed in 48-well sitting-drop plates (Hampton Research) at 289 K. The drop in each well was 2 µl in size, consisting of 1 µl reservoir solution plus 1 µl protein solution, and was equilibrated against 150 µl reservoir solution. The initial conditions were screened with the commercially available kits Crystal Screen, Crystal Screen 2, SaltRx and Index (Hampton Research), Wizard I, II and III (Emerald Bio). Crystallization information is summarized in Table 2 ▶.
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | 48-well sitting-drop plate |
| Temperature (K) | 289 |
| Protein concentration (mgml1) | 20 |
| Buffer composition of protein solution | 500mM KCl, 20mM Tris pH 8.3 |
| Composition of reservoir solution | 0.2M sodium formate, 20%(w/v) polyethylene glycol 3350 |
| Volume and ratio of drop | 1l protein solution:1l precipitant solution |
| Volume of reservoir (l) | 150 |
2.3. Data collection and processing
The crystals were soaked in a cryoprotectant consisting of the mother liquor plus 20% ethylene glycol (Sigma) before flash-cooling in liquid nitrogen. The data were collected using an ADSC Q315 CCD detector on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF) at a wavelength of 0.97915 Å. 180 images were collected with 1° oscillation per image. The data were processed using iMosflm v.7.0.9 and SCALA (Winn et al., 2011 ▶; Battye et al., 2011 ▶).
3. Results
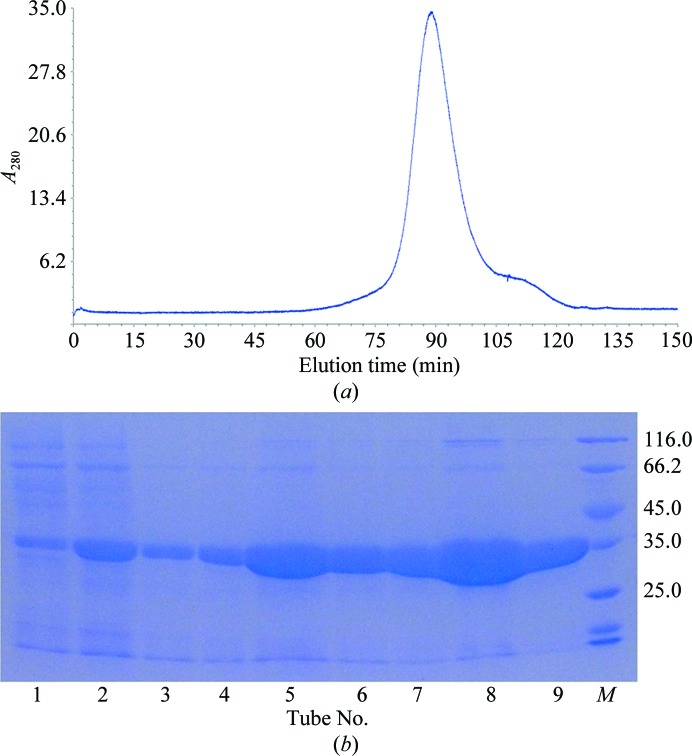
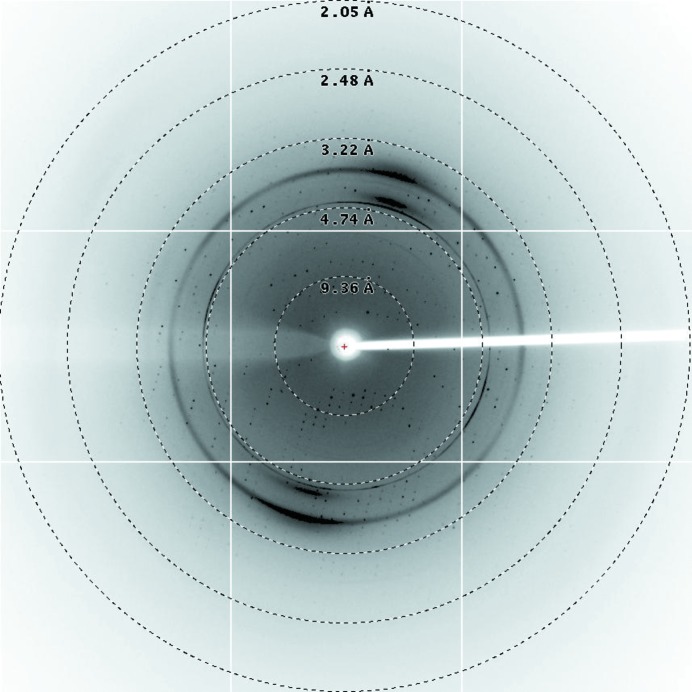
After purification by Ni–NTA resin and size-exclusion chromatography (Fig. 1 ▶ a), the purity of McbB was confirmed by SDS–PAGE (∼35 kDa; Fig. 1 ▶ b). It was then concentrated to 20 mg ml−1 for crystallization. Small crystals of McbB appeared after two weeks’ incubation in condition No. 90 of the Index crystallization kit [0.2 M sodium formate, 20%(w/v) polyethylene glycol 3350]. The crystals in the initial screens were too small to yield good diffraction. Thus, optimization of the crystallization conditions was carried out by altering the concentrations of the precipitant (polyethylene glycol) and the salt. After refinement of the crystallization condition, high-quality crystals (∼0.5 × 0.3 mm in size; Fig. 2 ▶) were obtained with good diffraction (Fig. 3 ▶). The optimized condition consisted of 0.1 M sodium formate, 30%(w/v) polyethylene glycol 3350. Both the sodium formate and the polyethylene glycol concentrations were changed in the optimization process in order to obtain good diffraction.
Figure 1.
Purification results of McbB by size-exclusion chromatography. (a) Size-exclusion chromatography on a Superdex 75 column. The peak fractions contained McbB with an estimated molecular weight of 35 kDa. (b) The eluted fractions from size-exclusion chromatography confirmed by SDS–PAGE. Lane M, unstained protein molecular-weight marker labelled in kDa (Thermo Scientific).
Figure 2.
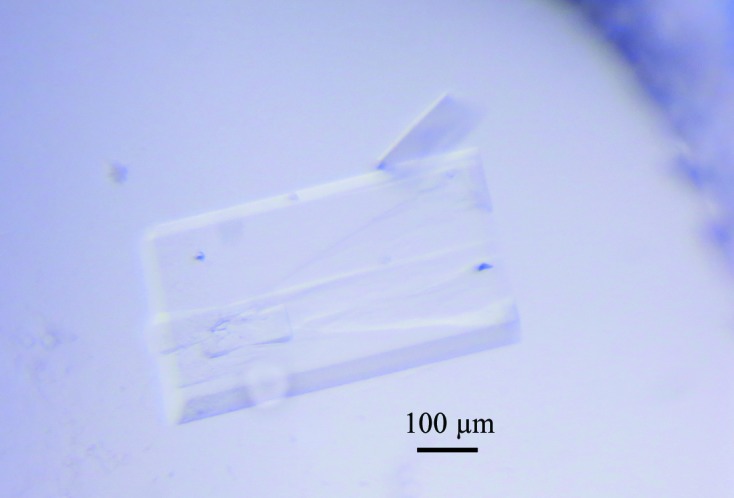
The crystal of McbB after crystallization-condition optimization. Crystals were obtained using the sitting-drop method. Initially, small crystals were found in condition No. 90 of the Index kit after two weeks’ incubation. Large rectangular crystals were obtained after refinement.
Figure 3.
A representative X-ray diffraction image. The diffraction images from an McbB crystal were collected using an ADSC Q315 CCD detector on beamline BL17U at the SSRF.
The best McbB crystal diffracted to 2.10 Å resolution. It belonged to the monoclinic space group P21, with unit-cell parameters a = 66.06, b = 85.48, c = 106.19 Å, α = 90.00, β = 106.77, γ = 90.00°. Cell-content analysis suggests that there are three molecules in the asymmetric unit. The solvent content of the crystal is 54.85% and the Matthews coefficient is 2.68 Å3 Da−1 (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶). The data-collection statistics are summarized in Table 3 ▶. Anomalous data collection is in progress.,
Table 3. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| Diffraction source | SSRF |
| Wavelength () | 0.97915 |
| Temperature (K) | 100 |
| Detector | ADSC Q315 CCD |
| Crystal-to-detector distance (mm) | 300.00 |
| Rotation range per image () | 1 |
| Total rotation range () | 55235 |
| Exposure time per image (s) | 0.8 |
| Space group | P21 |
| a, b, c () | 66.06, 85.48, 106.19 |
| , , () | 90.00, 106.77, 90.00 |
| Mosaicity () | 0.93 |
| Resolution range () | 41.762.10 (2.152.10) |
| Total No. of reflections | 151728 (22274) |
| No. of unique reflections | 63444 (9344) |
| Completeness (%) | 96.1 (97.3) |
| Multiplicity | 2.4 (2.4) |
| I/(I) | 7.3 (2.5) |
| R r.i.m. | 0.113 (0.561) |
| Overall B factor from Wilson plot (2) | 24.01 |
Acknowledgments
This research was financially supported by the National Key Basic Research Program of China (No. 2013CB933900) and the National Natural Science Foundation of China (No. 31000326). We thank the staff of beamline BL17U, SSRF for their support.
References
- Alomar, M. L., Rasse-Suriani, F. A. O., Ganuza, A., Cóceres, V. M., Cabrerizo, F. M. & Angel, S. O. (2013). BMC Res. Notes, 6, 193. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bernhardt, P., McCoy, E. & O’Connor, S. E. (2007). Chem. Biol. 14, 888–897. [DOI] [PMC free article] [PubMed]
- Bunsupa, S., Katayama, K., Ikeura, E., Oikawa, A., Toyooka, K., Saito, K. & Yamazaki, M. (2012). Plant Cell, 24, 1202–1216. [DOI] [PMC free article] [PubMed]
- Bunsupa, S., Yamazaki, M. & Saito, K. (2012). Front. Plant Sci. 3, 239. [DOI] [PMC free article] [PubMed]
- Cao, R., Peng, W., Wang, Z. & Xu, A. (2007). Curr. Med. Chem. 14, 479–500. [DOI] [PubMed]
- Chen, Q., Ji, C., Song, Y., Huang, H., Ma, J., Tian, X. & Ju, J. (2013). Angew. Chem. Int. Ed. Engl. 52, 9980–9984. [DOI] [PubMed]
- Huang, H., Yao, Y., He, Z., Yang, T., Ma, J., Tian, X., Li, Y., Huang, C., Chen, X., Li, W., Zhang, S., Zhang, C. & Ju, J. (2011). J. Nat. Prod. 74, 2122–2127. [DOI] [PubMed]
- Inoue, K. (2005). Yakugaku Zasshi, 125, 31–49. [DOI] [PubMed]
- Irmler, S., Schröder, G., St-Pierre, B., Crouch, N. P., Hotze, M., Schmidt, J., Strack, D., Matern, U. & Schröder, J. (2000). Plant J. 24, 797–804. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Ma, X., Koepke, J., Fritzsch, G., Diem, R., Kutchan, T. M., Michel, H. & Stöckigt, J. (2004). Biochim. Biophys. Acta, 1702, 121–124. [DOI] [PubMed]
- Ma, X., Panjikar, S., Koepke, J., Loris, E. & Stöckigt, J. (2006). Plant Cell, 18, 907–920. [DOI] [PMC free article] [PubMed]
- Mansoor, T. A., Ramalhete, C., Molnár, J., Mulhovo, S. & Ferreira, M. J. (2009). J. Nat. Prod. 72, 1147–1150. [DOI] [PubMed]
- Maresh, J. J., Giddings, L. A., Friedrich, A., Loris, E. A., Panjikar, S., Trout, B. L., Stöckigt, J., Peters, B. & O’Connor, S. E. (2008). J. Am. Chem. Soc. 130, 710–723. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Ritzeler, O., Castro, A., Grenier, L., Soucy, F., Hancock, W., Mazdiyasni, H., Palombella, V. & Adams, J. (2000). US Patent 20020099068 A1.
- Song, Y., Kesuma, D., Wang, J., Deng, Y., Duan, J., Wang, J. H. & Qi, R. Z. (2004). Biochem. Biophys. Res. Commun. 317, 128–132. [DOI] [PubMed]
- Song, Y., Wang, J., Teng, S. F., Kesuma, D., Deng, Y., Duan, J., Wang, J. H., Qi, R. Z. & Sim, M. M. (2002). Bioorg. Med. Chem. Lett. 12, 1129–1132. [DOI] [PubMed]
- Stöckigt, J., Antonchick, A. P., Wu, F. & Waldmann, H. (2011). Angew. Chem. Int. Ed. Engl. 50, 8538–8564. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, H. & Larock, R. C. (2001). Org. Lett. 3, 3083–3086. [DOI] [PubMed]
- Zou, H.-B., Zhu, H.-J., Zhang, L., Yang, L.-Q., Yu, Y.-P. & Stöckigt, J. (2010). Chem. Asian J. 5, 2400–2404. [DOI] [PubMed]