The SMase D from L. gaucho venom was expressed, purified, crystallized and diffraction data were collected to 2.6 Å resolution.
Keywords: sphingomyelinase D, Loxosceles gaucho
Abstract
Brown spider envenomation results in dermonecrosis, intravascular coagulation, haemolysis and renal failure, mainly owing to the action of sphingomyelinases D (SMases D), which catalyze the hydrolysis of sphingomyelin to produce ceramide 1-phosphate and choline or the hydrolysis of lysophosphatidylcholine to produce lysophosphatidic acid. Here, the heterologous expression, purification, crystallization and preliminary X-ray diffraction analysis of LgRec1, a novel SMase D from Loxosceles gaucho venom, are reported. The crystals belonged to space group P21212, with unit-cell parameters a = 52.98, b = 62.27, c = 84.84 Å and diffracted to a maximum resolution of 2.6 Å.
1. Introduction
Sphingomyelinase D activity has only been observed in the pathogenic bacteria Corynebacterium pseudotuberculosis, C. ulcernas, Archanobacterium haemolyticum and Vibrio damsel (McNamara et al., 1995 ▶) and in spider venoms of the genus Loxosceles. Except for the enzyme from V. damsel, the bacterial and spider venom SMases (sphingomyelinases) possess similar molecular masses (30–35 kDa) and display significant sequence homology in the N-terminal regions (Tambourgi et al., 1998 ▶; van Meeteren et al., 2004 ▶). Interestingly, infection by C. pseudotuberculosis or envenomation by Loxosceles results in similar pathologies (Binford et al., 2005 ▶), suggesting a possible evolutionary relationship (Bernheimer et al., 1985 ▶).
Spiders from the genus Loxosceles are widely distributed (Platnick, 2009 ▶) and envenomation results in severe dermonecrotic lesions. Sphingomyelinases D (SMases D; EC 3.1.4.41), also referred to as phospholipases D (PLDs), which are the principal causative agents of lesions, are the major components of the venoms of many spiders belonging to the genus Loxosceles (Gomes et al., 2011 ▶; Gremski et al., 2014 ▶). These enzymes catalyze the hydrolysis of sphingomyelin, which results in the concomitant production of ceramide 1-phosphate and choline or the hydrolysis of lysophosphatidylcholine, generating the lipid mediator lysophosphatidic acid (van Meeteren et al., 2004 ▶). Recently, the conversion of lysophosphatidylcholine and sphingomyelin to cyclic phosphates has been reported (Lajoie et al., 2013 ▶). The physiological symptoms of envenomation include dermonecrosis, intravascular coagulation, haemolysis and renal failure (Tambourgi et al., 1998 ▶; van Meeteren et al., 2004 ▶).
Based on their structural properties, spider venom SMases D have been divided into two classes (Murakami et al., 2006 ▶): class I enzymes possess a single disulfide bridge (Cys53–Cys57), whereas the class II members possess an additional disulfide bridge that stabilizes the flexible loop which partially occludes the catalytic site (Murakami et al., 2005 ▶; de Giuseppe et al., 2011 ▶; Ullah et al., 2011 ▶).
We have previously determined the structures of enzymes belonging to both classes I (Murakami et al., 2005 ▶, 2006 ▶) and II (de Giuseppe et al., 2011 ▶; Ullah et al., 2011 ▶) and the present structural study of SMase D from L. gaucho (LgRec1) was carried out to correlate structural differences with the observed variation in toxicity and enzymatic specificity.
2. Materials and methods
2.1. Macromolecule production
Mature LgRec1, cloned into the pAE vector (Ramos et al., 2004 ▶), was expressed in Escherichia coli BL21 Star (DE3) pLysS (Invitrogen) cells as described previously (Magalhães et al., 2013 ▶). Briefly, transformed bacteria were grown at 37°C in LB broth containing 100 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol until an optical density of 0.6 was attained at 600 nm. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM and expression was carried out for 4.5 h. Cells were harvested (5000g, 10 min, 4°C) and suspended in phosphate buffer (50 mM sodium phosphate pH 7.0, 300 mM NaCl, 10 mM imidazole) and intermittently sonicated on ice for 60 s, interspersed by intervals of 2 min for six cycles. Lysed cells were then centrifuged (7000g, 10 min, 4°C) and the recombinant protein was purified from the supernatant by immobilized metal-affinity chromatography (IMAC) using Ni–Sepharose resin (GE Healthcare). After purification, the recombinant protein was dialyzed against phosphate-buffered saline (PBS) and analyzed on a 12% SDS–PAGE gel under reducing conditions (Fig. 1 ▶). Macromolecule production is summarized in Table 1 ▶.
Figure 1.
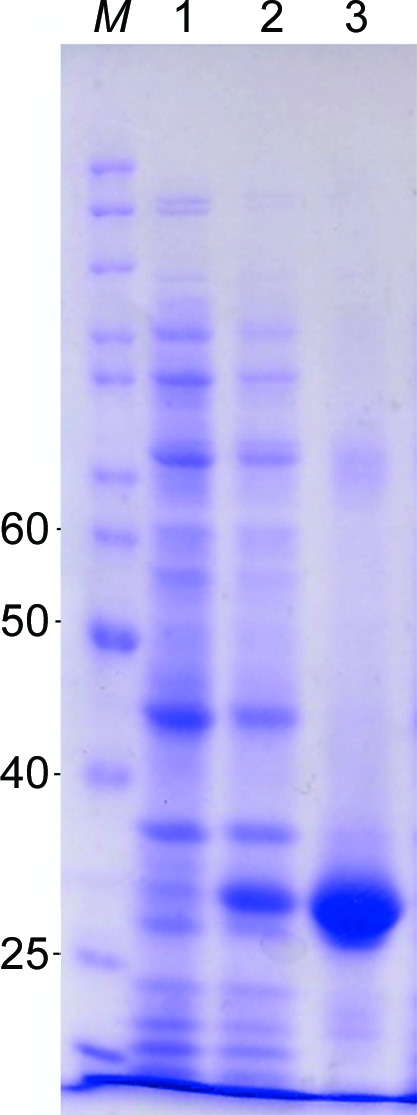
SDS–PAGE analysis of recombinant LgRec1 expression. Lane M, protein molecular-weight markers (Page Ruler, Thermo Scientific); lanes 1 and 2, extract from BL21 Star (DE3) pLysS without and with IPTG induction, respectively; lane 3, purified LgRec1 from Ni Sepharose (GE Healthcare). The numbers on the left correspond to the molecular masses in kDa of molecular-weight marker proteins in lane M.
Table 1. Macromolecule-production information.
| Source organism | L. gaucho |
| DNA source | L. gaucho venom gland |
| Forward primer† | GGTA GGATCCGCAGATAACCGGCGGCCTAT |
| Reverse primer‡ | ATTA AAGCTTTCCCTTGAACGTTTCCCAA |
| Cloning vector | pGEM-T Easy Vector Systems (Promega) |
| Expression vector | pAE |
| Expression host | E. coli BL21 Star(DE3)pLysS (Invitrogen) |
| Complete amino-acid sequence of the construct produced§ | MHHHHHH LEGSADNRRPIWVMGHMVNSLAQIDEFVGLGSNSIETDVSFDKQANPEYTYHGIPCDCGRACLHSTKFNDFLKGLRKVTTPGDSKYLEKLILVVFDLKTGSLYDNQAYDAGTKLAKNLLQHYWNNGNNGGRAYIILSIPNLNHYKLITGFKETLKNEGHEELLEKVGTDFSGNDDISDVQKTYNKAGVTGHVWQSDGITNCLLRGLTRVKAAVANRDSGSGIINKVYYWTVDKRQSTRDTLDANVDGIMTNYPDITVEILNEAAYKKKFRIATYEDNPWETFKG |
The BamHI restriction site is underlined and the four base pairs added on the side of the restriction site to improve cleavage performance are shown in italics.
The HindIII restriction site is underlined and the four base pairs added on the side of the restriction site to improve cleavage performance are shown in italics.
The histidine tag is shown in italics and the amino acids added owing to the sequence of the restriction enzymes present in the vector are underlined.
2.2. Crystallization
The LgRec1 sample was concentrated to 15 mg ml−1 and crystallization was carried out in 24-well tissue-culture plates by the hanging-drop vapour-diffusion method (Jancarik & Kim, 1991 ▶) using several commercial crystallization kits including Crystal Screen, Crystal Screen 2, Grid Screen PEG 6000, Grid Screen Ammonium Sulfate, PEG/Ion (Hampton Research) and The PEGs Suite (Qiagen). Typically, 1 µl drops of the LgRec1 solution were mixed with an equal volume of screening solution and equilibrated over a reservoir consisting of 0.5 ml of the latter solution. Small crystals were obtained from 0.1 M Tris–HCl pH 8.5, 20% polyethylene glycol 10 000. This condition was optimized by varying the pH of the buffer and concentration of polyethylene glycol 10 000; crystals suitable for X-ray diffraction experiments were obtained when 2 µl protein solution was mixed with an equal amount of reservoir solution consisting of 0.1 M Tris–HCl pH 8.2, 22% polyethylene glycol 10 000 (Fig. 2 ▶). Crystallization is summarized in Table 2 ▶.
Figure 2.
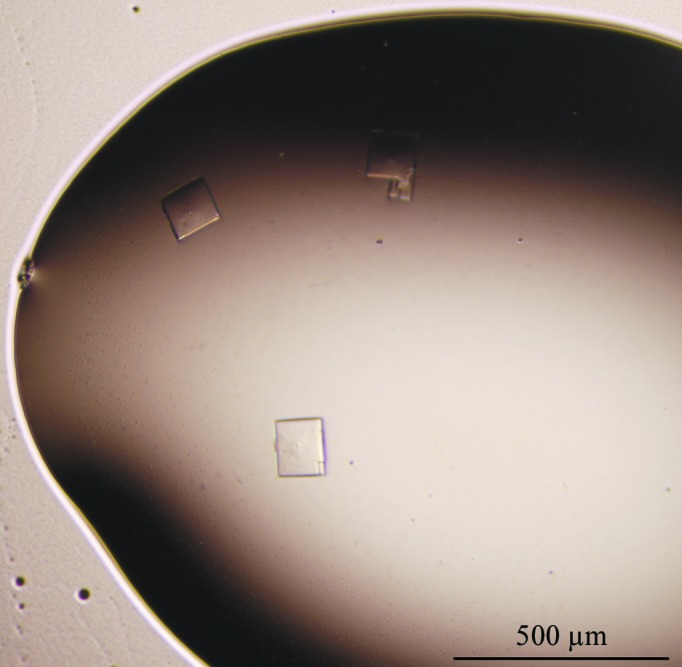
Photomicrograph of LgRec1 crystals.
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | Linbro 24-well tissue-culture plate |
| Temperature (C) | 18 |
| Protein concentration (mgml1) | 15 |
| Buffer composition of protein solution | Phosphate-buffered saline pH 7.0 |
| Composition of reservoir solution | 0.1M TrisHCl pH 8.2, 22% polyethylene glycol 10000 |
| Volume ratio of drop | 2:2 |
| Volume of reservoir (ml) | 0.5 |
2.3. Data collection, processing and structure determination
For X-ray diffraction data collection, an LgRec1 crystal was directly flash-cooled in a −173°C nitrogen-gas stream at the W01B-MX2 beamline at the Brazilian Synchrotron Light Laboratory (LNLS-Campinas, Brazil). The wavelength of the radiation source was set to 1.458 Å and a MAR CCD 165 mm detector was used to record the X-ray diffraction intensities. The LgRec1 crystal was exposed for 30 s per degree of rotation around ϕ and a total of 180 images were collected. The data were indexed, integrated and scaled using the DENZO and SCALEPACK programs from the HKL-2000 package (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are summarized in Table 3 ▶.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | W01B-MX2, LNLS |
| Wavelength () | 1.458 |
| Temperature (C) | 173 |
| Detector | MAR CCD 165mm |
| Crystal-to-detector distance (mm) | 90 |
| Rotation range per image () | 1 |
| Total rotation range () | 180 |
| Exposure time per image (s) | 30 |
| Space group | P21212 |
| a, b, c () | 52.98, 62.27, 84.84 |
| , , () | 90 |
| Mosaicity () | 1.65 |
| Resolution range () | 2.60 (2.72) |
| Total No. of reflections | 52611 (6720) |
| No. of unique reflections | 3919 (1193) |
| Completeness (%) | 97.8 (91.5) |
| Multiplicity | 3.3 (3.3) |
| I/(I) | 12.7 (2.0) |
| R r.i.m. | 4.0 (35.6) |
| Overall B factor from Wilson plot (2) | 64.4 |
3. Results and discussion
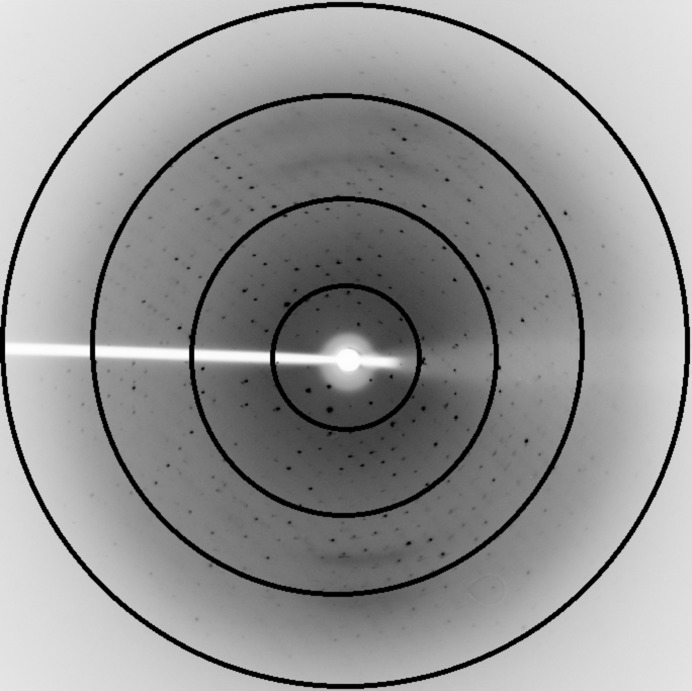
A homogeneous sample was obtained following a single affinity-purification step to yield approximately 10 mg active enzyme. The protein was monodisperse at a concentration of 15 mg ml−1 and crystallized in the presence of polyethylene glycol 10 000 at a slightly basic pH. Reduction of the pH to 8.2 was required to obtain large single crystals. The crystals of LgRec1 diffracted to a maximum resolution of 2.6 Å (Fig. 3 ▶) and were indexed in the orthorhombic system. Analysis of the systematic absences indicated that the most likely space group is P21212 (Table 3 ▶). The calculated Matthews coefficient of 2.0 Å3 Da−1 corresponds to a solvent content of 38.5% (Matthews, 1968 ▶); taking into consideration the molecular weight of the enzyme (30 000 Da), one molecule is present in the asymmetric unit. The structure of homologous SMases D from Loxosceles spp. available in the Protein Data Bank will be employed for molecular-replacement calculations. These structural results, along with mutagenesis and biochemical studies, are being carried out to shed light on the specific structural features related to functional differentiation among class I and II spider venom SMases D.
Figure 3.
X-ray diffraction pattern of an LgRec1 crystal. The concentric circles indicate resolutions of 7.5, 5.3, 3.2 and 2.6 Å, respectively.
Acknowledgments
This work was supported by grants from CAPES-Toxinologia, CAPES/DAAD, CNPq and FAPESP. AU is a FAPESP postdoctoral fellow.
References
- Bernheimer, A. W., Campbell, B. J. & Forrester, L. J. (1985). Science, 228, 590–591. [DOI] [PubMed]
- Binford, G. J., Cordes, M. H. & Wells, M. A. (2005). Toxicon, 45, 547–560. [DOI] [PubMed]
- Giuseppe, P. O. de, Ullah, A., Silva, D. T., Gremski, L. H., Wille, A. C. M., Chaves Moreira, D., Ribeiro, A. S., Chaim, O. M., Murakami, M. T., Veiga, S. S. & Arni, R. K. (2011). Biochem. Biophys. Res. Commun. 409, 622–627. [DOI] [PubMed]
- Gomes, M. T., Guimarães, G., Frézard, F., Kalapothakis, E., Minozzo, J. C., Chaim, O. M., Veiga, S. S., Oliveira, S. C. & Chávez-Olórtegui, C. (2011). Toxicon, 57, 574–579. [DOI] [PubMed]
- Gremski, L. H. et al. (2014). Toxicon, 83, 91–120. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Lajoie, D. M., Zobel-Thropp, P. A., Kumirov, V. K., Bandarian, V., Binford, G. J. & Cordes, M. H. (2013). PLoS One, 8, e72372. [DOI] [PMC free article] [PubMed]
- Magalhães, G. S., Caporrino, M. C., Della-Casa, M. S., Kimura, L. F., Prezotto-Neto, J. P., Fukuda, D. A., Portes-Junior, J. A., Neves-Ferreira, A. G., Santoro, M. L. & Barbaro, K. C. (2013). Biochimie, 95, 1773–1783. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McNamara, P. J., Cuevas, W. A. & Songer, J. G. (1995). Gene, 156, 113–118. [DOI] [PubMed]
- Meeteren, L. A. van, Frederiks, F., Giepmans, B. N., Pedrosa, M. F., Billington, S. J., Jost, B. H., Tambourgi, D. V. & Moolenaar, W. H. (2004). J. Biol. Chem. 279, 10833–10836. [DOI] [PubMed]
- Murakami, M. T., Fernandes-Pedrosa, M. F., de Andrade, S. A., Gabdoulkhakov, A., Betzel, C., Tambourgi, D. V. & Arni, R. K. (2006). Biochem. Biophys. Res. Commun. 342, 323–329. [DOI] [PubMed]
- Murakami, M. T., Fernandes-Pedrosa, M. F., Tambourgi, D. V. & Arni, R. K. (2005). J. Biol. Chem. 280, 13658–13664. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Platnick, N. I. (2009). The World Spider Catalog, v.10.0. New York: American Museum of Natural History. http://research.amnh.org/entomology/spiders/catalog/index.html.
- Ramos, C. R., Abreu, P. A., Nascimento, A. L. & Ho, P. L. (2004). Braz. J. Med. Biol. Res. 37, 1103–1109. [DOI] [PubMed]
- Tambourgi, D. V., Magnoli, F. C., van den Berg, C. W., Morgan, B. P., de Araujo, P. S., Alves, E. W. & Da Silva, W. D. (1998). Biochem. Biophys. Res. Commun. 251, 366–373. [DOI] [PubMed]
- Ullah, A., de Giuseppe, P. O., Murakami, M. T., Trevisan-Silva, D., Wille, A. C. M., Chaves-Moreira, D., Gremski, L. H., da Silveira, R. B., Sennf-Ribeiro, A., Chaim, O. M., Veiga, S. S. & Arni, R. K. (2011). Acta Cryst. F67, 234–236. [DOI] [PMC free article] [PubMed]