The C-terminal domain of C. trachomatis CdsD was purified and crystallized. The preliminary X-ray diffraction studies are presented.
Keywords: type III secretion system, Chlamydia, CdsD
Abstract
The inner membrane ring of the bacterial type III secretion system (TTSS) is composed of two proteins. In Chlamydia trachomatis this ring is formed by CdsD (gene name CT_664) and CdsJ (gene name CTA_0609). CdsD consists of 829 amino acids. The last 400 amino acids at its C-terminal end relate it to the type III secretion system YscD/HrpQ protein family. The C-terminal domain, consisting of amino acids 558–771, of C. trachomatis CdsD was overexpressed in Escherichia coli and purified using immobilized metal-affinity chromatography (IMAC) and size-exclusion chromatography. The protein was crystallized using the vapour-diffusion method. A data set was collected to 2.26 Å resolution. The crystals have the symmetry of space group C2, with unit-cell parameters a = 106.60, b = 23.91, c = 118.65 Å, β = 104.95°. According to the data analysis there is expected to be one molecule in the asymmetric unit, with a Matthews coefficient of 3.0 Å3 Da−1.
1. Introduction
Chlamydia trachomatis is a Gram-negative obligate intracellular bacterium and is the most common sexually transmitted pathogen. C. trachomatis infection can cause, for example, blindness and urogenital tract infections. Prolonged uterine infections can lead to ectopic pregnancies and infertility (reviewed by Bébéar & de Barbeyrac, 2009 ▶). Like many other pathogenic Gram-negative bacteria, C. trachomatis also utilizes the type III secretion system (Peters et al., 2007 ▶) to deliver its virulence proteins directly into the host cells.
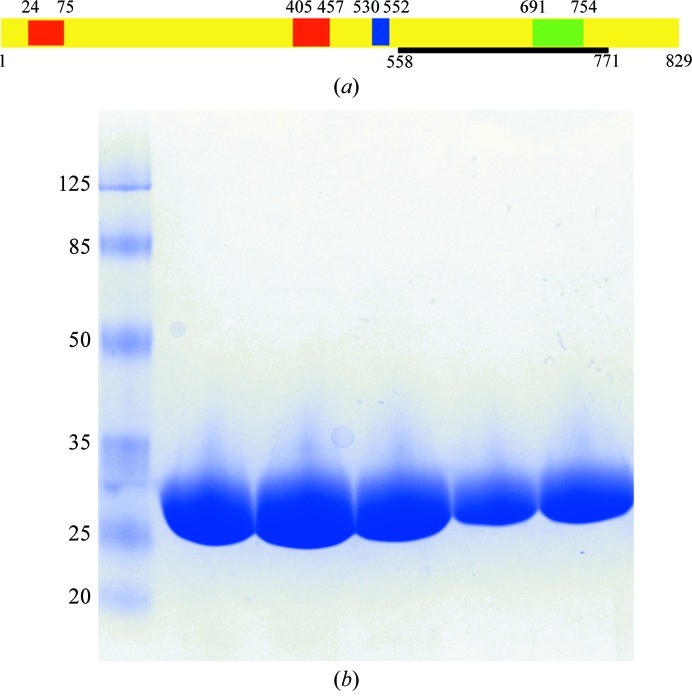
C. trachomatis CdsD is a structural protein of the inner membrane ring of the type III secretion system (TTSS). It is an 89 kDa protein with one predicted transmembrane domain corresponding to amino acids 530–552. It also has two forkhead-associated domains (FHA domains; residues 24–75 and 405–457) and a C-terminal BON domain (residues 691–754) (Fig. 1 ▶). CdsD forms higher oligomers which are mediated by disulfide bridges and the appearance of these complexes is strongly linked to the development cycle of the bacterium (Betts-Hampikian & Fields, 2011 ▶). CdsD is a putative substrate for the C. trachomatis protein kinase PknD (Johnson & Mahony, 2007 ▶).
Figure 1.
The CdsD(558–771) construct. (a) A schematic presentation of the domain structure of full-length CdsD. The two FHA domains are coloured red, the predicted transmembrane domain is shown in blue and the BON domain is shown in green. The black bar visualizes the domain that was crystallized. Residue numbers are presented at the beginning and the end of the corresponding sequences. (b) A Coomassie Blue-stained 4–20% Mini-Protean TGX Gel (Bio-Rad) of the purified CdsD(558–771) (24 kDa). The protein moves slightly more slowly than expected from its molecular weight compared with the molecular-weight standard (labelled in kDa).
The structure of one of the FHA domains of CdsD (residues 380–485) has previously been solved (PDB entry 3gqs; Midwest Center for Structural Genomics, unpublished work). In this study, we have crystallized the C-terminal fragment consisting of amino acids 558–771. This fragment shares comparatively low sequence identity with the previously described structures of the C-terminal domains of YscD of Yersinia enterocolitica (PDB entry 4alz) and PrgH of Salmonella enterica (PDB entry 4g1i) (Kudryashev et al., 2013 ▶; Spreter et al., 2009 ▶), with 18 and 8% sequence identity, respectively. The fragment that we have crystallized includes the BON domain (residues 691–754; Fig. 1 ▶), which is a putative membrane-binding domain (Yeats & Bateman, 2003 ▶). The location of the crystallized fragment in the inner membrane ring is expected to be in the periplasmic space.
2. Materials and methods
2.1. Macromolecule production
The synthetic codon-optimized gene encoding amino acids 558–771 of CdsD of C. trachomatis, cloned into the expression vector pET-28a (Novagen), was obtained from GenScript (Table 1 ▶). The proper length of the construct with minimal degradation was defined experimentally, initially using a longer construct which was found to be proteolytically degraded into a smaller fragment that was characterized by mass spectrometry (data not shown). For protein expression the plasmid was transformed into E. coli strain C41(DE3). An initial culture of 20 ml LB broth supplemented with 60 µg ml−1 kanamycin and 1%(w/v) glucose was started up with inoculation of one colony. This culture was grown overnight at 37°C for 16 h with shaking at 200 rev min−1. The next morning this culture was transferred into 4 l M9ZB medium (Studier et al., 1990 ▶) supplemented with 60 µg ml−1 kanamycin. Cells were grown at 37°C and 200 rev min−1 until the OD600 reached 0.6 and were subsequently induced with 400 µM isopropyl β-d-1-thiogalactopyranoside for 3 h at 37°C and 200 rev min−1. The cells were collected and suspended in 200 ml lysis buffer [50 mM Tris–HCl pH 8.0, 500 mM NaCl, 5%(v/v) glycerol] and frozen at −20°C.
Table 1. Production information for CdsD(558771).
| Source organism | C. trachomatis |
| DNA source | Synthetic gene (GenScript) |
| Expression vector | pET-28a (Novagen) |
| Cloning site | NdeIBamHI |
| Additional residues | N-terminal His tag, thrombin cleavage site |
| Expression host | E. coli C41(DE3) |
| UniProtKB entry | O84671 |
| Residues included in the construct | 558771 |
The cell suspension was thawed at room temperature and lysed by sonication. Cell remnants were removed by centrifugation and the protein was purified using a 2 ml Ni–NTA (Qiagen) column. The N-terminal His tag was removed using the Thrombin CleanCleave kit (Sigma–Aldrich). Purification was finalized by size-exclusion chromatography using a HiLoad 16/600 Superdex 75 PG column (GE Healthcare) using 10 mM Tris–HCl pH 8.0, 250 mM NaCl, 1 mM DTT as a buffer. The sample quality was assessed by SDS–PAGE analysis (Fig. 1 ▶).
2.2. Crystallization
The protein buffer was changed to 10 mM Tris–HCl pH 7.5 prior to crystallization and the protein was concentrated to 4.2 mg ml−1 as determined by measuring the absorbance at 280 nm. Initial crystallization conditions were screened using the ProPlex screen (Molecular Dimensions). Crystallizations were performed in 96-well plates using the sitting-drop vapour-diffusion technique. As the screen suggested many possible good crystallization conditions, several rounds of optimization were performed. The optimizations were performed by mixing 500 nl protein solution with 250 nl well solution and incubation at 295 K over 80 µl well solution consisting of 100 mM MES pH 6.0, 3% PEG 4000, 15% 2-propanol, 50 mM MgCl2 (Table 2 ▶).
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | Corning 3556 |
| Temperature (K) | 295 |
| Protein concentration (mgml1) | 4.2 |
| Buffer composition of protein solution | 10mM TrisHCl pH 7.5 |
| Composition of reservoir solution | 50mM MES pH 6.0, 3% PEG 4000, 15% 2-propanol, 50mM MgCl2 |
| Volume and ratio of drop | 750nl, 2:1 protein:well solution |
| Volume of reservoir (l) | 80 |
2.3. Data collection and processing
Crystals were flash-cooled in liquid nitrogen using mother liquor supplemented with 10% 2-methyl-2,4-pentanediol (MPD) as a cryoprotectant. Data were collected at 100 K on beamline I04-1 at Diamond Light Source, UK. Data were processed, integrated and scaled using the XDS package (Kabsch, 2010 ▶; Table 3 ▶).
Table 3. Data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Diffraction source | I04-1, Diamond Light Source |
| Wavelength () | 0.9 |
| Temperature (K) | 100 |
| Detector | PILATUS |
| Crystal-to-detector distance (mm) | 221.15 |
| Rotation range per image () | 0.5 |
| Total rotation range () | 360 |
| Space group | C2 |
| a, b, c () | 106.60, 23.91, 118.65 |
| , , () | 90.00, 104.95, 90.00 |
| Mosaicity () | 0.4 |
| Resolution range () | 28.662.26 (2.362.26) |
| Total No. of reflections | 91714 (10339) |
| No. of unique reflections | 13969 (1589) |
| Completeness (%) | 97.6 (92.2) |
| Multiplicity | 6.6 (6.5) |
| I/(I) | 11.8 (2.5) |
| R meas (%) | 13.7 (89.1) |
| Overall B factor from Wilson plot (2) | 40.4 |
3. Results and discussion
The CdsD domain (residues 558–771) was purified as described in §2 (Fig. 1 ▶). Initial crystallization screens using the ProPlex screen from Molecular Dimensions gave several good conditions which were further optimized until the optimal crystallization condition was found. The initial small crystals appeared in less than 30 min and stopped growing after 12–24 h (Fig. 2 ▶).
Figure 2.
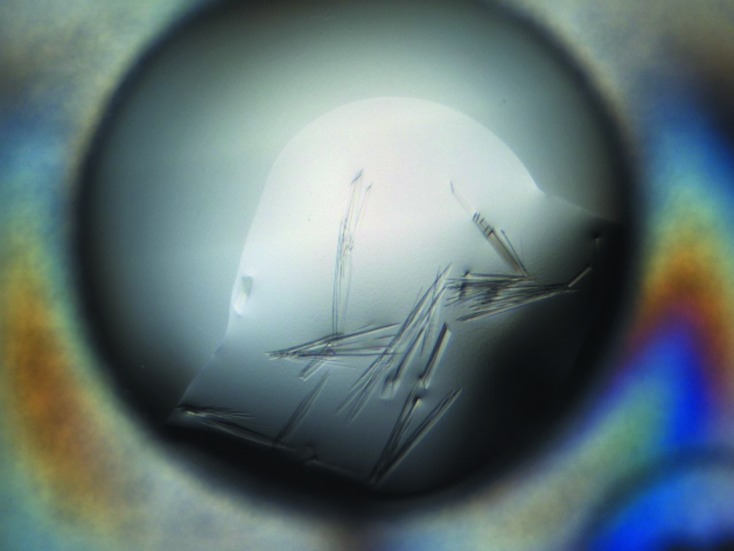
Crystallization of CdsD(558–771). The image was taken by a Rock Imager 54 (Formulatrix) 12 h after setting up the drop and no remarkable change was subsequently observed. The typical crystal size was about 400 × 10 × 5 µm. Owing to the high percentage of 2-propanol the crystals are rather fragile and difficult to handle. The data were collected from a fragment of a crystal similar to that shown here.
For cryoprotection, the crystal was transferred to a new drop consisting of well solution supplemented with 10% MPD. After a 2 min soak, the protein was flash-cooled in liquid nitrogen. A 360° data set with 0.5° oscillation was collected from a single crystal on beamline I04-1 at Diamond Light Source (Fig. 3 ▶). The crystal diffracted to 2.26 Å resolution and the data were processed with XDS (Kabsch, 2010 ▶). The crystal belonged to space group C2, with unit-cell parameters a = 106.60, b = 23.91, c = 118.65 Å, β = 104.95°. The calculated Matthews coefficient V M (Matthews, 1968 ▶) was found to be 3.0 Å3 Da−1, corresponding to a solvent content of 59.6% for one monomer per asymmetric unit.
Figure 3.
X-ray diffraction pattern measured from a C. trachomatis CdsD(558–771) crystal. The oscillation angle was 0.5°. Data were collected on beamline I04-1 at Diamond Light Source using a PILATUS detector. The reflections at the outer edge of the detector correspond to 2.26 Å resolution.
Currently, there are no suitable models that could be used to solve the structure by the molecular-replacement method. We are aiming to solve the structure using the single-wavelength anomalous dispersion (SAD) method. Initial phases have been obtained from a lead derivative.
Acknowledgments
The research leading to these results was funded by the Academy of Finland (Grant 251218) and the European Community’s Seventh Framework Programme (FP7/2007–2013) under Biostruct-X (grant agreement 283570). We are grateful to the beamline scientists at Diamond Light Source beamline I04-1 and to Dr Lari Lehtiö (University of Oulu, Finland) for collecting the data and to Ville Ratas for technical assistance.
References
- Bébéar, C. & de Barbeyrac, B. (2009). Clin. Microbiol. Infect. 15, 4–10. [DOI] [PubMed]
- Betts-Hampikian, H. J. & Fields, K. A. (2011). J. Bacteriol. 193, 6950–6959. [DOI] [PMC free article] [PubMed]
- Johnson, D. L. & Mahony, J. B. (2007). J. Bacteriol. 189, 7549–7555. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kudryashev, M., Stenta, M., Schmelz, S., Amstutz, M., Wiesand, U., Castaño-Díez, D., Degiacomi, M. T., Münnich, S., Bleck, C. K., Kowal, J., Diepold, A., Heinz, D. W., Dal Peraro, M., Cornelis, G. R. & Stahlberg, H. (2013). Elife, 2, e00792. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Peters, J., Wilson, D. P., Myers, G., Timms, P. & Bavoil, P. M. (2007). Trends Microbiol. 15, 241–251. [DOI] [PubMed]
- Spreter, T., Yip, C. K., Sanowar, S., André, I., Kimbrough, T. G., Vuckovic, M., Pfuetzner, R. A., Deng, W., Yu, A. C., Finlay, B. B., Baker, D., Miller, S. I. & Strynadka, N. C. J. (2009). Nature Struct. Mol. Biol. 16, 468–476. [DOI] [PMC free article] [PubMed]
- Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990). Methods Enzymol. 185, 60–89. [DOI] [PubMed]
- Yeats, C. & Bateman, A. (2003). Trends Biochem. Sci. 28, 352–355. [DOI] [PubMed]