Abstract
High‐sensitivity C‐reactive protein (hs‐CRP) levels in European populations are lower in patients with maturity‐onset diabetes of the young type 3 (MODY3) than in those with type 2 diabetes. hs‐CRP levels have been suggested to be useful for discriminating MODY3 from type 2 diabetes. As hs‐CRP levels are influenced by various factors including race and body mass index, it is worthwhile to examine whether hs‐CRP can serve as a biomarker for MODY3 in Japanese. Here we describe the case of a Japanese MODY3 patient with a nonsense mutation in the HNF1A gene. Two measurements showed consistently lower hs‐CRP levels (<0.05 and 0.09 mg/L) than in Japanese patients with type 1 and type 2 diabetes. Hepatic expression of Crp messenger ribonucleic acid was significantly decreased in Hnf1a knockout mice. The hs‐CRP level might be a useful biomarker for MODY3 in both Japanese and European populations.
Keywords: Hepatocyte nuclear factor 1α, High‐sensitivity C‐reactive protein, Maturity‐onset diabetes of the young type 3
Introduction
Maturity‐onset diabetes of the young (MODY) is a monogenic form of diabetes mellitus characterized by autosomal dominant inheritance and early onset. We previously reported that heterozygous mutations of the hepatocyte nuclear factor 1α (HNF1A) gene cause MODY31. We and others have shown that HNF1α controls β‐cell function by regulating Slc2a2, Tmem27, Hgfac and Hnf4a2.
Genetic testing, such as deoxyribonucleic acid (DNA) sequencing, is necessary for the diagnosis of MODY3. Selection of patients for genetic testing of MODY3 is based mainly on clinical features, such as family history and age of onset, but merely fulfilling clinical features does not provide effective selection criteria for genetic testing of MODY36. The C‐reactive protein (CRP) gene has two HNF1α binding sites in its promoter region, and HNF1α activates gene expression by binding to these sites7. Furthermore, common variants in the HNF1A gene are associated with circulating high‐sensitivity CRP (hs‐CRP) levels8. Recent studies have shown that hs‐CRP levels are lower in patients with MODY3 than in those with type 2 diabetes in European populations, and hs‐CRP has been suggested to be a useful prescreening tool for identifying patients for genetic testing9. However, hs‐CRP levels are influenced by various factors including race and body mass index (BMI), and hs‐CRP levels in Japanese people are notably lower than those in Western populations13. Therefore, it is unclear whether hs‐CRP has the potential to serve as a biomarker for Japanese MODY3.
Here we describe the case of a Japanese MODY3 patient where, interestingly, the patient's serum hs‐CRP level was markedly reduced. The hs‐CRP level could be a useful biomarker for MODY3 in both Japanese and European populations.
Materials and Methods
Participants
A 35‐year‐old man was diagnosed with diabetes at 8 years‐of‐age. He was first treated with diet therapy and started nateglinide at 22 years‐of‐age. Insulin therapy was started at Nissay Hospital at 33 years‐of‐age as a result of poor glycemic control. Fasting plasma C‐peptide immunoreactivity (CPR) level was 1.09 ng/mL, and antibody to glutamic acid decarboxylase was negative. His younger sister had also been diagnosed as having early‐onset diabetes, and his mother had been diagnosed as having gestational diabetes. He was not taking any medications, such as statins, aspirin, antihypertensive drugs or glucocorticoids.
The patient's serum hs‐CRP levels were compared with those of 65 Japanese patients with type 2 diabetes reported previously15 and 41 Japanese patients with type 1 diabetes measured in the present study (Table 1). Further information on the type 1 diabetes patients is provided in the Supporting Information.
Table 1. Clinical characteristics of type 1 diabetic participants.
| Variables | Mean ± SD |
|---|---|
| n | 41 |
| Sex (men/women) | 20/21 |
| Age (years) | 38.6 ± 18.8 |
| Duration of diabetes (years) | 7.1 ± 7.5 |
| BMI (kg/m2) | 20.7 ± 3.4 |
| Hypertension (yes/no) | 2/39 |
| Smoking (yes/no) | 12/29 |
| Insulin dose (IU/kg/day) | 0.67 ± 0.32 |
| HbA1c (NGSP%) | 10.5 ± 2.9 |
| Total cholesterol (mmol/L) | 4.61 ± 1.30 |
| Triglycerides (mmol/L) | 1.04 ± 0.41 |
| Creatinine (μmol/L) | 52.8 ± 17.2 |
| hs‐CRP (mg/L) | 0.26 (0.11–0.60)* |
BMI, body mass index; HbA1c, glycated hemoglobin; hs‐CRP, high‐sensitivity C‐reactive protein; NGSP, National Glycohemoglobin Standardization Program; SD, standard deviation. *Median (range).
Screening of HNF1α Gene Mutations
A detailed description of the DNA sequencing to detect HNF1α gene mutations is provided in the Supporting Information.
Biochemical Analysis
Biochemical data were measured by standard laboratory assays. Glycated hemoglobin (HbA1c) levels (National Glycohemoglobin Standardization Program) were calculated from HbA1c (Japan Diabetes Society) levels as described previously16. The serum hs‐CRP level was measured by latex‐enhanced immunonephelometrics on a BN II Analyzer (Dade Behring, Marburg, Germany)17. The range of determinants was 0.05–10 mg/L.
Quantitative Reverse Transcription Polymerase Chain Reaction
A detailed description of the quantitative reverse transcription polymerase chain reaction is provided in the Supporting Information.
Statistical Analysis
Significance was assessed with the unpaired t‐test at P < 0.05.
Results
Identification of HNF1A Gene Mutation
A nonsense mutation in codon 229, CGA (Arg) to TGA (stop; R229X, c.685C>T), was identified (Figure 1). This R229X mutation, which results in no transactivation domain, has previously been identified in other MODY patients18, and has been shown to be a loss‐of‐function mutation by reporter gene assay18. Other family members were not available for genetic testing.
Figure 1.
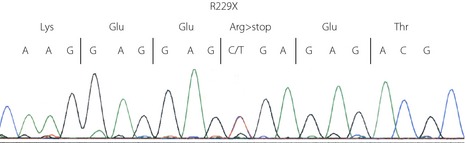
R229X mutation in the HNF1A gene.
Clinical and Biochemical Profiles of the Patient with HNF1A R229X Mutation
The patient's BMI was 22.1 kg/m2 and his HbA1c level was 6.1% (National Glycohemoglobin Standardization Program). He had simple diabetic retinopathy and microalbuminuria, and no previous history of cardiovascular disease. B‐mode ultrasound showed no thickening of the carotid arteries (maximum carotid intima‐media thickness of 0.65 mm). Brachial‐ankle pulse wave velocity was 1192 cm/s. Biochemical tests for liver function and kidney function were normal. Serum levels of low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol and triglyceride were also normal. Two measurements showed notably decreased serum hs‐CRP levels (<0.05, and 0.09 mg/L) and decreased plasma fibrinogen levels (184 and 215 mg/dL; normal, 220–435 mg/dL).
Serum Hs‐CRP Levels in Japanese Type 1 Diabetic Patients
The median hs‐CRP concentration in the 41 Japanese patients with type 1 diabetes was 0.26 mg/L (interquartile range 0.11–0.60 mg/L).
Expression of CRP and Fibrinogen Genes in the Liver of Hnf1a Knockout Mice
The expression levels of Hnf1a and Crp messenger ribonucleic acid (mRNA) in the liver of adult Hnf1a+/− mice were normal (Figure S1). Hnf1a−/− mice show severe liver dysfunction and die around the time of weaning22. As liver failure impairs CRP production23, we investigated the hepatic Crp mRNA expression in Hnf1a−/− mice on embryonic day 18.5. Expression of Pah, a known target gene of HNF1α22, was significantly decreased in the liver of Hnf1a−/− mice (Figure 2), and expression of Crp was also significantly decreased in the HNF1α knockout mice to 35.1% of the control level (P < 0.001). These results show the importance of HNF1α in the transcriptional regulation of the CRP gene in vivo. Fga mRNA expression was decreased in the liver of Hnf1a−/− mice to 56.4% of the control level, but the difference was not significant (P = 0.076).
Figure 2.
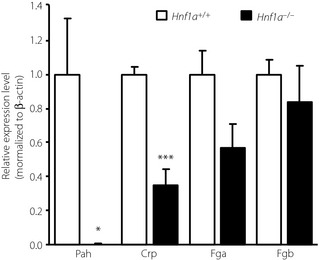
Gene expression of Pah,Crp,Fga, and Fgb messenger ribonucleic acid in the liver of Hnf1a+/+ (n = 4) and Hnf1a−/− (n = 4) mice on embryonic day 18.5. Expression is normalized to that of β‐actin. Data are expressed as means ± standard error (*P < 0.05, ***P < 0.001).
Discussion
We previously reported that the median concentration of hs‐CRP levels in 65 Japanese type 2 diabetic patients (BMI 24.4 ± 3.2 kg/m2) was 0.49 mg/L (interquartile range 0.26–0.87 mg/L)15. In contrast, the serum hs‐CRP level in the present Japanese MODY3 patient with the R229X mutation was notably lower (<0.05 and 0.09 mg/L). The patient was treated with insulin, but not with medications, such as statins, aspirin or steroids, which are known to reduce hs‐CRP levels15. These results suggest that hs‐CRP can be used as a marker for discriminating MODY3 from type 2 diabetes in Japanese patients. Using the same assay as ours, Thanabalasingham et al.12 reported that the cut‐off value of hs‐CRP for discriminating MODY3 from type 2 diabetes is 0.5 mg/L. However, this cut‐off value was similar to the median value of hs‐CRP in the Japanese patients with type 2 diabetes. Therefore, a further large cohort study is necessary to identify the appropriate hs‐CRP cut‐off levels in Japanese MODY3 patients. In the present study, we also measured the serum hs‐CRP levels in type 1 diabetic patients (BMI 20.7 ± 3.4 kg/m2, median 0.26 mg/L [interquartile range 0.11–0.60 mg/L]). The lack of glutamic acid decarboxylase antibodies has been reported as a useful criterion for discriminating MODY3 from type 1 diabetes26. The preset results results suggest that hs‐CRP might also be beneficial for distinguishing between MODY3 and type 1 diabetes.
Crp expression was significantly decreased to 35.1% of that of the controls in the liver of Hnf1a−/− mice, which is consistent with a previous DNA microarray analysis using Hnf1a knockout mice27. These findings suggest that HNF1α plays an important role in the expression of CRP in vivo. It has been reported that HNF1 is required for the optimal promoter function of the genes encoding the α and β chains of fibrinogen28. Although a previous small‐scale study found no significant difference in plasma fibrinogen concentration between MODY3 and MODY130, it is interesting that the plasma fibrinogen level was lower in our patient with MODY3. There is also a tendency for decreased Fga mRNA expression in the liver of Hnf1a−/− mice. Therefore, a combination of hs‐CRP and fibrinogen levels might serve as a useful biomarker for identifying MODY3 in the Japanese population.
Supplementary Material
Data S1 | Materials and methods.
Figure S1 | Gene expression of Hnf1a and Crp in the liver of 16‐week‐old female Hnfla+/+ (n = 3) and Hnfla+/− (n = 3) mice.
Acknowledgments
This work was supported by a Grant‐in‐Aid for Scientific Research (S), a Grant‐in‐Aid for Scientific Research (B), and grants from the Suzuken Memorial Foundation and Daiichi Sankyo Foundation of Life Science. The authors declare that there is no conflict of interests.
J Diabetes Invest 2014; 5: 513–516
References
- 1.Yamagata K, Oda N, Kaisaki PJ, et al Mutations in the hepatocyte nuclear factor‐1alpha gene in maturity‐onset diabetes of the young (MODY3). Nature 1996; 384: 455–458 [DOI] [PubMed] [Google Scholar]
- 2.Yamagata K, Nammo T, Moriwaki M, et al Overexpression of dominant‐negative mutant hepatocyte nuclear fctor‐1 alpha in pancreatic beta‐cells causes abnormal islet architecture with decreased expression of E‐cadherin, reduced beta‐cell proliferation, and diabetes. Diabetes 2002; 51: 114–123 [DOI] [PubMed] [Google Scholar]
- 3.Fukui K, Yang Q, Cao Y, et al The HNF‐1 target collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab 2005; 2: 373–384 [DOI] [PubMed] [Google Scholar]
- 4.Ohki T, Sato Y, Yoshizawa T, et al Identification of hepatocyte growth factor activator (Hgfac) gene as a target of HNF1α in mouse β‐cells. Biochem Biophys Res Commun 2012; 425: 619–624 [DOI] [PubMed] [Google Scholar]
- 5.Boj SF, Parrizas M, Maestro MA, et al A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci USA 2001; 98: 14481–14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stride A, Hattersley AT. Different genes, different diabetes: lessons from maturity‐onset diabetes of the young. Ann Med 2002; 34: 207–216 [PubMed] [Google Scholar]
- 7.Toniatti C, Demartis A, Monaci P, et al Synergistic trans‐activation of the human C‐reactive protein promoter by transcription factor HNF‐1 binding at two distinct sites. EMBO J 1990; 9: 4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiner AP, Barber MJ, Guan Y, et al Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor‐1α are associated with C‐reactive protein. Am J Hum Genet 2008; 82: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen KR, Thanabalasingham G, James TJ, et al Assessment of high‐sensitivity C‐reactive protein levels as diagnostic discriminator of maturity‐onset diabetes of the young due to HNF1A mutations. Diabetes Care 2010; 33: 1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald TJ, Shields BM, Lawry J, et al High‐sensitivity CRP discriminates HNF1A‐MODY from other subtypes of diabetes. Diabetes Care 2011; 34: 1860–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco J, Costa A, Giménez M, et al Comment on: McDonald et al. High‐sensitivity CRP discriminates HNF1A‐MODY from other subtypes of diabetes. Diabetes Care 2011;34:1860‐1862. Diabetes Care 2011; 34: e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanabalasingham G, Shah N, Vaxillaire M, et al A large multi‐centre European study validates high‐sensitivity C‐reactive protein (hsCRP) as a clinical biomarker for the diagnosis of diabetes subtypes. Diabetologia 2011; 54: 2801–2810 [DOI] [PubMed] [Google Scholar]
- 13.Arima H, Kubo M, Yonemoto K, et al High‐sensitivity C‐reactive protein and coronary heart disease in a general population of Japanese: the Hisayama study. Arterioscler Thromb Vasc Biol 2008; 28: 1385–1391 [DOI] [PubMed] [Google Scholar]
- 14.Matsushita K, Yatsuya H, Tamakoshi K, et al High‐sensitivity C‐reactive protein is quite low in Japanese men at high coronary risk. Circ J 2007; 71: 820–825 [DOI] [PubMed] [Google Scholar]
- 15.Motomura T, Okamoto M, Kitamura T, et al Effects of pitavastatin on serum lipids and high sensitivity C‐reactive protein in type 2 diabetic patients. J Atheroscler Thromb 2009; 16: 546–552 [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi A, Kasuga M, Araki E, et al International clinical harmonization of glycated hemoglobin in Japan: From Japan Diabetes Society to National Glycohemoglobin Standardization Program values. J Diabetes Invest 2012; 3: 39–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto K, Kasayama S, Yamamoto H, et al Strong association of C‐reactive protein with body mass index and 2‐h post‐challenge glucose in non‐diabetic, non‐smoker subjects without hypertension. Diabet Med 2004; 21: 581–585 [DOI] [PubMed] [Google Scholar]
- 18.Yamada S, Tomura H, Nishigori H, et al Identification of mutations in the hepatocyte nuclear factor‐1alpha gene in Japanese subjects with early‐onset NIDDM and functional analysis of the mutant proteins. Diabetes 1999; 48: 645–648 [DOI] [PubMed] [Google Scholar]
- 19.Kaisaki PJ, Menzel S, Lindner T, et al Mutations in the hepatocyte nuclear factor‐1alpha gene in MODY and early‐onset NIDDM: evidence for a mutational hotspot in exon 4. Diabetes 1997; 46: 528–535 [DOI] [PubMed] [Google Scholar]
- 20.Pruhova S, Ek J, Lebl J, et al Genetic epidemiology of MODY in the Czech republic: new mutations in the MODY genes HNF‐4alpha, GCK and HNF‐1alpha. Diabetologia 2003; 46: 291–295 [DOI] [PubMed] [Google Scholar]
- 21.Furuzawa GK, Giuffrida FM, Oliveira CS, et al Low prevalence of MODY2 and MODY3 mutations in Brazilian individuals with clinical MODY phenotype. Diabetes Res Clin Pract 2008; 81: e12–e14 [DOI] [PubMed] [Google Scholar]
- 22.Pontoglio M, Barra J, Hadchouel M, et al Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 1996; 84: 575–585 [DOI] [PubMed] [Google Scholar]
- 23.Pepys MB, Hirschfield GM. C‐reactive protein: a critical update. J Clin Invest 2003; 111: 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridker PM, Rifai N, Clearfield M, et al Measurement of C‐reactive protein for the targeting of statin therapy in the primary prevention of acute coronary events. N Engl J Med 2001; 344: 1959–1965 [DOI] [PubMed] [Google Scholar]
- 25.Kasayama S, Tanemura M, Koga M. Asthma is an independent risk for elevation of plasma C‐reactive protein levels. Clin Chim Acta 2009; 399: 79–82 [DOI] [PubMed] [Google Scholar]
- 26.Lehto M, Tuomi T, Mahtani MM, et al Characterization of the MODY3 phenotype. Early‐onset diabetes caused by an insulin secretion defect. J Clin Invest 1997; 99: 582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih DQ, Bussen M, Sehayek E, et al Hepatocyte nuclear factor‐1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 2001; 27: 375–382 [DOI] [PubMed] [Google Scholar]
- 28.Hu CH, Harris JE, Davie EW, et al Characterization of the 5′‐flanking region of the gene for the alpha chain of human fibrinogen. J Biol Chem 1995; 270: 28342–28349 [DOI] [PubMed] [Google Scholar]
- 29.Courtois G, Baumhueter S, Crabtree GR, et al Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte‐specific promoters. Proc Natl Acad Sci USA 1988; 85: 7937–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasaki N, Ogata M, Tomonaga O, et al Liver and kidney function in Japanese patients with maturity‐onset diabetes of the young. Diabetes Care 1998; 21: 2144–2148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 | Materials and methods.
Figure S1 | Gene expression of Hnf1a and Crp in the liver of 16‐week‐old female Hnfla+/+ (n = 3) and Hnfla+/− (n = 3) mice.


