Abstract
Although autism spectrum disorder (ASD) shows a high degree of heritability, only a few mutated genes and mostly de novo copy number variations (CNVs) with a high phenotypic impact have as yet been identified. In families with multiple ASD patients, transmitted CNVs often do not appear to cosegregate with disease. Therefore, also transmitted single nucleotide variants which escape detection if genetic analyses were limited to CNVs may contribute to disease risk. In several studies of ASD patients, CNVs covering at least one gene of the contactin gene family were found. To determine whether there is evidence for a contribution of transmitted variants in contactin genes, a cohort of 67 ASD patients and a population-based reference of 117 healthy individuals, who were not related to the ASD families, were compared. In total, 1,648 SNPs, spanning 12.1 Mb of genomic DNA, were examined. After Bonferroni correction for multiple testing, the strongest signal was found for a SNP located within the CNTN5 gene (rs6590473 [G], p = 4.09 × 10-7; OR = 3.117; 95% CI = 1.603-6.151). In the ASD cohort, a combination of risk alleles of SNPs in CNTN6 (rs9878022 [A]; OR = 3.749) and in CNTNAP2 (rs7804520 [G]; OR = 2.437) was found more frequently than would be expected under random segregation, albeit this association was not statistically significant. The latter finding is consistent with a polygenic disease model in which multiple mutagenic mechanisms, operating concomitantly, elicit the ASD phenotype. Altogether, this study corroborates the possible involvement of contactins in ASD, which has been indicated by earlier studies of CNVs.
Key Words: Autism, Candidate gene association study, Contactin gene family, CNTN5, CNTN6, CNTNAP2, Copy number variation
Autism spectrum disorders (ASDs) represent a group of neurodevelopmental disorders involving impaired social interactions, restricted interests and behaviors, and language impairment and communication. ASDs occur either sporadically or as familial cases, with an estimated prevalence of 1/150 children and boys being 4 times more frequently affected than girls [Freitag, 2007]. Comparisons of monozygotic and dizygotic twins suggested heritability as high as 90% [Skuse, 2007]. ASD may either be part of Mendelian syndromes, such as the Rett and the Smith-Lemli-Opitz syndrome; result from genomic rearrangements, or results from mutations in genes such as DISC1, NRXN1, RELN, CNTNAP2, and MECP2 [Abrahams and Geschwind, 2008]. However, nonsyndromic cases may have a multifactorial genomic architecture.
Hypotheses on the genetics of neurobehavioral disorders such as ASDs either assume rare variants with a high phenotypic impact, such as copy number variations (CNVs), or common variants with a lesser effect [State and Levitt, 2011]. Several large-scale studies have identified de novo CNVs in sporadic patients and cosegregating CNVs in families with multiple affected children (multiplex families) [Marshall et al., 2008; Glessner et al., 2009; Holt et al., 2010]. In addition to recurrent de novo CNVs in 5 genomic regions [Sanders et al., 2011], hundreds of distinct rare variants have each been observed only once [Bucan et al., 2009]. In multiplex ASD families in particular, CNVs covering plausible candidate genes often did not cosegregate with disease in the family [Marshall et al., 2008; Bucan et al., 2009; Salyakina et al., 2011; van Daalen et al., 2011]. For instance, in the study by van Daalen et al. [2011], the loss of part of CNTN5 in family M2 was found in all 3 affected boys (i.e. cosegregated), while the loss of part of chromosome band 3p26 in family M3 was found in only 1 of the 2 affected boys (i.e. was not cosegregating). Therefore, such not cosegregating CNVs are by themselves not sufficient to cause ASD, but may in concert with other, as yet unidentified, alleles contribute to disease risk in a particular family. This further underscores the complex genomic architecture of ASD.
Large-scale genome-wide association studies have detected common variants in genes such as CDH9, CDH10, NRXN1 and others, which often could not be independently replicated [Autism Genome Project Consortium et al., 2007; Ma et al., 2009; Wang et al., 2009; Curran et al., 2011]. Overall, individual common variants exert weak effects on the risk for ASD [Anney et al., 2012] but by acting additively may explain as much as 60% of the narrow-sense heritability in ASD individuals from multiplex families and approximately 40% for simplex families [Klei et al., 2012]. Thus, assuming a polygenic model, combinations of 2 or more variants may be necessary to cause the full ASD phenotype, as has been found with intellectual delay (ID) [Girirajan et al., 2012].
Whole exome sequencing studies of over 1,000 ASD families revealed hundreds of de novo single nu-cleotide variants (SNVs). Only 4 genes, SCN2A, NTNG1, KATNAL2, and CHD8 carried more than a single SNV [Neale et al., 2012; O'Roak et al., 2012a; Sanders et al., 2012]. In addition, single de novo SNVs were found in genes such as FOXP1, GRIN2B, SCN1A, and LAMC3, which have been associated with a variety of neurodevelopmental disorders, including ID [Endele et al., 2010; O'Roak et al., 2011]. By targeted sequencing, O'Roak et al. [2012b] found recurrent mutations in CHD8, DYRK1A, GRIN2B, TBR1, PTEN, and TBL1XR1. Losses of gene function due to gene disrupting mutations such as nonsense, coding indels, and splice acceptor/donor site mutations have been found at significantly elevated frequencies in severe ID, epilepsy and ASD [Petrovski et al., 2013]. On the other hand, functional enrichment analysis of CNVs in a cohort of patients with ID and mutation analysis in patients with ID highlighted GRIN2B as a candidate gene for both ID and ASD [Webber, 2011; Endele et al., 2010; Poot et al., 2010a]. Altogether, the exome sequencing studies point towards hundreds of potential ASD risk genes, of which some overlap with disorders such as ID and epilepsy.
Genome-wide analyses of CNVs in ASD patients showed a preponderance of genes of the contactin gene family (CNTN5, CNTN4, CNTN6) [van Daalen et al., 2011; Nava et al., 2013]. Due to the relatively small patient cohort, of these studies, the overrepresentation of these contactin genes was not statistically significant [van Daalen et al., 2011; Nava et al., 2013]. Another limitation of these studies was their focus on CNVs, such that other mutagenic mechanisms independent of gene dosage will be neglected [Poot et al., 2011; Poot, 2013]. The hypothesis that contactin genes may be involved in ASD by non-CNV-dependent mechanisms can be tested by performing an association study of all contactin and contactin-associated genes in the same patient cohort. In addition, all CNV genes with transcription in the human brain found in the same patient cohort were included. In view of the genetic overlap between ASD and ID (e.g. GRIN2B, see above), the genes that emerged after functional enrichment analysis from a CNV study of ID patients from the same population base also were included [Poot et al., 2010a]. Thus, all members of the contactin and the contactin-associated protein families, and the ATXN7, ATP1A2, GIRDIN, GRIA2, GRIA4, GRIK2, GRIN2A, GRIN2B, IMMP2L, KCNJ3 (GIRK3), KCNJ9, KCNJ10, KCNK5, KCNK17, KCNMB3, KCNMB4, KDELR2, MCPH1, PIK3CA, and ZNF12 genes were included in the present association study (online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000362891).
Materials and Methods
An initial cohort of 210 referrals with at least one sign of ASD before their third birthday was assessed according to DSM-IV criteria, and only those patients who conformed to a strict ‘best estimate diagnosis of autism’ and did not show any evidence for a syndromic form of autism were further investigated [van Daalen et al., 2011]. Upon extensive medical genetic investigation and reassessment using ADI-R and ADOS-G, those patients with above cut-off scores on all 3 psychiatric evaluation tools and at least 3 dysmorphic or developmental comorbidities, as assessed by standard medical genetic evaluation, were selected. Thus, a patient cohort, consisting of 67 children, 61 male and 6 female index patients, 16 from multiplex ASD families and 51 from families with a single patient, was included in this study. To avoid confounding effects of CNVs, all 9 patients who had a CNV covering one of the genes included in this study were excluded. A reference cohort composed of 117 healthy, mostly adult individuals from families not being related to the ASD families, but from the same population base, was compiled (age range 20-49 years, gender distribution 50:50, all of native Dutch origin). Both cohorts were genotyped using Illumina Infinium HumanHap300 Genotyping BeadChip SNP arrays [van Daalen et al., 2011].
For the candidate gene association study, all SNPs located on the BeadChip Array within, and the first 2 SNPs immediately bordering genes with clear transcription in the brain from the 9 previously detected CNVs were included [van Daalen et al., 2011]. Also, all genes of the contactin and contactin-associated protein gene families, except for CNTNAP1 and CNTNAP3, which did not contain enough SNPs, and the genes found to be overrepresented by functional enrichment analysis of the CNVs in patients with ID from the same population base were selected [Poot et al., 2010a] (for a full list of all SNPs, see online suppl. table 1).
Using PLINK [Purcell et al., 2007], SNPs were filtered according to the following quality criteria: proportion of genotypes not called per SNP <0.1, proportion of genotypes not called per individual <0.1, Hardy-Weinberg equilibrium with a p value >0.00001, and a minor allele frequency >0.03. After filtering, 1,648 SNPs were included in the analysis. Allele frequency differences between patients and references were tested with a χ2 test for independence. The threshold for significance after Bonferroni correction for multiple testing, was set at p = 3.0 × 10-5. ORs were recorded (online suppl. table 1). Plots showing linkage disequilibrium (LD) in regions with statistically significant association were created with LocusZoom [Pruim et al., 2010] using LD data from the HapMap CEU data set.
To identify genes that are significantly associated with ASD, all SNPs were ranked according to ascending p values, and the risk alleles were recorded. To test for possible interaction between the genes, the top 20 SNPs on the base of their p values were taken for a pairwise analysis (table 1). Since some genes carried several SNPs, which may be in LD with each other, only the top SNP for each of the top 6 genes was used for the pairwise co-occurrence analysis. Fisher exact tests were used to determine whether co-occurrence of these selected SNPs within the patient and the reference cohorts significantly deviated from random segregation.
Table 1.
The top 20 SNPs ranked acording to p value for association
| Chromosome number | SNP | Position | Allele 1 | Frequency in |
Allele 2 | χ2 | p value | OR | Gene | |
|---|---|---|---|---|---|---|---|---|---|---|
| ASD cohort | reference cohort | |||||||||
| 11 | rs6590473 | 99,852,154 | G | 0.5769 | 0.3043 | A | 25.650 | 4.09E-07 | 3.117 | CNTN5 |
| 3 | rs9878022 | 1,143,423 | C | 0.1940 | 0.0603 | A | 15.590 | 7.85E-05 | 3.749 | CNTN6 |
| 11 | rs1035339 | 99,866,659 | A | 0.5352 | 0.3421 | G | 14.279 | 1.58E-04 | 2.214 | CNTN5 |
| 3 | rs7641285 | 2,247,856 | G | 0.1667 | 0.3462 | A | 13.440 | 2.46E-04 | 0.378 | CNTN4 |
| 11 | rs1453576 | 99,818,828 | C | 0.5493 | 0.3779 | A | 10.994 | 9.14E-04 | 0.498 | CNTN5 |
| 11 | rs7942402 | 99,593,787 | C | 0.2769 | 0.4522 | A | 10.730 | 1.05E-03 | 0.464 | CNTN5 |
| 6 | rs1415482 | 102,157,813 | A | 0.1642 | 0.0598 | C | 10.510 | 1.19E-03 | 3.087 | GRIK2 |
| 3 | rs2872338 | 1,162,040 | G | 0.1567 | 0.0560 | A | 10.220 | 1.39E-03 | 3.131 | CNTN6 |
| 3 | rs1504076 | 1,154,558 | A | 0.1418 | 0.0474 | G | 10.050 | 1.52E-03 | 3.319 | CNTN6 |
| 6 | rs2852564 | 102,130,208 | A | 0.1716 | 0.0664 | G | 9.874 | 1.68E-03 | 2.915 | GRIK2 |
| 11 | rs6590446 | 99,811,891 | A | 0.5282 | 0.3664 | G | 9.871 | 1.68E-03 | 1.936 | CNTN5 |
| 3 | rs908487 | 2,768,541 | A | 0.1385 | 0.0470 | G | 9.533 | 2.02E-03 | 3.258 | CNTN4 |
| 12 | rs918168 | 14,078,633 | A | 0.2197 | 0.3761 | G | 9.489 | 2.07E-03 | 0.467 | GRIN2B |
| 6 | rs10485271 | 102,017,665 | G | 0.0149 | 0.0983 | A | 9.352 | 2.23E-03 | 0.139 | GRIK2 |
| 3 | rs6797539 | 1,147,200 | G | 0.1308 | 0.0431 | A | 9.277 | 2.32E-03 | 3.340 | CNTN6 |
| 7 | rs7804520 | 146,719,194 | A | 0.2059 | 0.0962 | G | 9.274 | 2.32E-03 | 2.437 | CNTNAP2 |
| 2 | rs2576696 | 55,505,680 | A | 0.2239 | 0.1068 | C | 9.182 | 2.44E-03 | 2.412 | GIRDIN |
| 7 | rs10262822 | 146,567,638 | G | 0.2353 | 0.3837 | A | 8.840 | 2.95E-03 | 0.494 | CNTNAP2 |
| 7 | rs1608628 | 146,708,043 | A | 0.3603 | 0.5154 | C | 8.641 | 3.29E-03 | 0.530 | CNTNAP2 |
| 2 | rs6721614 | 125,424,552 | A | 0.2385 | 0.3870 | G | 8.241 | 4.09E-03 | 0.496 | CNTNAP5 |
The SNP with significant association after Bonferroni correction is shown in bold type. Frequency = Frequency of allele 1; p value = p value of the χ2 test; OR = odds ratio ASD vs. reference cohort.
Results
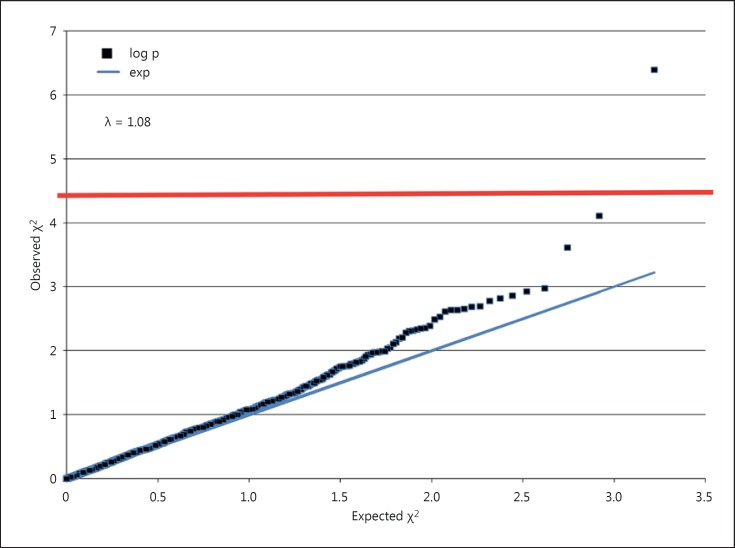
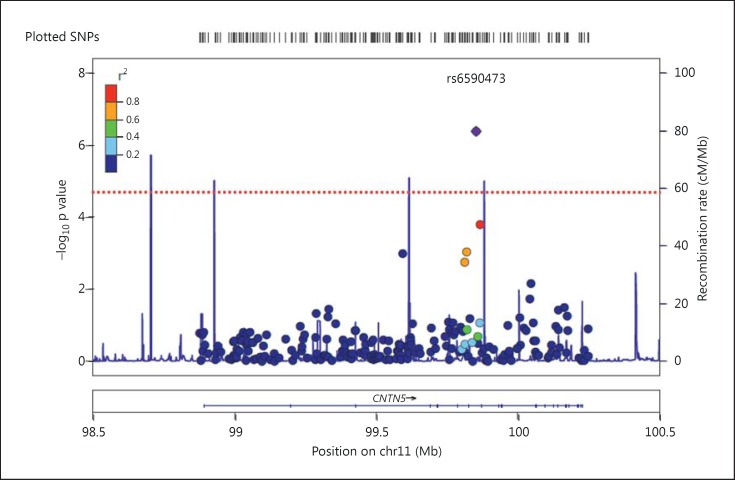
Allele distributions of SNPs covering 24 genes from a patient and a reference cohort were examined. After quality control (see Materials and Methods), 1,648 SNPs were retained (online suppl. table 2). QQ plots of the observed versus the expected χ2 values, showed a significant deviation from λ = 1.00 (λ = 1.08) (fig. 1). The slight deviation from λ = 1.00 is consistent with some, albeit not a significant association of several SNPs with disease. Therefore, the assumption of mainly neutral variants, necessary to obtain λ = 1.00, may not be met. Upon Bonferroni correction for multiple testing (threshold p = 3.0 × 10-5), the strongest signal was found for a SNP located within the CNTN5 gene (rs6590473 [G], p = 4.09 × 10-7; OR = 3.117; 95% CI = 1.603-6.151) (fig. 2; table 1).
Fig. 1.
QQ plots of expected versus observed χ2 values for all SNPs. For further explanations, see text. The horizontal red bar indicates the Bonferroni-corrected threshold of significance for this study (p = 3.0 × 10-5).
Fig. 2.
LocusZoom plot of the recombination rate data of the European HapMap sample and the p values of the SNPs for the region of 11q22.1 containing CNTN5. The horizontal dotted line indicates the Bonferroni-corrected threshold of significance for this study (p = 3.0 × 10-5).
Table 1 shows the top 20 SNPs ranked on ascending p values (the complete set of SNPs is given in online suppl. table 2). These data are consistent with a model in which several risk alleles, which individually did not associate significantly with disease, may elicit disease if occurring together. The distribution of OR-weighted risk alleles was apparently normal, and the maximum of this distribution for the ASD patients was higher than that of the reference cohort (results not shown). Since the risk alleles were normally distributed, segregation that deviates from independence may reflect gene interaction. This hypothesis was tested by determining whether the number of individuals carrying the risk allele for a given SNP occurred more frequently than expected if they would segregate independently in the patient and the reference cohorts (using the Fisher exact test). As shown in tables 1 and 2, the combination of SNPs rs9878022 [A], in CNTN6 on chromosome 3, and rs7804520 [G], in CNTNAP2 on chromosome 7, showed a slight departure from independent segregation among ASD patients (p = 0.018). In the reference cohort, risk alleles for all pairs of SNPs appeared to segregate randomly.
Table 2.
p values of the pairwise occurrence of risk alleles in the indicated genes (2-sided Fisher exact test)
| CNTN5 | CNTN6 | CNTN4 | GRIK2 | GRIN2B | CNTNAP2 | |
|---|---|---|---|---|---|---|
| ASD cohort | ||||||
| CNTN5 | ||||||
| CNTN6 | 1.000 | |||||
| CNTN4 | 0.139 | 0.290 | ||||
| GRIK2 | 0.050 | 0.400 | 0.549 | |||
| GRIN2B | 0.220 | 0.273 | 1.000 | 0.551 | ||
| CNTNAP2 | 0.768 | 0.018 | 0.269 | 1.000 | 1.000 | |
| CNTNAP5 | 0.774 | 1.000 | 1.000 | 0.588 | 0.626 | 1.000 |
|
Reference cohort | ||||||
| CNTN5 | ||||||
| CNTN6 | 1.000 | |||||
| CNTN4 | 0.383 | 0.643 | ||||
| GRIK2 | 0.409 | 0.661 | 0.375 | |||
| GRIN2B | 0.096 | 1.000 | 0.395 | 0.429 | ||
| CNTNAP2 | 0.167 | 1.000 | 0.070 | 0.734 | 0.738 | |
| CNTNAP5 | 0.268 | 0.153 | 0.770 | 0.781 | 1.000 | 0.815 |
Discussion
ASD is a complex neurobehavioral disorder with a high degree of heritability [Freitag, 2007; Skuse, 2007]. Individual common variants identified by large-scale genome-wide association studies appeared to exert weak effects on the risk for ASD, such that all variants together may explain only a modest part of the genetic component of the disease [Anney et al., 2012]. While recent exome sequencing studies uncovered hundreds of de novo SNVs in ASD patients, only 4 genes, SCN2A, NTNG1, KATNAL2, and CHD8 carried more than a single SNV [Neale et al., 2012; O'Roak et al., 2012a; Sanders et al., 2012]. In addition, SNVs were found in genes such as FOXP1, GRIN2B, SCN1A, and LAMC3, which were previously associated with a variety of neurodevelopmental disorders [O'Roak et al., 2011]. Although these SNVs appear to converge on genes related to synapsis formation and synaptic function, a single pathway or biological mechanism did not emerge [Huguet et al., 2013]. Taken together, these genome-wide and targeted exome sequencing studies highlight the complex genomic architecture of ASD and indicate a considerable genetic overlap between ASD and neurodevelopmental disorders [O'Roak et al., 2011, 2012a, b; Neale et al., 2012; Sanders et al., 2012].
CNTNAP2 has repeatedly been implicated in ASD [Peñagarikano and Geschwind, 2012]. In the present study, none of the 247 SNPs within CNTNAP2 tested here associated significantly with ASD (online suppl. table 1). It should be stressed, however, that only one of the previously reported SNPs in CNTNAP2 (rs2710102, OR = 1.04; p = 0.84) [Stein et al., 2011] was available on our array. In the present study, none of the SNPs in LD with rs1718101 [Anney et al., 2012], located within intron 1 of CNTNAP2, reached a p value of association below 0.05. Also, the SNP rs2518261, which is located within intron 1 of GRIK2 and for which a linkage to ASD has been reported, was not available on our array [Holt et al., 2010]. Yet, none of the SNPs on our array within intron 1 of GRIK2 and being in LD with rs2518261 showed a p value of association below 0.05. Interestingly, rs1415482 in intron 6 of GRIK2 returned a p value for association of 0.00119 (table 1). Also the surrounding SNPs that were in LD with rs1415482 showed p values below 0.05. Conceivably, these SNPs may return a significant signal in larger cohorts of ASD patients. Finally, no significant association with GRIN2B was found, although SNVs in this gene have been reported [O'Roak et al., 2012]. These negative findings may be due to the limited size of the patient cohort in the present study.
A previous CNV study showed a cosegregating loss of part of CNTN5 in a single family, which, because of its segregation pattern within this family, is likely to be pathogenic by causing haploinsufficiency for this gene [van Daalen et al., 2011, Poot, 2013]. In the present candidate gene association study of the same patient cohort, from which this particular patient was removed, the SNP with the strongest signal was located in CNTN5. Although the p value for association, 4,09 × 10-7, falls short of genome-wide significance, this finding is consistent with a further possible involvement of CNTN5 in ASD. The contactin-5 protein is a neural cell recognition molecule of the contactin/F3 subgroup, which promotes neurite outgrowth of the cerebral cortical neurons in the auditory pathway, suggesting that it plays specific roles in the development and maturation of certain regions of the brain [Ogawa et al., 2001]. The contactin-5 protein also binds to the amyloid precursor-like protein 1 (APLP1), thus, possibly forming a network by physical interactions of encoded proteins [Shimoda et al., 2012]. Since involvement of CNTN5 in ASD has been supported by both the CNV and the present candidate gene association study, further neurobiological and, in particular, neurobehavioral studies of this gene appear warranted [Poot, 2013].
It is conceivable that CNTN5, in analogy to recently reported findings with SHANK2 in patients with ASD and ID [Leblond et al., 2012; Chilian et al., 2013], may be part of a gene interaction network consisting of physical interactions between the encoded proteins [Poot et al., 2011]. Alternatively, genes may form a network of epistatic regulation consisting of transcription factors and their cognate target sites. Such a network has been suggested for CNTNAP2 and FOXP2 [Vernes et al., 2008; Poot et al., 2010b]. Interestingly, 15 of the top 20 SNPs ranked on ascending p values were located within genes of the contactin and the contactin-associated protein gene families (table 1). This finding may indicate a genetic network of interaction among contactins. The nominally significant departure from independent segregation of 2 SNPs, rs9878022 [C] in CNTN6 and rs7804520 [A] in CNTNAP2, may reflect a synergism of risk alleles for CNTN6 and CNTNAP2 [Toro et al., 2010]. Given the appreciable body of evidence indicating involvement of CNTNAP2 in ASD and related neurobehavioral disorders [Poot et al., 2011; Peñagarikano and Geschwind, 2012; Rodenas-Cuadrado et al., 2014], this gene merits further scrutiny [Peñagarikano et al., 2011].
The present data do corroborate a possible involvement of contactins in ASD. Second, the findings indicate that within a patient cohort, certain genes may harbor variants with both major effects, such as the transmitted CNV of CNTN5, and with a lesser degree of effect for the same phenotype [Girirajan and Eichler, 2010]. The latter is consistent with a polygenic model for autism in which multiple mutagenic mechanisms may be operating simultaneously. Such a model may explain different degrees of phenotypic impact of inherited and de novo CNVs and SNVs both in families with multiple affected children [Bucan et al., 2009; Salyakina et al., 2011; van Daalen et al., 2011] and in sporadic patients [O'Roak et al., 2012a, b]. Plausible molecular hypotheses for such a model may include either physical interaction of encoded proteins or regulatory networks of transcription factors [Poot et al., 2010b, 2011; Leblond et al., 2012; Huguet et al., 2013], which all merit further study. If specific polygenic constellations would be uncovered in individual patients, this may pave the way to distinguishing molecular subtypes of ASD, which eventually may become targets for individualized treatment approaches [Poot, 2013].
Supplementary Material
Supplementary data
Acknowledgements
I am grateful to Drs. E. van Daalen, N.E. Verbeek, M.V. de Jonge, and E.H. Brilstra for providing clinical data, and to the patients and parents for their kind cooperation and their consent to publish these findings. I also wish to thank Dr. C.G.F. de Kovel for helpful advice with the SNP association study. This research was supported by a grant of the Dutch Foundation for Brain Research (Hersenstichting) grant 2008(1).34 to M. Poot.
References
- 1.Abrahams B, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autism Genome Project Consortium. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chilian B, Abdollahpour H, Bierhals T, Haltrich I, Fekete G, et al. Dysfunction of SHANK2 and CHRNA7 in a patient with intellectual disability and language impairment supports genetic epistasis of the two loci. Clin Genet. 2013;84:560–565. doi: 10.1111/cge.12105. [DOI] [PubMed] [Google Scholar]
- 6.Curran S, Bolton P, Rozsnyai K, Chiocchetti A, Klauck SM, et al. No association between a common single nucleotide polymorphism, rs4141463, in the MACROD2 gene and autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:633–639. doi: 10.1002/ajmg.b.31201. [DOI] [PubMed] [Google Scholar]
- 7.Endele S, Rosenberger G, Geider K, Popp B, Tamer C, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 8.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 9.Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:176–R187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, et al. Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet. 2010;18:1013–1019. doi: 10.1038/ejhg.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- 14.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, et al. Genetic and functional analyses of SHANK2mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nava C, Keren B, Mignot C, Rastetter A, Chantot-Bastaraud S, et al. Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. Eur J Hum Genet. 2014;22:71–78. doi: 10.1038/ejhg.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa J, Lee S, Itoh K, Nagata S, Machida T, et al. Neural recognition molecule NB-2 of the contactin/F3 subgroup in rat: specificity in neurite outgrowth-promoting activity and restricted expression in the brain regions. J Neurosci Res. 2001;65:100–110. doi: 10.1002/jnr.1133. [DOI] [PubMed] [Google Scholar]
- 21.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012a;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012b;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peñagarikano O, Geschwind DH. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med. 2012;18:156–163. doi: 10.1016/j.molmed.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poot M. Towards identification of individual etiologies by resolving genomic and biological conundrums in patients with autism spectrum disorders. Mol Syndromol. 2013;4:213–226. doi: 10.1159/000350041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poot M, Eleveld MJ, van't Slot R, Ploos van Amstel HK, Hochstenbach R. Recurrent copy number changes in mentally retarded children harbour genes involved in cellular localization and the glutamate receptor complex. Eur J Hum Genet. 2010a;18:39–46. doi: 10.1038/ejhg.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poot M, Beyer V, Schwaab I, Damatova N, Van't Slot R, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010b;11:81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 30.Poot M, van der Smagt JJ, Brilstra EH, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011;135:228–240. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]
- 31.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodenas-Cuadrado P, Ho J, Vernes SC. Shining a light on CNTNAP2 complex functions to complex disorders. Eur J Hum Genet. 2014;22:171–178. doi: 10.1038/ejhg.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salyakina D, Cukier HN, Lee JM, Sacharow S, Nations LD, et al. Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS One. 2011;6:26049. doi: 10.1371/journal.pone.0026049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoda Y, Koseki F, Itoh M, Toyoshima M, Watanabe K. A cis-xcomplex of NB-2/contactin-5 with amyloid precursor-like protein 1 is localized on the presynaptic membrane. Neurosc Let. 2012;510:148–153. doi: 10.1016/j.neulet.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 39.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein MB, Yang BZ, Chavira DA, Hitchcock CA, Sung SC, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 is associated with increased risk for selective mutism and social anxiety-related traits. Biol Psychiatry. 2011;69:825–831. doi: 10.1016/j.biopsych.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 42.van Daalen E, Kemner C, Verbeek NE, van der Zwaag B, Dijkhuizen T, et al. Social Responsiveness Scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics. 2011;12:315–323. doi: 10.1007/s10048-011-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webber C. Functional enrichment analysis with structural variants: pitfalls and strategies. Cytogenet Genome Res. 2011;135:277–285. doi: 10.1159/000331670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data