Abstract
Nager syndrome, or acrofacial dysostosis type 1 (AFD1), is a rare multiple malformation syndrome characterized by hypoplasia of first and second branchial arches derivatives and appendicular anomalies with variable involvement of the radial/axial ray. In 2012, AFD1 has been associated with dominant mutations in SF3B4. We report a 22-week-old fetus with AFD1 associated with diaphragmatic hernia due to a previously unreported SF3B4 mutation (c.35-2A>G). Defective diaphragmatic development is a rare manifestation in AFD1 as it is described in only 2 previous cases, with molecular confirmation in 1 of them. Our molecular finding adds a novel pathogenic splicing variant to the SF3B4 mutational spectrum and contributes to defining its prenatal/fetal phenotype.
Key Words: Acrofacial dysostosis, AFD1, Diaphragmatic hernia, Nager syndrome, SF3B4
Nager syndrome, or acrofacial dysostosis type 1 (AFD1; OMIM #154400), is a well-known pleiotropic malformation syndrome defined by the association of hypoplasia of first and second branchial arches derivatives (i.e. downslanting palpebral fissures, malar hypoplasia/flattening, malformed ears, and micrognathia) and appendicular anomalies with variable involvement of the radial/axial ray. The former feature may be so severe in specific cases that it partly overlaps with otocephaly, which, in turn, may be distinguished from AFD1 by total agenesis of the mandible (i.e. agnathia) and midline fusion of the external ears below the maxillae. Appendicular involvement, usually limited to the upper limbs, is considered the hallmark of AFD1 and allows differentiation from other AFDs, such as Miller syndrome [Halal et al., 1983; Opitz, 1987]. For decades, the molecular basis of AFD1 remained unknown, with most cases occurring sporadically and rarely clustering in families and therefore being compatible with both autosomal dominant and recessive transmission [Chemke et al., 1988; Hall, 1989; Aylsworth et al., 1991; McDonald and Gorski, 1993; Kennedy and Teebi, 2004]. Bernier et al. [2012] identified 18 different heterozygous mutations in the SF3B4 gene in 20 families from a total of 35 pedigrees with a clinical diagnosis of AFD1. Sixteen additional families with molecularly confirmed AFD1 were subsequently reported [Czeschik et al., 2013; Petit et al., 2013]. In all but 4 cases, the diagnosis was established after birth. Here, we report a 22-week-old fetus with AFD1 and diaphragmatic hernia carrying a novel SF3B4 mutation.
Case Report
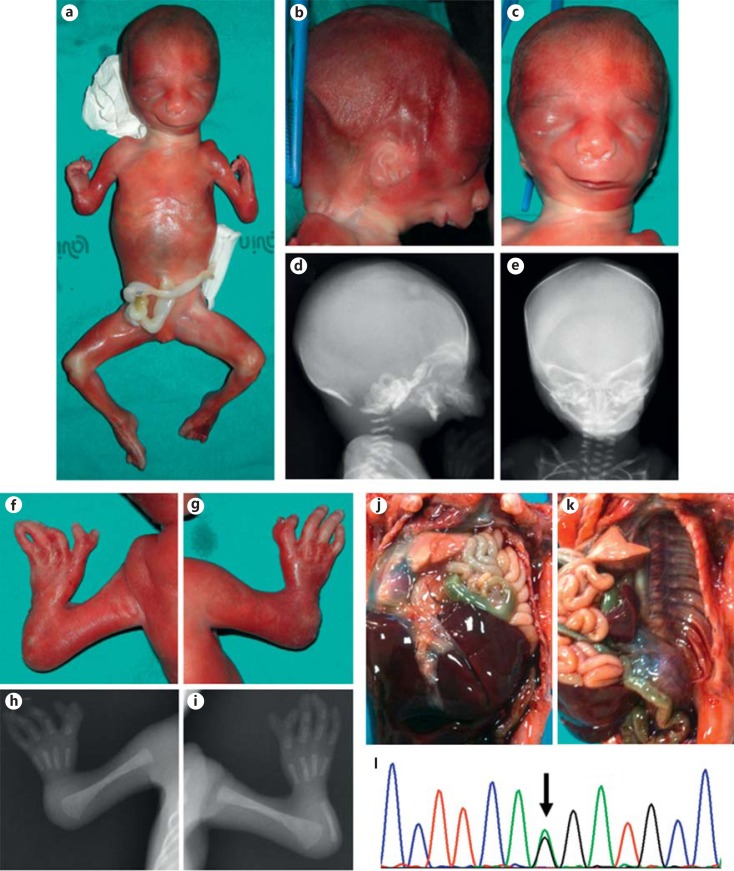
The fetus was the second child of a 33-year-old healthy woman and her unrelated 37-year-old husband. The pregnancy progressed uneventful, without exposure to any known teratogen. Standard second trimester ultrasound screening, performed at the 21st week of gestation, revealed multiple malformations, comprising intra-thoracic visualization of the gastric bubble and a severe reduction defect of the upper limbs. Two days later, a further ultrasound examination better characterized the observed defects as diaphragmatic hernia with thoracic translocation of the stomach and hypoplasia of the left lung and marked shortening of the upper limbs with predominant involvement of the radial ray. Hypoplasia of the maxillae and mandibula was also noted. Amniocentesis was not performed, and the couple chose for termination of pregnancy at 22 weeks of gestation. External examination was consistent with a female fetus showing apparent hypertelorism, downslanting palpebral fissures with lateral flares of the eyebrows, hypoplasia of the middle face, and severe micrognathia. The ears appeared small and slightly low-set and lacked an external auditory canal. The upper limbs were hypoplastic with marked shortening of the forearms and radial clubhands. Both hands were 4-fingered, and the first radial fingers showed complete distal duplication. Evisceration confirmed left diaphragmatic agenesis with intra-thoracic localization of the stomach and revealed agenesis of the uterus. Fetal radiographs showed marked retro-micrognathia, reduced volume of both maxillary bones, aplasia of both radii and right ulna, severe hypoplasia of the left ulna, oligodactyly with 4 metacarpals in both hands, broad proximal phalanx of the first fingers, which were distally duplicated with rudimentary middle phalanges, and double distal phalanges. A clinical diagnosis of Nager syndrome/AFD1 was established (fig. 1a–k).
Fig. 1.
a Whole-body picture of the fetus showing typical appendicular and craniofacial features. b-e Comparison of the clinical (b right lateral view, c frontal view) and radiographic (d right lateral view, e frontal view) presentation of the craniofacial anomalies. f-i Comparison of the clinical (f left hand, g right hand) and radiographic (h left hand, i right hand) presentation of the hand anomalies. j Cranial dislocation of the gut through a large defect of the left diaphragm. k Partial evisceration showing complete agenesis of the left hemi-diaphragm. l Direct sequencing electropherogram of the acceptor splicing junction of SF3B4 exon 2, showing the heterozygous mutation c.35-2A>G.
Material and Methods
Genomic DNA was extracted from the aborted fetus and peripheral blood of both parents by standard methods. Fetal DNA was analyzed for SF3B4 mutations by Sanger sequencing. The entire SF3B4 coding region and splicing junctions were amplified in 7 PCR fragments using the following primer pairs: F: 5′-TCTTCGCCCCGGAAGT-3′ and R: 5′-CCCACCCCTGCAGAGAC-3′ (exon 1), F: 5′-TGTTTCCATCACTTCCTTCCTT-3′ and R: 5′-TGAATACTGCTGGGACCCT-3′ (exon 2), F: 5′-TCAGTTTATTTCTGGAACCATTCTT-3′ and R: 5′-ACAGAGGTACTGCCCATTCA-3′ (exon 3), F: 5′-GCCTTTGGGGTCATCTTACA-3′ and R: 5′-CCTCAGTCCTCTTCCCATCA-3′ (exon 3), F: 5′-AGGCCAGATCAGGACAGG-3′ and R: 5′-CTGTTGAGGAACAAAGGGCA-3′ (exon 4), F: 5′-GCTGTTCTTGAACTCCTGGG-3′ and R: 5′-AAGTTAGTAAGGGCACGGGA-3′ (exon 5), and F: 5′-TCTAGCCACCTCCCTCATCT-3′ and R: 5′-GGATTAGTACCTTTGCCCCA-3′ (exon 6). Primers were designed using the NCBI/Primer-BLAST software [Ye et al., 2012], and the pairs with partial or total identity with the retro-SF3B4 pseudogene (chr14: 67665808-67666576) were excluded. PCR reactions were carried out under standard conditions. Direct sequencing was performed using a BigDye Terminator v1.1 Cycle Sequencing Kit (Life Technologies, Foster City, Calif., USA) and the standard protocol. Electropherograms were compared with the SF3B4 mRNA reference sequence (NM_005850).
Results and Discussion
A novel heterozygous splicing variant c.35-2A>G was identified in the fetus and confirmed in 2 separately extracted samples of DNA (fig. 1l). This nucleotide change was found neither in the parents' DNA, indicating a de novo nature, nor in 200 control chromosomes from unaffected, ethnicity-matched individuals. Non-paternity was excluded by microsatellite analysis. The dbSNP (www.ncbi.nlm.nih.gov/SNP/) and 1000 Genomes (www.1000genomes.org) databases were searched for the same variant with negative results. In silico prediction using the online NNSPLICE 0.9 version web interface (www.fruitfly.org/seq_tools/splice.html) showed that the identified c.35-2A>G variant abolishes the canonical acceptor splice site of SF3B4 exon 2 while it creates a new acceptor spice site a single nucleotide upstream. This variation was then assumed to retain the last nucleotide of SF3B4 intron 1 into the exon 2 coding sequence, resulting in a frameshift with the creation of a premature stop codon at residue 22 of the coded protein. The presence of the same nucleotide change in the retro-SF3B4 pseudogene (located at chr14:67665811G) pointed out gene conversion as a potential mutagenic mechanism for the identified mutation, but no further study was carried out in order to test this hypothesis.
To date, a total of 46 AFD1 patients from 36 pedigrees (8 familial, 28 sporadic) with a mutation in SF3B4 have been identified [Bernier et al., 2012; Czeschik et al., 2013; Petit et al., 2013]. Thirty distinct SF3B4 mutations are described, including 20 frameshift, 6 nonsense, 2 splice, and 2 initiator codon nucleotide changes. All mutations are predicted to cause loss-of-function, although any functional study or the description of AFD1 patients with a complete gene deletion have not been published to date. Our molecular finding is in line with previous observations and adds a novel pathogenic splicing variant (c.35-2A>G) to the SF3B4 mutation spectrum associated with AFD1. In consideration of the severity of the observed phenotype, our findings permitted an accurate genetic counseling for the couple and, subsequently, the opportunity of offering early prenatal diagnosis.
Genotype-phenotype correlations can not be established with certainty in SF3B4 mutations [Petit et al., 2013]. Among patients with a clinical diagnosis of AFD1, preliminary comparisons suggest a significant increased rate of hearing loss, cleft palate, midface hypoplasia, and thumb aplasia (as opposed to thumb hypoplasia of triphalangeal thumb) in subjects carrying a SF3B4 alteration, compared to those not displaying any mutation [Czeschik et al., 2013]. Our patient showed full-blown characteristics of AFD1, but circumstances did not permit a better characterization of the phenotype, particularly concerning subtle or functional features that are not easily testable in fetuses. In addition to the facial phenotype, the diagnosis was supported by the evocative hand involvement with pre-axial oligodactyly with 4 three-phalangeal, not opposable fingers and distal duplication of the first one. Such a recognizable pattern, well documented in at least 2 additional AFD1 patients with mutations in SF3B4 [Czeschik et al., 2013; Petit et al., 2013], supports the concept that the encoded protein is not only involved in ubiquitous mechanisms of mRNA splicing but may specifically affect bone morphogenesis with different mechanisms, such as inhibition of BMP-mediated osteochondral cell differentiation [Watanabe et al., 2007]. However, specific influence of SF3B4 perturbations on limb patterning is likely an inconstant mechanism in AFD1, as many patients do not display the recognizable digital phenotype. In turn, it is presumed that limb anomalies arise in AFD1 non-specifically, secondary to abnormalities in cell division, as illustrated, for example, by the mouse oligosyndactylism mutants [Pravtcheva and Wise, 2001].
Our patient presented lethal left diaphragmatic hernia. Defective diaphragmatic development is a rare occurrence in AFD1, as it is reported in only 2 previously reported cases, with molecular confirmation in 1 of them [Czeschik et al., 2013; Petit et al., 2013]. The 2 mutations, including ours, reported in AFD1 patients with diaphragmatic hernia do not suggest any consistent genotype-phenotype correlation. However, the repeated observation of diaphragmatic hernia in AFD1 seems to be an association and might further expand the spectrum of non-causal morphogenetic consequences of SF3B4 perturbations. The mechanisms leading to such a low-frequency association remain unknown and may include a variable genetic background, environmental factors, and stochastic events.
References
- 1.Aylsworth AS, Lin AE, Friedman PA. Nager acrofacial dysostosis: male-to-male transmission in 2 families. Am J Med Genet. 1991;41:83–88. doi: 10.1002/ajmg.1320410121. [DOI] [PubMed] [Google Scholar]
- 2.Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, et al. Haploinsufficiency of SF3B4 a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chemke J, Mogilner BM, Ben-Itzhak I, Zurkowski L, Ophir D. Autosomal recessive inheritance of Nager acrofacial dysostosis. J Med Genet. 1988;25:230–232. doi: 10.1136/jmg.25.4.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czeschik JC, Voigt C, Alanay Y, Albrecht B, Avci S, et al. Clinical and mutation data in 12 patients with the clinical diagnosis of Nager syndrome. Hum Genet. 2013;132:885–898. doi: 10.1007/s00439-013-1295-2. [DOI] [PubMed] [Google Scholar]
- 5.Halal F, Herrmann J, Pallister PD, Opitz JM, Desgranges MF, Grenier G. Differential diagnosis of Nager acrofacial dysostosis syndrome: report of four patients with Nager syndrome and discussion of other related syndromes. Am J Med Genet. 1983;14:209–224. doi: 10.1002/ajmg.1320140203. [DOI] [PubMed] [Google Scholar]
- 6.Hall BD. Nager acrofacial dysostosis: autosomal dominant inheritance in mild to moderately affected mother and lethally affected phocomelic son. Am J Med Genet. 1989;33:394–397. doi: 10.1002/ajmg.1320330321. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy SJ, Teebi AS. Newly recognized autosomal recessive acrofacial dysostosis syndrome resembling Nager syndrome. Am J Med Genet A. 2004;129A:73–76. doi: 10.1002/ajmg.a.30113. [DOI] [PubMed] [Google Scholar]
- 8.McDonald MT, Gorski JL. Nager acrofacial dysostosis. J Med Genet. 1993;30:779–782. doi: 10.1136/jmg.30.9.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opitz JM. Nager ‘syndrome’ versus ‘anomaly’ and its nosology with the postaxial acrofacial dysostosis syndrome of Genee and Wiedemann. Am J Med Genet. 1987;27:959–963. doi: 10.1002/ajmg.1320270425. [DOI] [PubMed] [Google Scholar]
- 10.Petit F, Escande F, Jourdain A, Porchet N, Amiel J, et al. Nager syndrome: confirmation of SF3B4 haploinsufficiency as the major cause. Clin Genet doi: 10.1111/cge.12259. [Epub ahead of print] (2013) [DOI] [PubMed]
- 11.Pravtcheva DD, Wise TL. Disruption of Apc10/Doc1 in three alleles of oligosyndactylism. Genomics. 2001;72:78–87. doi: 10.1006/geno.2001.6474. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe H, Shionyu M, Kimura T, Kimata K, Watanabe H. Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. J Biol Chem. 2007;282:20728–20738. doi: 10.1074/jbc.M703292200. [DOI] [PubMed] [Google Scholar]
- 13.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]