Abstract
Kobuviruses are member of the family Picornaviridae. Initially, members in Kobuvirus genus were named according to the basis of their host species. The viruses found in humans called “Aichi virus”, the viruses from cattle called “bovine kobuvirus”, and the viruses isolated from pigs called “porcine kobuvirus”. Currently, taxonomy of kobuviruses has been proposed and the virus species have been renamed. The “Aichi virus” has been renamed as “Aichivirus A”, “bovine kobuvirus” has been renamed as “Aichivirus B”, and “porcine kobuvirus” has been changed to “Aichivirus C”. Among Aichivirus A, three distinct members, including Aichi virus 1 (Aichivirus in human), canine kobuvirus 1, and murine kobuvirus 1, have been described. Aichi virus 1 in human is globally distributed and has been identified at low incidence (0–3 %) in sporadic acute gastroenteritis cases. Aichi virus 1 has been reported to be associated with variety types of clinical illnesses including diarrhea, vomiting, fever, purulent conjunctivitis, and respiratory symptoms. The studies from Japan, Spain, Germany, and Tunisia demonstrated that high antibody prevalence against Aichi virus 1 were found in the populations. Aichivirus B or previously known as bovine kobuvirus was first reported in 2003. Since then, Aichivirus B has also been reported from several countries worldwide. An overall prevalence of Aichivirus B varies from 1 to 34.5 %, and the highest prevalence was found in cattle with diarrhea in Korea. Aichivirus C or porcine kobuvirus is widely distributed in pigs. Aichivirus C has been found in both diarrhea and healthy pigs and the positive rate of this virus varies from 3.9 up to 100 %. It was reported that Aichivirus C was found with high prevalence in wild boars in Hungary. The accumulated data of the biological, pathological, as well as epidemiological studies of kobuviruses are still limited. Comprehensive global investigations of the prevalence and diversity are required and will be helpful for providing further insight into pathogenicity, genetic heterogeneity, interspecies transmission, and global distribution of kobuviruses.
Keywords: Kobuvirus, Aichivirus, Diarrhea, Epidemiology
Introduction
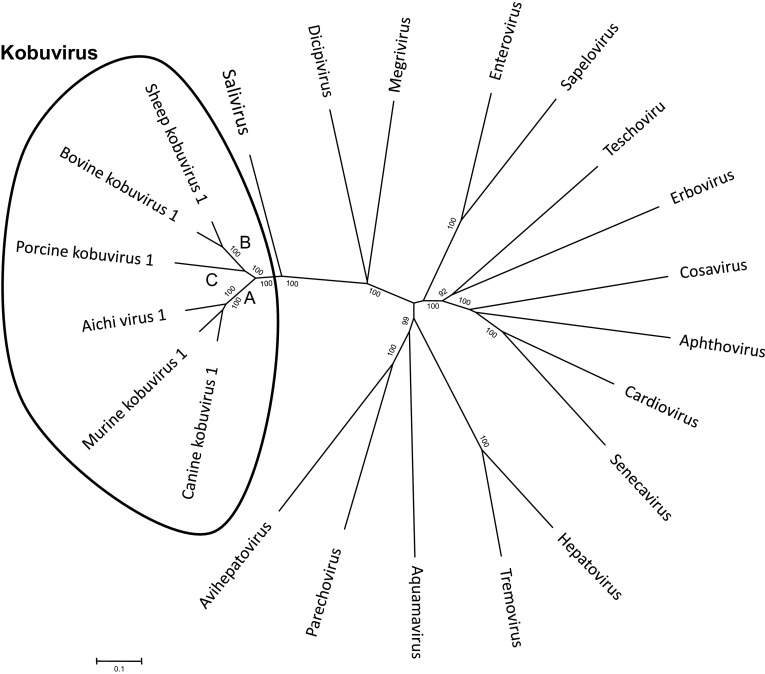
Acute gastroenteritis is one of the most common diseases in children and adults, and continues to be a significant cause of morbidity and mortality worldwide [3]. Most recently, several viruses related to diarrhea have been discovered in human stool samples, mostly by viral metagenomic strategy. Once emerging viruses related to diarrheal diseases are discovered, there is a need to constantly monitor the prevalence of these viruses in the community for clarification of their clinical significance. Initially, members in Kobuvirus genus, Picornaviridae family, were named according to the basis of their host tropisms. The viruses found in humans called “Aichi virus”, the viruses from cattle called “bovine kobuvirus”, and the viruses isolated from pigs called “porcine kobuvirus” [21]. However, currently taxonomy of kobuviruses has been proposed by International Committee on Taxonomy of Viruses (ICTV: http://www.ictvonline.org/virusTaxonomy.asp) and the virus species have been renamed [1]. The “Aichi virus” has been renamed as “Aichivirus A”, “bovine kobuvirus” has been renamed as “Aichivirus B”, and “porcine kobuvirus” has been changed to “Aichivirus C” (Fig. 1). In addition to these three host species, kobuviruses have also been found in other animal species. The murine kobuvirus 1 and canine kobuvirus 1 are found to be related to the human Aichi virus in Aichivirus A cluster, while sheep kobuvirus 1 is the member of Aichivirus B.
Fig. 1.
Relationship between kobuviruses and other picornaviruses based on nucleotide sequences of complete coding region
History
The Aichivirus prototype strain was found initially in Aichi prefecture, Japan, in 1989 [30]. The spherical virus-like particles of approximately 30 nm in diameter were detected by electron microscopy (EM) in fecal specimens of oyster-associated nonbacterial gastroenteritis patients. The agents from fecal samples could produce cytopathic effect (CPE) in BS-C-1 cells. By reverse transcription-polymerase chain reaction (RT-PCR) or the enzyme-linked immunosorbent assay (ELISA), this new agent was not related to previously known acute gastroenteritis viruses [31]. Therefore, this agent was initially called “Aichi virus” as it was firstly detected in Aichi prefecture in Japan. Later, complete nucleotide sequence of Aichi virus strain A846/88 was characterized and the results leaded to the conclusion that Aichi virus should be classified as a new virus in the genus separated from other known genera of the Picornaviridae family and so-called genus “Kobuvirus” [33]. The word “Kobu” means bump or knob in Japanese, because under EM the morphological characteristic of the virus particle appears to be bumpy [34].
A prototype strain of bovine kobuvirus or Aichivirus B, named U-1, was first recognized as a new species of kobuvirus in 2003 as a cytopathic agent contaminated in culture medium of HeLa cells that had been used for more than 30 years in the laboratory [29]. The genome organization of the U-1 strain is analogous to that of picornaviruses. Morphologically, the U-1 strain resembles the Aichi virus, while genetically it is relatively distinct [29]. Therefore, the U-1 strain is classified as another species of kobuvirus genus in the Picornaviridae family, and it is called “bovine kobuvirus” or “Aichivirus B”.
Porcine kobuvirus or Aichivirus C has been discovered recently in pigs by two different groups of investigators from Hungary and China [24, 37]. By RT-PCR for the detection of caliciviruses in domestic pigs, the unexpected band of PCR products of about 1,100 bp were found in these samples. The nucleotide sequence of the non-specific PCR fragments were determined and found to be similar to the 3CD region of human Aichi virus and bovine kobuvirus [24, 37]. Later, complete nucleotide sequences of porcine kobuvirus strain S-1-HUN from Hungary and Y-1-CHI from China were performed and demonstrated that they were considerably distinct from human Aichi virus and bovine kobuvirus, and then were considered as the third species in the genus Kobuvirus [19, 36].
Virus classification and biology
The family Picornaviridae is genetically highly diverse, currently comprising 17 genera, many of which consist of several species, subspecies, and genotypes [1]. Kobuvirus is one of a newly recognized genus in the family Picornaviridae. The kobuvirus particle is a small round non-enveloped, approximately 30 nm in diameter. The kobuvirus genome is a single-stranded RNA molecule of about 8.2–8.3 kb long and contains a large open reading frame coding for a single polyprotein [19, 33, 34, 36]. The kobuvirus genome can be divided into three distinct functional protein-encoded regions termed P1 (encoded structural proteins), P2 and P3 (encoded non-structural proteins). The organization of kobuvirus genome consists of VPg, 5′UTR, leader protein, 3 structural viral proteins (VP0, VP3, and VP1), 7 nonstructural proteins (2A–2C and 3A–3D), 3′UTR, and poly(A) tail.
The kobuvirus strains have been isolated from various host species and genetic variability between strains allows the classification of kobuvirus into three different species, Aichivirus A, B, and C (Fig. 1). According to the 9th ICTV report, members in the Aichivirus species should be shared greater than 70 % amino acid (aa) identity in the polyprotein, 70 % aa identity in P1, 80 % aa identity in 2C and 3CD regions [1]. Current taxonomy of kobuvirus demonstrated that Aichivirus A comprises of three distinct members, including Aichi virus 1 (found in humans), canine kobuvirus 1, and murine kobuvirus 1 [12, 18, 30]. Aichivirus B consists of two members, bovine kobuvirus 1 and sheep kobuvirus 1 [20, 29]. For Aichivirus C, only a single type of porcine kobuvirus 1 is identified [24, 37].
The prototype strain of Aichi virus 1 (A846/88) is cytopathic in African green monkey kidney epithelial cell lines such as BS-C-1 and Vero cell lines. However, bovine kobuvirus and porcine kobuvirus cannot readily be propagated in cell cultures. Up to now, a formal genotype classification system of kobuviruses has not yet been well established, excepted for Aichi virus 1 that found in humans. Based on nucleotide and deduced amino acid sequence analyses, at least 3 genotypes (genotypes A, B, and C) of Aichi virus 1 have been described [2].
For the viral pathogenesis and evolution, although kobuviruses are thought to be gastrointestinal pathogens as they have been found in fecal samples, the target cells, specific receptors, cell entry process, as well as viral evolutionary mechanism are yet to be understood. One of the evolutionary mechanisms of Aichi virus 1 is thought to be due to viral genome recombination, as the mosaic genomes of Aichi virus 1 can be found in several strains from different countries [5]. Most of the studies on kobuvirus replication have also been conducted for Aichi virus 1. Sasaki et al. [26] demonstrated that a stem-loop structure at the 5′ end of the genome and L protein play the essential role in the viral RNA replication and encapsidation. In addition, it has been reported that the interaction of 3ABC of P3 protein with the 5′ terminal region of the genome is involved in negative strand synthesis [14].
Several studies demonstrated that kobuvirus is transmitted through fecal-oral route or consumption of contaminated food or water. Kobuvirus has been suspected as a causative agent of gastroenteritis in humans and other animal species. Aichi virus 1 was first isolated in stool samples of acute gastroenteritis patients in Japan after consumption of raw oysters [30]. Later, several outbreaks caused by Aichi virus 1 are found to be linked with oyster or seafood consumption [2, 34]. In addition, contaminations of Aichi virus 1 have been found in untreated sewage water [10]. Presumably, the kobuviruses are transmitted primarily via the fecal-oral route, either by person-to-person or by ingestion of contaminated food and water. This hypothesis is supported by the finding of high Aichi virus 1 RNA copy numbers of up to 1.32 × 1012 per gram of stool in patients suffering from gastroenteritis [4].
Epidemiology of kobuviruses
Aichivirus A (Aichi virus)
Among Aichivirus A, three distinct members, including Aichi virus 1 (Aichivirus in human), canine kobuvirus 1, and murine kobuvirus 1, have been described. Aichi virus 1 in human is globally distributed and has been identified at low incidence in sporadic acute gastroenteritis cases. Transmission route thought to be fecal-oral transmission. Prevention of Aichivirus A infection, therefore, relies primarily on the provision of clean drinking water, improving personal hygiene, and avoiding consumption of raw, uncooked food. Aichi virus 1 has been reported to be associated with a wide range of clinical illnesses including diarrhea, vomiting, fever, purulent conjunctivitis, and respiratory symptoms. However, serological studies demonstrated that antibody prevalence were rising by age in Japanese population. Approximately 30 % of young adulthood are seropositive for Aichi virus 1 and rapidly increases to 80 % by middle age [31]. The studies in Spain, Germany, and Tunisia also confirm that 70, 76, and 92 % of the populations across all age groups are seropositive against Aichi virus 1 [15, 25, 27]. Since it discovery, Aichi virus 1 has been detected from several outbreaks or acute gastroenteritis patients both in young children and adults. It was reported that Aichi virus 1 antigen was found in stool samples of travelers who returned to Japan from the Southeast Asian countries at about 0.7 % [32]. A report from China reveals that 1.8 % of the specimens collected from hospitalized children in Shanghai were positive for this viral agent [35]. In addition, 28 of 912 fecal specimens from acute gastroenteritis pediatric patients from several countries in Asia including Japan, Thailand, Vietnam, and Bangladesh were positive for Aichi virus 1 at about 3 % [17]. In Europe, the prevalence of Aichi virus 1 has been reported in Finland, France, Hungary, Germany, and Sweden, ranging from 0.5 to 1.5 % [15]. In addition to Asia and Europe, Aichi virus 1 related to acute gastroenteritis in human was also reported from Brazil and Tunisia [15, 28]. Recently, Aichi-like virus 1 have been found in other animal species, such as canine kobuvirus (Canine kobuvirus 1) was found from both healthy and diarrhea domestic dogs [12], while mouse kobuvirus (Murine kobuvirus 1) was found widely in wild rodents [18]. These three viruses formed the same genetic cluster within Aichivirus A. It is likely possible that kobuviruses are widely distributed among many animal species.
Aichivirus B (Bovine kobuvirus)
Aichivirus B or previously known as bovine kobuvirus was first identified in Japan in cell culture medium when the infected cattle serum was used and the initial molecular epidemiology of Aichivirus B in stool samples of healthy cattle was performed in Japan. It was found that 16.7 % were positive for this viral agent [29]. Since then, Aichivirus B has also been reported from several countries worldwide, including Thailand, Korea, Belgium, Hungary, the Netherlands, Italy, and Brazil [8, 13, 21, 22]. Using RT-PCR technique, an overall prevalence of Aichivirus B varies from 1 to 34.5 %, and the highest prevalence was found in cattle with diarrhea in Korea. In addition, the bovine-like kobuviruses have also been detected recently in domestic sheep in Hungary and black goats in Korea [11, 20], suggesting that Aichivirus B has a worldwide distribution in ruminants including cattle, sheep, and goat populations.
Aichivirus C (Porcine kobuvirus)
Aichivirus C or porcine kobuvirus is widely distributed in pigs in several countries such as the USA, Hungary, Italy, the Czech Republic, the Netherlands, Brazil, China, Korea, Japan, and Thailand [21]. The presence of Aichivirus C in pigs has been evaluated primarily in stool samples of both diarrhea and healthy animals. However, recent studies have demonstrated that Aichivirus C genomes have also found in serum samples, indicating that viremia has occurred in domestic pigs [21]. In addition to direct fecal-oral transmission, the possibility of Aichivirus C transmission through breast-feeding, bloodborne, foodborne, and zoonotic infections, as well as the clinical significance of this viral agent remain unclear. Aichivirus C has been found in both diarrhea and healthy pigs and the positive rate of this virus varies from 3.9 up to 100 %. It was reported that Aichivirus C was found with high prevalence in wild boars in Hungary. The finding suggests that wild boars could also be an important host and reservoir for kobuvirus [23].
The evidences of zoonotic infection or interspecies transmission of bovine kobuvirus to pig was documented recently [6, 7, 16]. The factors that promote interspecies transmission are poorly understood, however, it could be possible that close contact between different animal species may increase chance of the occurrence of interspecies transmission. The study from Japan demonstrated that one specimen (H023/2009/JP) from 6 months old pig in Hokkaido was positive for kobuvirus by RT-PCR. Sequence and phylogenetic analysis of the H023/2009/JP revealed that this virus strain was most closely related to bovine kobuvirus than with porcine kobuvirus. The finding implies that interspecies transmission of bovine kobuvirus to pigs or vice versa might occur in nature [6, 16].
Kobuvirus detection
For the detection of kobuviruses, several methods are available. These include EM, EIA, ELISA, RT-PCR, real-time PCR, and nucleic acid sequence analysis. Among these methods, RT-PCR, and nucleic acid sequence analysis are widely used for the detection and genotyping of kobuviruses. Initially, screening primers specific for Aichivirus A and Aichivirus B have been designed separately. Later, consensus kobuvirus primers have been designed based on the conserved sequences of the three species including Aichivirus A, B, and C [19]. The oligonucleotide primers used for the detection of Aichiviruses are summarized in Table 1.
Table 1.
Oligonucleotide primers for the detection of kobuviruses
| Primer name | Region | Sequence 5′-3′ | Sense | PCR Product size | Species specific | Reference | |
|---|---|---|---|---|---|---|---|
| 6261 | 3C–3D | First PCR | ACACTCCCACCTCCCGCCAGTA | + | Aichivirus A | [34] | |
| 6779 | 3C–3D | First PCR | GGAAGAGCTGGGTGTCAAGA | – | 519 bp | Aichivirus A | [34] |
| C94b | 3C–3D | Second PCR | GACTTCCCCGGAGTCGTCGTCT | + | Aichivirus A | [34] | |
| 246k | 3C–3D | Second PCR | GACATCCGGTTGACGTTGAC | – | 223 bp | Aichivirus A | [34] |
| C94b | 3C | – | GACTTCCCCGGAGTCGTCGTCT | + | Aichivirus A | [34] | |
| AiMP-R | 3C | – | GCRGAGAATCCRCTCGTRCC | – | 158 | Aichivirus A | [9] |
| 10f | 3D | – | GATGCTCCTCGGTGGTCTCA | + | Aichivirus B | [29] | |
| 10r | 3D | – | GTCGGGGTCCATCACAGGGT | – | 631 bp | Aichivirus B | [29] |
| UNIV-kobu-F | 3D | – | TGGAYTACAARTGTTTTGATGC | + | Aichivirus A, B, C | [19] | |
| UNIV-kobu-R | 3D | – | ATGTTGTTRATGATGGTGTTGA | – | 216 bp | Aichivirus A, B, C | [19] |
Conclusions and future perspectives
Diarrhea is one of the most common illnesses in humans and animals worldwide. In recent years, several novel viruses have been discovered. The recent applications of molecular methods for screening and characterization of kobuviruses reveal the greater prevalence, genetic diversity, and heterogeneity in their epidemiology. The discovery of kobuviruses, including Aichivirus A, Aichivirus B, and Aichivirus C reveal the linkage or may play a role as the causative agents of these viruses to gastroenteritis diseases. The accumulated data of the biological, pathological, as well as epidemiological studies of the viruses in the genus kobuvirus are still limited. Comprehensive global investigations of the prevalence and diversity are required and will be helpful for providing further insight into pathogenicity, genetic heterogeneity, interspecies transmission, and global distribution of kobuviruses.
Acknowledgments
This research was supported by grant for Medical Research, Faculty of Medicine, Chiang Mai University, Thailand and the JSPS Grant-in-Aid for Scientific Research (B), Grant Numbers 23406036 and 24390266, Japan.
References
- 1.Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambert-Balay K, Lorrot M, Bon F, Giraudon H, Kaplon J, Wolfer M, Lebon P, Gendrel D, Pothier P. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J Clin Microbiol. 2008;46:1252–1258. doi: 10.1128/JCM.02140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennehy PH. Viral gastroenteritis in children. Pediatr Infect Dis J. 2011;30:63–64. doi: 10.1097/INF.0b013e3182059102. [DOI] [PubMed] [Google Scholar]
- 4.Drexler JF, Baumgarte S, de Souza Luna LK, Eschbach-Bludau M, Lukashev AN, Drosten C. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg Infect Dis. 2011;17:1544–8. [DOI] [PMC free article] [PubMed]
- 5.Han X, Zhang W, Xue Y, Shao S. Sequence analysis reveals mosaic genome of Aichi virus. Virol J. 2011;8:390. doi: 10.1186/1743-422X-8-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khamrin P, Maneekarn N, Hidaka S, Kishikawa S, Ushijima K, Okitsu S, Ushijima H. Molecular detection of kobuvirus sequences in stool samples collected from healthy pigs in Japan. Infect Genet Evol. 2010;10:950–954. doi: 10.1016/j.meegid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Khamrin P, Maneekarn N, Kongkaew A, Kongkaew S, Okitsu S, Ushijima H. Porcine kobuvirus in piglets, Thailand. Emerg Infect Dis. 2009;15:2075–2076. doi: 10.3201/eid1512.090724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khamrin P, Maneekarn N, Peerakome S, Okitsu S, Mizuguchi M, Ushijima H. Bovine kobuviruses from cattle with diarrhea. Emerg Infect Dis. 2008;14:985–986. doi: 10.3201/eid1406.070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khamrin P, Okame M, Thongprachum A, Nantachit N, Nishimura S, Okitsu S, Maneekarn N, Ushijima H. A single-tube multiplex PCR for rapid detection in feces of 10 viruses causing diarrhea. J Virol Methods. 2011;173:390–393. doi: 10.1016/j.jviromet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Kitajima M, Haramoto E, Phanuwan C, Katayama H. Prevalence and genetic diversity of Aichi viruses in wastewater and river water in Japan. Appl Environ Microbiol. 2011;77:2184–2187. doi: 10.1128/AEM.02328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MH, Jeoung HY, Lim JA, Song JY, Song DS, An DJ. Kobuvirus in South Korean black goats. Virus Genes. 2012;45:186–189. doi: 10.1007/s11262-012-0745-6. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauroy A, Scipioni A, Mathijs E, Thys C, Thiry E. Molecular detection of kobuviruses and recombinant noroviruses in cattle in continental Europe. Arch Virol. 2009;154:1841–1845. doi: 10.1007/s00705-009-0518-2. [DOI] [PubMed] [Google Scholar]
- 14.Nagashima S, Sasaki J, Taniguchi K. Interaction between polypeptide 3ABC and the 5′-terminal structural elements of the genome of Aichi virus: implication for negative-strand RNA synthesis. J Virol. 2008;82:6161–6171. doi: 10.1128/JVI.02151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh DY, Silva PA, Hauroeder B, Diedrich S, Cardoso DD, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 16.Okitsu S, Khamrin P, Thongprachum A, Hidaka S, Kongkaew S, Kongkaew A, Maneekarn N, Mizuguchi M, Hayakawa S, Ushijima H. Sequence analysis of porcine kobuvirus VP1 region detected in pigs in Japan and Thailand. Virus Genes. 2012;44:253–257. doi: 10.1007/s11262-011-0692-7. [DOI] [PubMed] [Google Scholar]
- 17.Pham NT, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Ushijima H. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45:2287–2288. doi: 10.1128/JCM.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter G, Boldizsár A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 20.Reuter G, Boros A, Pankovics P, Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis. 2010;16:869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter G, Boros A, Pankovics P. Kobuviruses: a comprehensive review. Rev Med Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- 22.Reuter G, Egyed L. Bovine kobuvirus in europe. Emerg Infect Dis. 2009;15:822–823. doi: 10.3201/eid1505.081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter G, Nemes C, Boros A, Kapusinszky B, Delwart E, Pankovics P. Porcine kobuvirus in wild boars (Sus scrofa) Arch Virol. 2013;158:281–282. doi: 10.1007/s00705-012-1456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter G, Boldizsár A, Kiss I, Pankovics P. Candidate new species of Kobuvirus in porcine hosts. Emerg Infect Dis. 2008;14:1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribes JM, Montava R, Téllez-Castillo CJ, Fernández-Jiménez M, Buesa J. Seroprevalence of Aichi virus in a Spanish population from 2007 to 2008. Clin Vaccine Immunol. 2010;17:545–549. doi: 10.1128/CVI.00382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki J, Nagashima S, Taniguchi K. Aichi virus leader protein is involved in viral RNA replication and encapsidation. J Virol. 2003;77:10799–10807. doi: 10.1128/JVI.77.20.10799-10807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sdiri-Loulizi K, Hassine M, Bour JB, Ambert-Balay K, Mastouri M, Aho LS, Gharbi-Khelifi H, Aouni Z, Sakly N, Chouchane S, Neji-Guédiche M, Pothier P, Aouni M. Aichi virus IgG seroprevalence in Tunisia parallels genomic detection and clinical presentation in children with gastroenteritis. Clin Vaccine Immunol. 2010;17:1111–1116. doi: 10.1128/CVI.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sdiri-Loulizi K, Hassine M, Gharbi-Khelifi H, Sakly N, Chouchane S, Guediche MN, Pothier P, Aouni M, Ambert-Balay K. Detection and genomic characterization of Aichi viruses in stool samples from children in Monastir, Tunisia. J Clin Microbiol. 2009;47:2275–2278. doi: 10.1128/JCM.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84:3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 31.Yamashita T, Sakae K, Ishihara Y, Isomura S, Utagawa E. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J Clin Microbiol. 1993;31:2938–2943. doi: 10.1128/jcm.31.11.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita T, Sakae K, Kobayashi S, Ishihara Y, Miyake T, Mubina A, Isomura S. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol Immunol. 1995;39:433–435. doi: 10.1111/j.1348-0421.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Miyamura T, Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol. 1998;72:8408–8412. doi: 10.1128/jvi.72.10.8408-8412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita T, Sugiyama M, Tsuzuki H, Sakae K, Suzuki Y, Miyazaki Y. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J Clin Microbiol. 2000;38:2955–2961. doi: 10.1128/jcm.38.8.2955-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S, Zhang W, Shen Q, Yang Z, Zhu J, Cui L, Hua X. Aichi virus strains in children with gastroenteritis, China. Emerg Infect Dis. 2009;15:1703–1705. doi: 10.3201/eid1510.090522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu JM, Xu ZQ, Li BW, Zhang Q, Cui SX, Jin M, Duan ZJ. Analysis and characterization of the complete genome of a member of a new species of kobuvirus associated with swine. Arch Virol. 2011;156:747–751. doi: 10.1007/s00705-010-0907-6. [DOI] [PubMed] [Google Scholar]
- 37.Yu JM, Jin M, Zhang Q, Li HY, Li DD, Xu ZQ, Li JS, Cui SX, Yang SH, Liu N, Duan ZJ. Candidate porcine Kobuvirus, China. Emerg Infect Dis. 2009;15:823–825. doi: 10.3201/eid1505.081518. [DOI] [PMC free article] [PubMed] [Google Scholar]