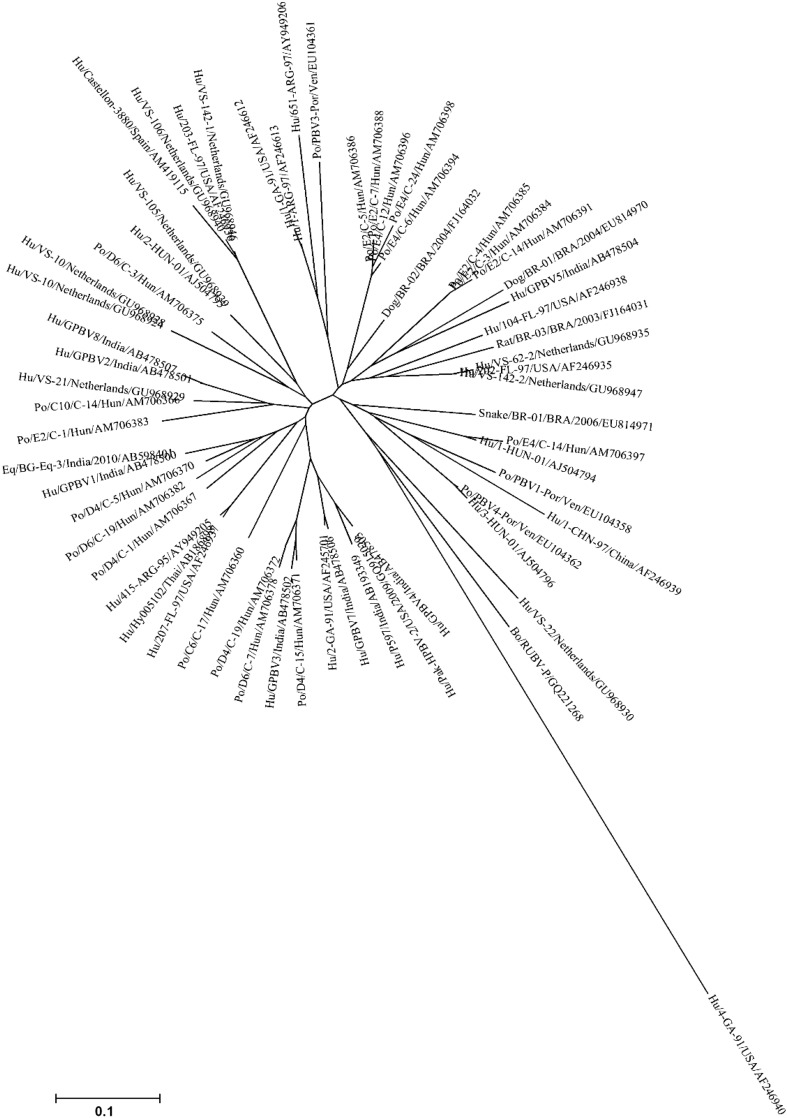
Fig. 6.
The phylogenetic tree showing the overall genetic diversity and evolutionary differences of genogroup I PBV strains detected from different hosts and from different geographical locales. The genogroup I PBV picobirnavirus strains reported from human, porcine, bovine, canine, murine and serpentine hosts, based on partial amino acid sequence [56 amino acids (aa)] of genomic segment 2 were taken from the public DNA Databank NCBI/DDBJ/EMBL. The tree was rooted with cognate stretch of gene segment 2 of genogroup II prototype strain Hu/4-GA-91 (USA) defined as the outgroup strain. The phylogenetic tree was inferred using the MEGA software (Version 4.1) program in combination with the neighbor-joining method, with evolutionary distances computed using the p-distance method. A bootstrap value >90% was indicated for the corresponding nodes based on a resampling analysis of 1000 replicates. The scale bar represents 0.1 substitution per 100 nucleotide residues