Abstract
Pseudocowpox virus (PCPV) infects cattle throughout the world and has zoonotic potential. However, it is not known to infect cattle in Turkey. In August 2013, we observed ulcerative nodular swelling and pustules on udder and teats of a cow in a small village nearly Lake of Bafa, Milas, Mugla locate in southwestern part of Turkey. Interestingly, the similar lesions were also observed on skin of milkier women’s hand at the same time. A PCPV strain was characterized based on the major envelop gene sequence. The phylogenetic analysis showed that the isolated strain was closely related to the members of other parapoxvirus genus. This study provides the first description of PCPV infection in Turkey.
Keywords: Pseudocowpox virus, Cattle, Turkey, Zoonose
The pseudocowpox virus (PCPV) is a member of the genus Parapoxvirus that infects vertebrates, and is classified into the Chordopoxvirinae subfamily, under to the family Poxviridae [3]. The virus causes contagious ecthyma in sheep goats and dairy cattle. The parapoxviruses cause zoonotic infections, and these occupational exanthematic skin diseases are important public health concerns as they can infect humans via direct contact.
The PCPV infection presents with sporadic or endemic dissemination in many regions of the world [1, 11, 13]. Turkey has reported parapoxvirus infections in different animals and humans [7, 8, 12]; however, before this study, there were no record of infection caused by the parapoxvirus genus in Turkey, especially PCPV in cattle.
In this study, we have identified and diagnosed the Turkish pseudocowpox virus infection in cattle, based on the analysis of the major envelope gene region of parapoxviruses.
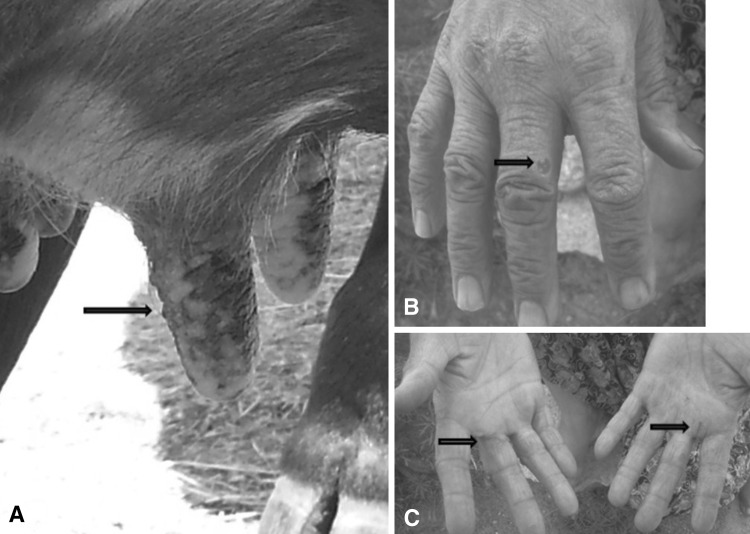
The dried exudate material (scab) used in this study originated from a cow in a small village (Karahayıt, 37′29′09.03″N, 27′34′19.38″E) in Muğla, Turkey. It was clinically observed that some exanthematic lesions, which began with nodular swelling, becoming a papule, and then developing into an ulcerative pustule, were present on the udders and teats of an 8 year-old cow. According to the owner of the animals, all of the cattle (n = 6) were between 4 and 10 years old and in lactation periods. Additionally, three animals in this herd had the same lesions on their udders. Secondary infections were not reported and the lesions healed in 20–25 days without any scars.
In this village, the same lesions have been described on the hands of women milkmaids (Fig. 1). These lesions in humans were characterized by reddish, sensitive nodules, a dry crust in the center of the nodules, and an erythematous zone surrounding the nodules.
Fig. 1.
a The clinical presentation of udders and teats of the cow whose scab material was obtained in this study. b Milker woman’s hand affected from stated cow. c Milker woman’s healed palms
In the laboratory, the dried exudate sample was mechanically homogenized with phosphate buffered saline (PBS) after a freeze-thawing period at −20 °C. The conventional method [9] based on phenol-chloroform-isoamyl alcohol (24:25:1) was applied for viral DNA extraction, and the obtained pellet was solubilized with 20 µl sterile distilled water. The primers and conditions used for the PCR are described by Guo et al. [4], with DNA from Madin–Darby bovine kidney cells used as the negative control. The 1170 bp amplicon of the major envelope protein gene of the PCPV genome was purified using a commercial purification kit (Invitrogen, Germany).
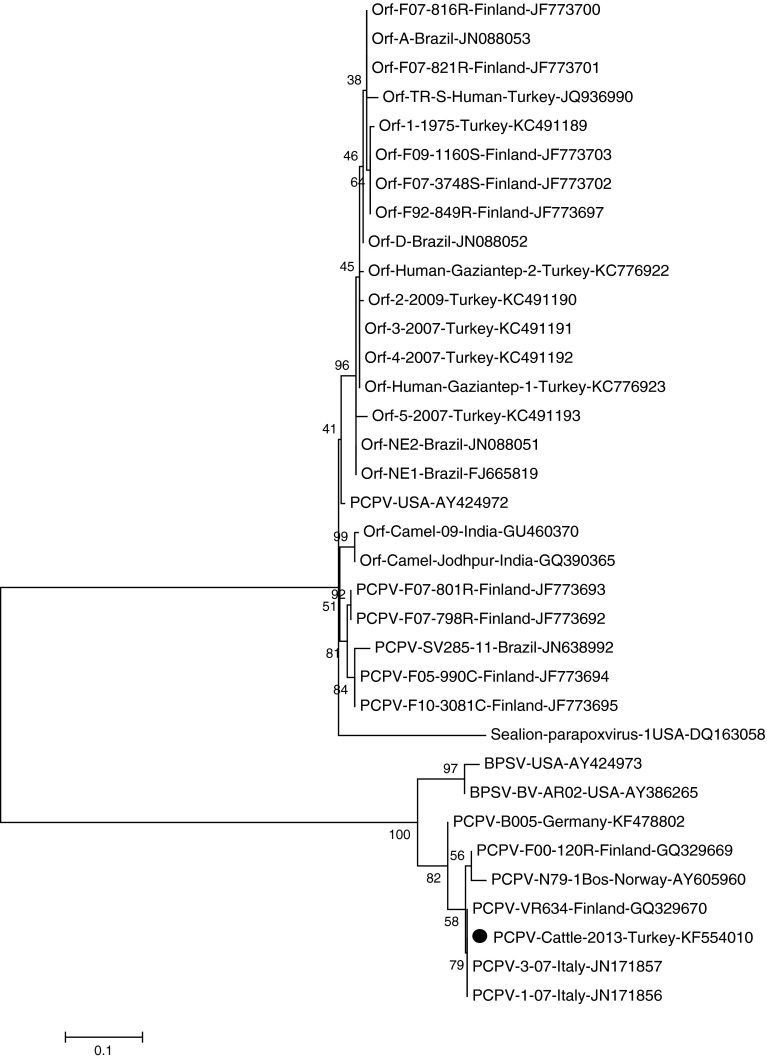
The sequence analysis of the purified PCR product was performed with a genetic analyzer (Applied Biosystems, Ankara). The data were analyzed using the BioEdit (v.7.2.4) [5] and MEGA 5 (v.5.2.2) software [10] (Fig. 2), and the phylogenetic tree was constructed using the Maximum Likelihood method. The significance of the deduced tree was verified by the bootstrapping analysis of 1,000 replicates, and the B2L sequence of the sea lion parapoxvirus was used as an out group.
Fig. 2.
The molecular phylogenetic analysis was inferred by using the Maximum Likelihood method based on the Tamura-Nei model in MEGA5. The tree was constructed from the nucleotide sequences of the major envelope protein genes from different parapoxviruses. A reference PCPV strain VR634 used in alignment which all sequences were compared against nucleotide numbers 17,847–18,983 of their complete genome. The phylogenetic tree with bootstrapping with 1,000 replicates was constructed with the highest log likelihood (−1088.2 199). The analysis involved 35 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding
The phylogenetic analysis results of the obtained nucleic acid from the exudate sample material verified that it contained PCPV (Fig. 2). Thus, the clinical diagnosis confirmed (Fig. 1) the infection with the sequence results. The one PCPV strain from this study has the accession number KF554010 in GenBank.
We have compared our sequence with the widespread orf virus, PCPV, and BPSV from different animals and humans available in the GenBank database. The results of this comparison were variable; for example, the sequence of the Turkish PCPV strain indicated 43 % heterology with reindeer sequences from Finland. However, it also indicated a low divergence rate with a major envelope gene comparison of the complete genome of another reindeer PCPV isolate, VR634, 1.5 %. Additionally, a Turkish PCPV strain from cattle was found to have a low similarity (49 %) with a cattle PCPV strain from Germany, and was located in a separate branch. Interestingly, in the phylogenetic tree, numbers AY424972 (USA) and JF773693 (Finland) are non-ovine in origin, but are placed closer to ovine parapoxviruses. It was apparent that the Turkish PCPV strain obtained from cattle was closer to the bovine parapoxviruses than to ovine parapoxviruses.
The objective of the present study was to diagnose the PCPV infection in exudate material from cattle by PCR. The 1,170 bp amplicon from the major envelope gene of parapoxviruses was sequenced and compared with other parapoxviruses obtained from the GenBank database. This study is the first to describe the presence of the PCPV infection in Turkey.
The genetic analysis results indicated that the PCPV in Turkey is closely related to the other parapoxviruses, especially those infecting sheep and goats. Although PCPV and orf viruses infect different animal hosts, they cause similar clinical lesions in humans. This similarity may be important for epidemiology and public health. Until this study, the orf virus has only been reported as a causative agent of parapoxvirus infections in humans in Turkey. In accordance with the obtained history from the village where the infected animal was found, humans can also be infected with the same PCPV strain. Therefore, PCPV infections should be considered in conjunction with orf virus infections in epidemiological surveillance studies, and in differential diagnosis by clinicians in hospitals.
Fairley et al. [2] has reported a PCPV infection in a cat, which was transferred from a cow, and that possibly, this may have been associated with some deficiency in the immune system of the cat. The crossing of the species barrier can cause mutations, and PCPV can infect other animals and humans [6]. Thus, these findings have expanded the host range of PCPV, and suggest that new mutated viruses could be isolated from animal species which have never been diagnosed with PCPV infections. Once the species barrier is crossed, the development of new mutant viruses is possible, and gives rise to the likelihood of human to human and/or animal to animal transmission.
In this study, we have reported the presence of a PCPV infection in Turkey for the first time. Additionally, the Turkish PCPV strain has been molecularly characterized using the partial major envelope protein gene sequence.
Zoonotic PCPV is present in the cattle population of Turkey, and it is important to include this agent in bovine and human vesicular disease diagnoses. Due to PCPV’s zoonotic potential and animal movements, when its risk of dissemination is taken into consideration, it is important to inform farmers, milkmaids, animal keepers, butchers, veterinarians, and other humans who may come in contact with these animals in Turkey.
PCPV or milker’s nodules affects milkmaids and farmers, and therefore, we recommend wearing gloves in the milking shed, thoroughly washing teats and udders before the milking process, and the disinfection of contaminated milking equipment.
References
- 1.Cargnelutti JF, Flores MM, Teixeira FRM, Weiblen R, Flores EF. An outbreak of pseudocowpox in fattening calves in southern Brazil. J Vet Diagn Invest. 2012;24(2):437–441. doi: 10.1177/1040638711435408. [DOI] [PubMed] [Google Scholar]
- 2.Fairley A, Mercer AA, Copland CI, Craig SM, Heslip PA. Persistent pseudocowpox virus infection of the skin of a foot in a cat. New Zeal Vet J. 2013;61(4):242–243. doi: 10.1080/00480169.2012.757728. [DOI] [PubMed] [Google Scholar]
- 3.Fauquet CM, Mayo MA. The viruses. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA editors. Eighth report of the International Committee on the taxonomy of viruses. Amsterdam: Elsevier Academic Press; 2005.
- 4.Guo J, Rasmussen J, Wünschmann A, de la Concha-Bermejillo A. Genetic characterization of orf viruses isolated from various ruminant species of a zoo. Vet Microbiol. 2004;99:81–92. doi: 10.1016/j.vetmic.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/Nt. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 6.Hautaniemi M, Ueda N, Tuimala J, Mercer AA, Lahdenperä J, McInnes CJ. The genome of pseudocowpoxvirus: comparison of a reindeer isolate and a reference strain. J Gen Virol. 2010;91:1560–1576. doi: 10.1099/vir.0.018374-0. [DOI] [PubMed] [Google Scholar]
- 7.Karakaş A, Oğuzoğlu TC, Coskun O, Artuk C, Mert G, Gul HC, Sener K, Ozkul A. First molecular characterization of the Turkish Orf virus strain from a human based on a partial B2L sequence. Arch Virol. 2013;158:1105–1108. doi: 10.1007/s00705-012-1575-5. [DOI] [PubMed] [Google Scholar]
- 8.Midilli K, Erkılıc A, Kuşkucu M, Analay H, Erkılıc S, Benzonana N, Yıldırım MS, Mulayim K, Acar H, Ergonul O. Nosocomial outbreak of disseminated Orf infection in a burn unit, Gaziantep, Turkey, October to December 2012. Euro Surveill. 2013;18(11):20425. [DOI] [PubMed]
- 9.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 10.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tikkanen MK, McInnes CJ, Mercer AA, Büttner M, Tuimala J, Hirvelä-Koski V, Neuvonen E, Huovilainen A. Recent isolates of parapoxvirus of Finnish reindeer (Rangifer tarandus tarandus) are closely related to bovine pseudocowpox virus. J Gen Virol. 2004;85:1413-8. [DOI] [PubMed]
- 12.Toplu N, Aydogan A, Oguzoglu TC. Visceral leishmaniosis and parapoxvirus infection in a mediterranean monk seal (Monachus Monachus) J Comp Pathol. 2007;136:283–287. doi: 10.1016/j.jcpa.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Vikøren T, Lillehaug A, Âkerstedt J, Bretten T, Haugum M, Trylande M. A severe outbreak of contagious ecthyma (orf) in a free-ranging musk ox (Ovibos moschatus) population in Norway. Vet Microbiol. 2008;127:10–20. doi: 10.1016/j.vetmic.2007.07.029. [DOI] [PubMed] [Google Scholar]