Abstract
The forkhead box O (FOXO) family has been extensively investigated in aging and metabolism, but its role in tissue-repair processes remains largely unknown. Herein, we clarify the molecular aspect of the FOXO family in skin wound healing. We demonstrated that Foxo1 and Foxo3a were both up-regulated during murine skin wound healing. Partial knockout of Foxo1 in Foxo1+/− mice throughout the body led to accelerated skin wound healing with enhanced keratinocyte migration, reduced granulation tissue formation, and decreased collagen density, accompanied by an attenuated inflammatory response, but we observed no wound phenotype in Foxo3a−/− mice. Fibroblast growth factor 2, adiponectin, and notch1 genes were significantly increased at wound sites in Foxo1+/− mice, along with markedly altered extracellular signal–regulated kinase 1/2 and AKT phosphorylation. Similarly, transient knockdown of Foxo1 at the wound site by local delivery of antisense oligodeoxynucleotides enhanced skin wound healing. The link between FOXO1 and scarring extends to patients, in particular keloid scars, where we see FOXO1 expression markedly increased in fibroblasts and inflammatory cells within the otherwise normal dermis. This occurs in the immediate vicinity of the keloid by comparison to the center of the mature keloid, indicating that FOXO1 is associated with the overgrowth of this fibrotic response into adjacent normal skin. Overall, our data indicate that molecular targeting of FOXO1 may improve the quality of healing and reduce pathological scarring.
CME Accreditation Statement: This activity (“ASIP 2014 AJP CME Program in Pathogenesis”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint sponsorship of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“ASIP 2014 AJP CME Program in Pathogenesis”) for a maximum of 48 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
The skin plays a major role in protecting us against all extrinsic traumatic factors (ie, microbes, UV radiation, heat, and chemicals). Damage to the skin immediately triggers tissue repair mechanisms alongside a robust inflammatory response for host defense.1 Skin wound healing is generally considered to consist of three phases: inflammation, proliferation/migration, and maturation. During an acute wound inflammatory response, many neutrophils rapidly migrate into damaged tissues to protect against microbes, followed by macrophages that contribute to formation of an associated granulation tissue, including the wound angiogenic response. Unfortunately, this wound inflammatory response also contributes to the final fibrotic outcome of adult tissue repair.2 In parallel with connective tissue repair, epithelial cells migrate over the newly forming granulation tissue to cover the wound site in a process known as re-epithelialization.3 Finally, the wound tissues are partially remodeled, including some removal of excess extracellular matrix at the scar site by proteolytic degradation.4
Tissue repair speed and quality are dependent on aging and metabolic status at a whole-body level, in addition to local immunity and cellular responses at the wound site.5,6 The skin is one of the clearest indicators of aging, and skin healing is highly associated with the aging process. Skin repair occurs perfectly, without scarring, until fairly late in gestation (embryonic 14 days in the mouse and the end of the second trimester in humans).7 By contrast, elderly persons are known to exhibit impaired healing. To date, several studies have, therefore, focused on the molecular mechanisms linking tissue repair and skin biological features with age- and/or metabolic-related genes.8–12 A worldwide increase in patients with delayed skin wound healing due to an abnormal healing process is linked with aging, diabetes, malnutrition, chemotherapy, and hereditary diseases.
The mammalian forkhead box O (FOXO) is a family of transcription factors consisting of FOXO1, FOXO3A, FOXO4, and FOXO6. These proteins remain transcriptionally active in the nucleus in the absence of environmental and growth factors.13 Modification of FOXO leads to its translocation to the cytoplasm and/or its degradation, resulting in the suppression of transcriptional activity. Foxo1 deficiency (Foxo1−/−) in embryonic mice has been shown to be lethal, causing abnormal vascular development.14,15 We have previously reported that FOXO1 plays a key role in aging and in caloric restriction and exhibits antineoplastic characteristics.16 Several lines of evidence suggest that FOXO proteins may play several key roles during tissue repair. Sarcopenia is an age-associated degenerative condition resulting in the loss of skeletal muscle mass and muscle tissue repair.17 Activation of the FOXO family is implicated in skeletal muscle regeneration.18 In skin wounds, Foxo1 and Foxo4 are overly expressed in the transcription factor binding sites of promoters from many differentially expressed genes in the epidermis.19 In diabetic mice, impaired skin wound healing is associated with enhanced activation of FOXO1.20 Other studies have recently reported that re-epithelialization during scalp wound healing is impaired in keratinocyte-specific Foxo1−/− mice.21 However, the full involvement of FOXO family members and their mechanism of action in all cell lineages involved in skin wound healing and scarring in vivo remains largely unknown.
Herein, we investigated the molecular functions for FOXO family gene members in skin wound healing, and their potential application in a clinical setting. We find that heterozygous Foxo1-deficient (Foxo1+/−) mice exhibit accelerated skin wound healing, decreased scarring, and enhanced keratinocyte migration. Acute knockdown of the FOXO1 protein at wound sites [using Foxo1 antisense oligodeoxynucleotides (AS ODNs)] improved the quality of healing. We also suggest that FOXO1 may be implicated in the development of human keloids, because the altered expression of FOXO1 during this process is race dependent. Our findings suggest that modulating expression of FOXO1 may regulate wound healing and scar formation and, thus, are potential therapeutic targets for improving wound healing in the clinic.
Materials and Methods
Wound Model
All experiments were conducted according to the Ethics Review Committee for Animal Experimentation (numbers 1108010940-7 and 1311121101) at Nagasaki University (Nagasaki, Japan). The generation of Foxo1+/− and Foxo3a−/− mice has been described previously.16,22 The wound model was performed as previously described.23 In brief, four full-thickness excisional (4-mm biopsy punch; Kai Industries, Tokyo, Japan) wounds or two full-thickness incisional wounds (1 cm) in the dorsal skin (after shaving under anesthesia) were performed in 7- to 12-week-old mice. Wounds were then harvested using a 6-mm biopsy punch (Kai Industries) and recorded using a digital camera. Areas were calculated using PhotoshopCS4 (Adobe Systems, San Jose, CA).
Human Samples
Human keloid tissue samples were harvested from Japanese and African American patients at the time of surgery, and diagnosis was confirmed by routine pathological examination (Supplemental Table S1). Normal skin tissue samples were harvested from the immediate vicinity of the keloid site. All experiments were conducted with the approval of the ethics committee of Nagasaki University Hospital (number 09062523-2), and in accordance with the Declaration of Helsinki principles. Written informed consent was obtained from each individual.
Histological Features
Tissue was fixed in 4% paraformaldehyde for paraffin embedding. Sections (6 μm thick) were used for various staining techniques: H&E, Masson’s trichrome, Picrosirius red, and immunohistochemistry (IHC) for FOXO1, neutrophils, F4/80, proliferating cell nuclear antigen (PCNA), and phospho–extracellular signal–regulated kinase (pERK). Observations were made via microscopy [polarized light, epifluorescence, or confocal (C2+ system; Nikon Corp., Tokyo, Japan)]. NIS-Elements C software version 4.13 or AR software version 4.0 (Nikon Corp.) was used for data analysis. Immunostaining procedures and antibody information are listed in Table 1.
Table 1.
List of Antibodies
| Primary antibody∗ (manufacturer) | Species | Dilution | Blocking (manufacturer)† | Secondary antibody (manufacturer)‡ | Dilution |
|---|---|---|---|---|---|
| AKT (pan) (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO Life Science, Osaka, Japan) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare Japan, Tokyo, Japan) |
1:10,000 |
| Phospho-AKT (pan) (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody |
1:10,000 |
| CD31 (BD Pharmingen, San Jose, CA) |
Rat | 1:200 (IHC-f) |
Blocking One Histo (Nacalai Tesque, Kyoto, Japan) | Goat α-rabbit IgG Alexa Fluor 568 (Molecular Probes; Life Technologies, Carlsbad, CA) |
1:400 |
| F4/80 (Abcam, Cambridge, UK) |
Rat | 1:400 (IHC-p) |
Blocking One Histo (Nacalai Tesque) | Goat α-rabbit IgG Alexa Fluor 568 (Molecular Probes) |
1:800 |
| FOXO1 (Abcam) |
Rabbit | 1:400 (IHC-p) |
Blocking One Histo (Nacalai Tesque) | Goat α-rabbit IgG Alexa Fluor 488 or 568 (Molecular Probes) Biotinylated α-rabbit IgG (Vector Lab, Burlingame, CA) |
1:800 1:200 |
| FOXO1 (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody |
1:10,000 |
| Phospho-FOXO1 (Thr24) (CST) | Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| FOXO3A (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| Phospho-FOXO3A (Ser318/321) (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| FOXO4 (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| p44/42 MAPK (ERK1/2) (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| Phospho-p44/42 MAPK (ERK1/2) | Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| (CST) | 1:200 (ICC) |
Blocking One Histo (Nacalai Tesque) | Goat α-rabbit IgG Alexa Fluor 568 (Molecular Probes) |
1:1000 | |
| PCNA (Abcam) |
Mouse | 1:400 (IHC-p and IHC-f) |
Blocking One Histo (Nacalai Tesque) | Histofine mouse stain kit (Nichirei Biosciences Inc., Tokyo, Japan) |
|
| Myosin IIb (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody |
1:10,000 |
| NF-κB p65 (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| Phospho-NF-κB p65 (Ser536) (CST) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| Neutrophil (Abcam) |
Rat | 1:400 (IHC-p) |
Blocking One Histo (Nacalai Tesque) | Goat α-rabbit IgG Alexa Fluor 488 (Molecular Probes) |
1:800 |
| α-Tubulin (Abcam) |
Rabbit | 1:1000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked whole antibody (GE Healthcare) |
1:10,000 |
| β-Actin (Abcam) |
Rabbit | 1:5000 (WB) |
PVDF Blocking Reagent (TOYOBO) | α-Rabbit IgG HRP-linked Whole antibody (GE Healthcare) |
1:10,000 |
CST, Cell Signaling Technology, Danvers, MA; f, frozen section; HRP, horseradish peroxidase; p, paraffin section; WB, Western immunoblotting.
Overnight incubation of primary Abs at 4°C (IHC-p, IHC-f, ICC, and WB).
Blocking time: 2 hours at room temperature.
Incubation of second Abs for 1 hour at room temperature.
Analysis of Epithelial Tongue and Area of Granulation Tissue
Measurement of epithelial tongue and area of granulation tissue were performed as previously described.23 In brief, the epithelial tongue on H&E-stained wound sections and areas of granulation tissue on Masson’s trichrome staining sections were measured using NIS-Elements AR software.
Analysis of Angiogenesis
Wounded skin was fixed in 4% paraformaldehyde for 16 hours, then exposed to 10%, 20%, and 30% sucrose (each percentage for 16 hours), and frozen in Tissue-Tek O.C.T. Compound (Sakura Finetek Japan, Tokyo, Japan). Sections (50 μm) were permeabilized with histological blocking reagent (Blocking One Histo; Nacalai Tesque, Kyoto, Japan) and 0.3% Triton X-100 for 2 hours. IHC for CD31, including antibody information, is listed in Table 1. Evaluation of three-dimensional imaging for blood vessels and vascular density were obtained by confocal microscopy, NIS-Elements C software, and IMARIS software version 7.7.1 (BITPLANE, Zurich, Switzerland).
TEM and Morphological Analysis of Collagen
Transmission electron microscopy (TEM) and morphological analysis of collagen were performed as previously described.23,24
Measurement of Macrophages and FOXO1-Positive Cells at Human Intact Skin and Keloid Sites
F4/80-positive cells (indicative of macrophages) and FOXO1-positive cells in the wound bed (defined as the area surrounded by unwounded skin, fascia, regenerated epidermis, and eschar), keloids, or intact skin were counted from three random fields (0.14 mm2).
RNA Isolation and Quantitative Polymerase Chain Reaction
RNA isolation and quantitative PCR were performed as previously described.24 The gene-specific primers and probes for quantitative PCR analysis were obtained from TaqMan gene expression assays (Applied Biosystems, Foster City, CA) and gene-specific primer sets (Takara Bio, Shiga, Japan).
Extraction of Nuclear Protein and Measurement of FOXO1 Activity
Nuclear proteins were extracted by the Nuclear Extract kit (Active Motif Japan, Tokyo), according to the manufacturer’s instructions. Harvested skin wound sites were homogenized using TissueLyzer II (Qiagen, Hilden, Germany).
FOXO activity was measured with the TransAM FKHR (FOXO1/4; Active Motif Japan), according to the manufacturer’s instructions. FOXO1 consensus oligonucleotide-treated extracts were used as the negative control. Absorbance was read by the spectrophotometer (model LS-PLATEmanager 2004; Wako Pure Chemical Industries, Osaka, Japan). The degree of FOXO1 binding activity was calculated as follows:
Total Protein Extraction and Western Immunoblot Analysis
Harvested skin wound sites were homogenized using TissueLyzer II (Qiagen) and were added to T-PER Reagent (Thermo Fisher Scientific, Waltham, MA), consisting of proteinase and dephosphorylation inhibitor (Thermo Fisher Scientific). Supernatant debris was eliminated using an Ultrafree-MC 0.45-μm filter (Millipore, Bedford, MA). Filtered protein samples were separated on a 4% to 12% NuPAGE Novex Bis-Tris gel (Life Technologies, Carlsbad, CA), transferred to polyvinylidene difluoride (PVDF), and blotted according to standard protocols (antibody details are listed in Table 1). Protein bands were visualized by chemiluminescence (Thermo Fisher Scientific), and band intensity was calculated using ImageJ software version 1.47a (NIH, Bethesda, MD).
Cell Culture, Foxo1 siRNA Treatment, Wound Scratch Assay, and Measurement of pERK Activity
Mouse primary keratinocytes (PKs; Cell Lines Service, Eppelheim, Germany) were transfected with 100 nmol/L Stealth RNAi Negative Control Medium GC Duplex 2 or Stealth Foxo1 siRNA (Life Technologies) using the Neon Transfection System (1400 V, 20 ms, two pulses) (Life Technologies). The wound scratched assay was performed as previously described.25 Recombinant mouse fibroblast growth factor 2 (FGF2; Cell Signaling Technology, Danvers, MA) (100 ng/mL) was used. The intensity of pERK fluorescence was measured by NIS-Elements AR software (Nikon Corp.).
LPS Challenge
Lipopolysaccharide (LPS) (from Escherichia coli serotype O55, phenol extraction) was obtained from Wako Pure Chemical Industries and was reconstituted in saline. Mice (aged 8 to 12 weeks; weight, approximately 30 g) were i.p. injected with 1.0 mg of LPS, and their survival was monitored.
ELISA
Extracted proteins were measured by the myeloperoxidase (MPO) mouse ELISA kit (Abcam, Cambridge, UK), according to the manufacturer’s instructions.
Microarray Analysis
Cyanine 3–labeled complementary RNA was generated from 200 ng of total RNA using the Low Input Quick Amp Labeling Kit, one color (Agilent Technologies, Santa Clara, CA), and was purified by the RNeasy mini kit (Qiagen), according to the manufacturer’s instructions. Fragmented cyanine 3–labeled complementary RNA (600 ng) was hybridized to SurePrint G3 mouse GE microarray, 8 × 60 K (Agilent Technologies), at 65°C for 17 hours. The microarray was then washed and scanned using the DNA microarray scanner (Agilent Technologies).
Analysis of microarray data was performed using Ingenuity iReport (Ingenuity System, Redwood City, CA). Probe set intensities were summarized and normalized using robust multi-array average. Significant differential expression was determined by a moderated t-test (Limma) using a P value cutoff of 0.05 and a fold-change cutoff of 1.5. All raw data are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; accession number GSE48473).
Screening AS ODN Candidates for Knockdown Studies
AS ODNs and the in vitro AS ODN cleavage experiments were both designed as previously described.23 Basic Local Alignment Search Tool (BLAST) searches for AS ODN sequences (Table 2) were conducted to exclude any sequences that were non-specific for Foxo1 mRNA using GenBank (http://www.ncbi.nlm.nih.gov; accession number NM_019739). Foxo1 mRNA was transcribed from the Riken FANTOM FLS Clone (clone E430027H20; DNAFORM, Kanagawa, Japan).
Table 2.
Sequences for Control and AS ODNs
| Name | Sequence |
|---|---|
| Control ODN | 5′-GTGTAACACGTCTATACG-3′ |
| Foxo1 AS (1273) | 5′-CTGGAGAGATGCTTTTTT-3′ |
| Foxo1 AS (1341) | 5′-GACCCAGGACTCGCAGGC-3′ |
| Foxo1 AS (1686) | 5′-GAATTTAGACTGGTGTTT-3′ |
| Foxo1 AS (1692) | 5′-CTGGGTGAATTTAGACTG-3′ |
| Foxo1 AS (1717) | 5′-GTATGTGTACTTTGAGTA-3′ |
| Foxo1 AS (1914) | 5′-AGGACCCGACTGTTGGGT-3′ |
| Foxo1 AS (1986) | 5′-ATTTTGTTATGAGATGCC-3′ |
| Foxo1 AS (2070) | 5′-TTCACCACATGGGGCAGG-3′ |
| Foxo1 AS (2095) | 5′-CATGGCAGATGTGTGAGG-3′ |
| Foxo1 AS (2138) | 5′-ACAGAGGCACTTGTAAAG-3′ |
| Foxo1 AS (2208) | 5′-ATCCTACCATAGCCATTG-3′ |
For in vivo experiments involving ODN delivery, ODNs [1 or 10 μmol/L in 50 μL 30% Pluronic F-127 gel (Sigma-Aldrich, St. Louis, MO), which acts as a slow-release vehicle23] were topically applied immediately after wounding (50 μL; 1 or 10 μmol/L of ODNs).
Statistical Analysis
All data are expressed as the means ± SEM. Statistical significance was assessed by analysis of variance, followed by: i) Tukey’s post hoc test for multiple comparisons, ii) Dunnett’s post hoc test for comparisons of all columns versus control, or iii) paired or unpaired Student’s t-test. Survival curves were analyzed using Kaplan-Meier survival analysis and were compared with the log-rank test. Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). Significance was reached at values of P < 0.05.
Results
Skin Wound Healing Accelerates in Foxo1+/− Mice
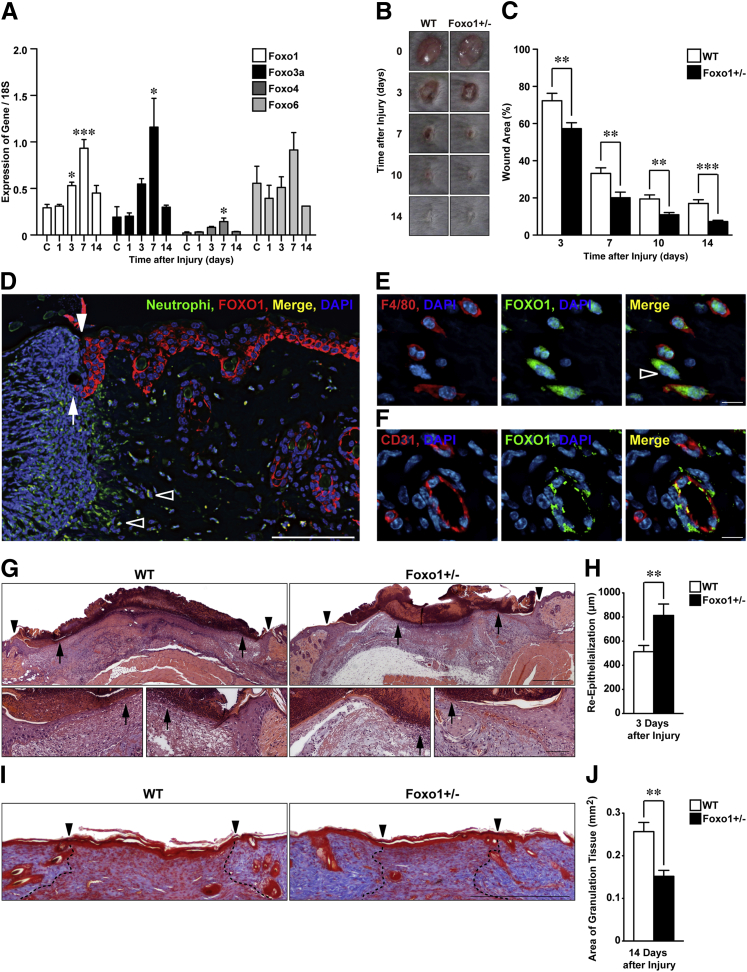
We investigated the expression of Foxo in wild-type (WT) mice after dorsal aseptic skin wounding by quantitative PCR. Gene expression of Foxo1 was significantly increased 3 and 7 days after injury and expression of Foxo3a was significantly increased 7 days after injury by comparison with unwounded skin (Figure 1A). Gene expression of Foxo4 was markedly low, and Foxo6 was not significantly induced. These results indicated that Foxo1 and Foxo3a genes are the Foxo family members predominantly expressed during the skin repair process.
Figure 1.
Skin wound healing is accelerated in Foxo1+/− mice. A: Gene expression of murine Foxo family in skin wound healing measured by quantitative PCR relative to 18S ribosomal RNA (n = 4 to 6). B: Representative images for gross appearances of excisional wound in WT (white bars) and Foxo1+/− (black bars) mice. C: The proportion of the wound remaining open relative to the initial wound area at each time point (n = 12). D: IHC for FOXO1 and neutrophils shows neutrophils, epithelium, and hair follicle expressing FOXO1 1 day after injury in WT mice. Nuclei were counterstained with DAPI [wound margin (white arrowhead), leading edge of epithelia (arrow), and FOXO1-expressing neutrophils (open arrowheads)]. IHC of FOXO1 and F4/80 or CD31 shows wound-infiltrated macrophages (E), fibroblasts (open arrowhead, E), and endothelial cells (F) at 7 days after injury in WT mice. G: H&E staining of re-epithelialization [wound margin (arrowheads) and the leading edge of epithelia (arrows)]. Top panels: The higher-power field of the bottom panels. H: Measurement of epithelial tongue at 3 days after injury (n = 13). I: Middle of wound tissue at 14 days after injury stained with Masson’s trichrome, and the extent (cross-sectional area) of granulation tissue visualized and quantified at the midpoint of the wound (dotted lines) [wound margin (arrowheads)]. J: Quantification of the area of granulation tissue 14 days after injury (n = 6). All values represent means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars: 100 μm (D and bottom panels in G); 10 μm (E and F); 500 μm (high-power fields in G and I).
We then explored the role of FOXO1 and FOXO3A in skin wound healing in Foxo1+/− and Foxo3a−/− male mice, respectively. Foxo1+/− male mice are viable despite expressing <50% of both FOXO1 in intact skin (data not shown), and Foxo1 mRNA in liver, spleen, muscle, adipose tissue, and hippocampus.16 After 3 days of injury, Foxo1+/− mice exhibited a significantly smaller wound area (57% ± 3.2%) by comparison to time-matched WT mice (72% ± 4.0%) (Figure 1, B and C). By contrast, wound closures in Foxo3a−/− mice were not altered compared with WT mice (data not shown). These results prompted us to further analyze the function of FOXO1 in skin wound healing.
To determine which cells express FOXO1 protein during skin wound healing, we performed IHC analysis. By only 1 day after injury, FOXO1 was markedly present in the leading edge and basal layer of keratinocytes and hair follicles, and in recruited neutrophils (Figure 1D and Supplemental Figure S1). Seven days after injury, FOXO1 was present in macrophages, fibroblasts, and endothelial cells at the wound site (Figure 1, E and F). Histological analysis allowed us to quantify both the extent of re-epithelialization and the area of granulation tissue at various time points during repair. The length of epithelial wound tongues in Foxo1+/− mice 3 days after injury was markedly higher (813 ± 94 μm) than for WT mice (513 ± 52 μm) (Figure 1, G and H). IHC for the proliferation marker, PCNA, showed that the percentage of proliferating cells in the epithelial tongue of Foxo1+/− mice 3 days after injury was markedly increased compared with WT mice (55% versus 40%). Masson’s trichrome staining of sections of excisional wounds revealed that the area of granulation tissue in the midwound region of Foxo1+/− mice was significantly reduced (0.15 ± 0.014 mm2) compared with WT mice (0.26 ± 0.021 mm2) (Figure 1, I and J).
Because angiogenesis is crucial for granulation tissue formation, and FOXO1 is known to be involved in vasculogenesis in embryonic and fetal development,14 we investigated vessel outgrowth during the repair process in Foxo1+/− versus WT mice. The three-dimensional blood vessel network of the wound was reconstructed via confocal microscopy of sections stained for platelet endothelial cell adhesion molecule/CD31, which is a marker for endothelial cells.26 These studies revealed no difference in the vessel network in intact skin of Foxo1+/− and WT mice (0.033 ± 0.0024 and 0.026 ± 0.0057 μm3/μm3, respectively), and the same was true at both 7- and 14-day wound sites (Supplemental Figure S2).
Our analyses suggest that attenuated expression of FOXO1 protein leads to accelerated repair and improved quality of skin wound healing, owing to enhanced migration of keratinocytes in the early stage of wound healing, followed by decreased area of granulation tissue formation. However, this is not because of differences in wound angiogenesis.
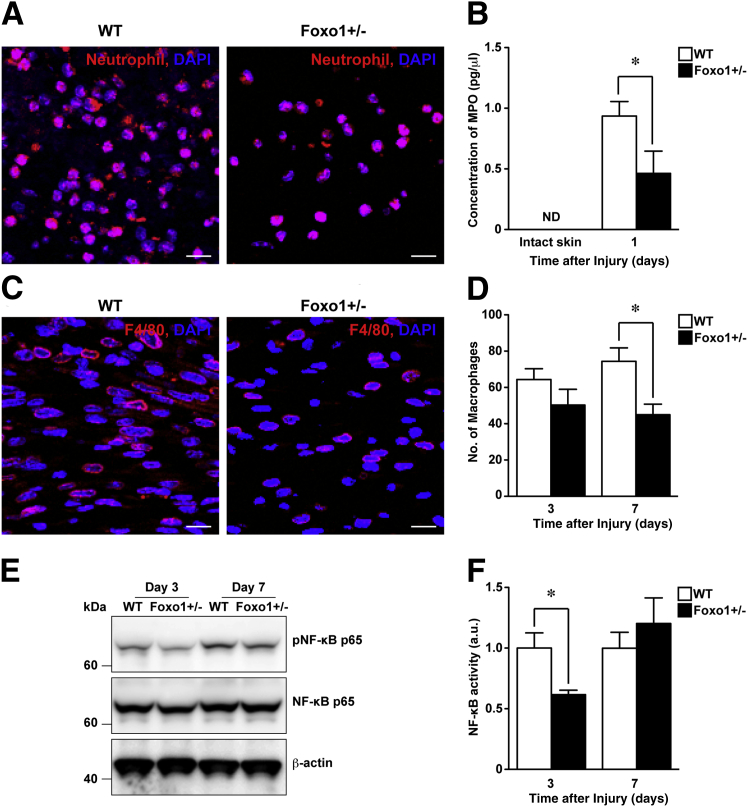
The Inflammatory Response Attenuates at Wound Sites of Foxo1+/− Mice
Several leukocyte lineages infiltrate wound sites at various time points during the skin repair process.2 Our IHC staining showed that wound-infiltrated neutrophils and macrophages expressed FOXO1 protein (Figure 1, D and E). Neutrophils, revealed by either neutrophil IHC or measurement of MPO, were reduced in Foxo1+/− wounds versus WT controls (Figure 2, A and B). F4/80 IHC for macrophages27 confirms a similar reduction (by 40%) in macrophage numbers at wound sites of Foxo1+/− mice (Figure 2, C and D).
Figure 2.
Attenuation of neutrophil and macrophage infiltration and inflammatory responses at wound sites of Foxo1+/− mice. A: IHC for neutrophils (red) showing the number of wound-infiltrated neutrophils at wound sites 1 day after injury, which is reduced in Foxo1+/− mice compared with WT mice (nuclei were counterstained with DAPI). B: MPO concentration using ELISA reveals that MPO levels at wound sites of Foxo1+/− mice are significantly reduced compared with WT mice (n = 8 per group). C: IHC for macrophages using F4/80 at the middle area of wound sites 7 days after injury. D: Quantification of F4/80-positive macrophages at wound sites of WT mice (n = 6 per group for 3 days, and n = 5 per group for 7 days) and Foxo1+/− mice (n = 6 per group for both 3 and 7 days). E: Western immunoblot analysis of pNF-κB p65 and total NF-κB. Full scans for Western immunoblots in Supplemental Figure S5A. F: Densitometric analysis for pNF-κB activity over total NF-κB (n = 4 to 5). All values represent means ± SEM. ∗P < 0.05. Scale bar = 10 μm (A and C). ND, not detected.
Because NF-κB plays a pivotal role in the inflammatory response,28 we performed Western immunoblot analysis and showed that phosphorylation levels of NF-κB p65 (Ser536) at wound sites of Foxo1+/− mice 3 days after injury were markedly reduced (by 38%) compared with WT mice (Figure 2, E and F). In addition, we found that Foxo1+/− mice exhibited significant resistance to high-dose LPS-induced endotoxin shock that leads to activation of NF-κB signaling via Toll-like receptor 4 in vivo (Supplemental Figure S3).29 Collectively, these data suggest that FOXO1 may regulate inflammatory cell recruitment to wound sites, and the reduced FOXO1 in Foxo1+/− mice dampens down this inflammatory response.
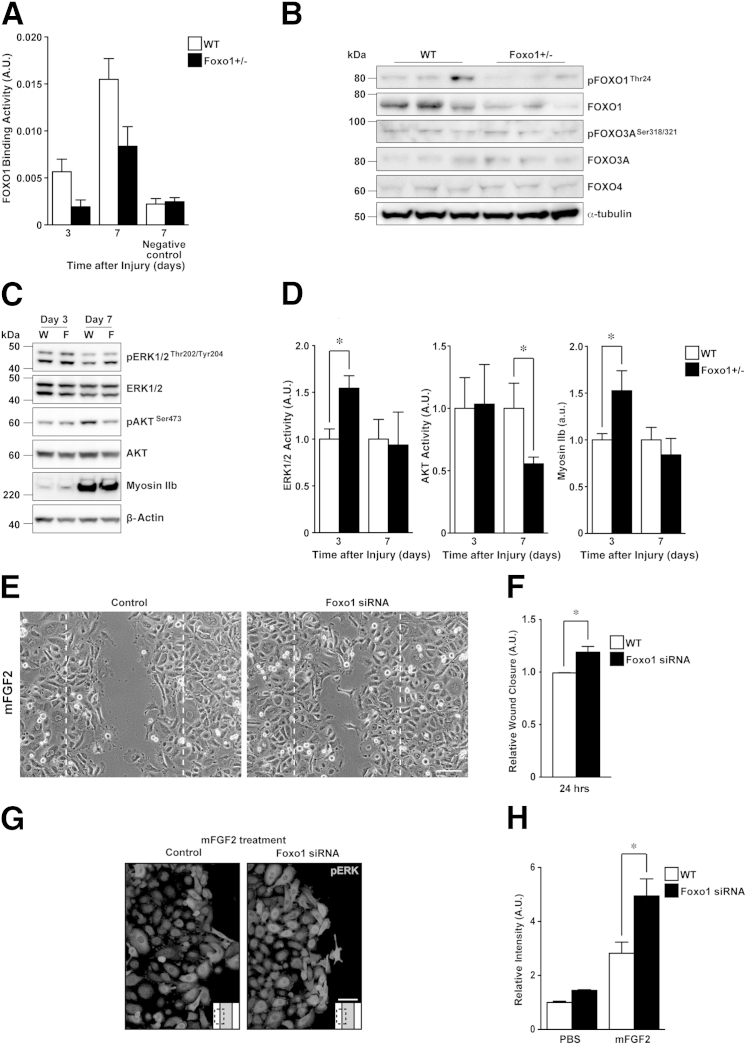
Expression and Phosphorylation of the FOXO Family at Wound Sites
Our findings provide clear evidence that Foxo1+/− mice exhibit accelerated skin wound healing and enhanced re-epithelialization at the early stage of skin wound healing and a diminished inflammatory response. We next performed a comprehensive gene expression profile at wound sites of Foxo1+/− and WT mice, after first confirming reduced FOXO1 DNA binding activity. ELISA studies indicate reduced binding of FOXO1 in cells from wound sites of Foxo1+/− mice 3 and 7 days after injury (66% ± 13% and 56% ± 15%, respectively) compared with WT mice (Figure 3A). Similar results were found for both gene (Table 3) and protein (Figure 3B) expression levels of FOXO1.
Figure 3.
ERK1/2 activation and myosin IIb expression are enhanced at wound sites of Foxo1+/− mice. A: FOXO1 binding activity in the nuclear wound tissue extract was measured by immobilized oligonucleotide ELISA. FOXO1 consensus oligonucleotide-treated extracts were used as negative control (n = 2 to 4). B: Western immunoblot analysis of wound tissue at 3 days shows weak expression of total FOXO1 and pFOXO1 (Thr24) at wound sites of Foxo1+/− mice. Bands for total FOXO3A, pFOXO3A (Ser318/321), and FOXO4 remain unchanged in both groups. Full scans for Western immunoblot analysis in Supplemental Figure S5B. C: Western immunoblot analysis of pERK1/2 (Thr202/Tyr204), total ERK1/2, pAKT (Ser473), total AKT, and myosin IIb. Full scans for Western immunoblots in Supplemental Figure S5C. D: Densitometric analysis of pERK1/2 (Thr202/Tyr204) and pAKT (Ser473) over total ERK1/2 and AKT, respectively, and myosin IIb (n = 4 to 5). E: Wound scratch assay 24 hours after treatment of mFGF2 in PKs with control (left panel) or Foxo1 (right panel) siRNAs. F: Wound closure ratio (n = 4). G: Confocal images of pERK in mFGF2-treated control PKs (left panel) and Foxo1 siRNA-treated PKs (right panel) 24 hours after scratching (n = 3). H: Fluorescence intensity of pERK by mFGF2 24 hours after scratching in control PKs and Foxo1 siRNA-treated PKs (n = 3). All values represent means ± SEM. ∗P < 0.05. Scale bars: 100 μm (E); 50 μm (G).
Table 3.
Expression of Skin Wound-Related Genes
| Gene symbol: name | Ratio | Molecular function | Reference |
|---|---|---|---|
| Il24: interleukin 24 | 2.478 | Cytokine | 30 |
| Ifna1/Ifna13: interferon, α 1 | 2.199 | Cytokine | 31 |
| Itga1: integrin, α 1 | 2.316 | Receptor | 32 |
| Notch1 | 1.944 | Receptor | 33 |
| Adipoq: adiponectin, C1Q, and collagen domain containing | 1.824 | Cytokine | 8,9 |
| Npy2r: neuropeptide Y receptor Y2 | 1.821 | Receptor | 34 |
| Lif: leukemia inhibitory factor | 1.777 | Cytokine | 35 |
| Fgf2: fibroblast growth factor 2 (basic) | 1.647 | Growth factor | 36–38 |
| Myh10: myosin, heavy chain 10, nonmuscle | 1.641 | Actin-based motor protein | 39 |
| Adora1: adenosine A1 receptor | 1.637 | Receptor | 40 |
| Foxo1: forkhead box O1 | −1.92 | Transcription factor |
Gene expressions from the microarray data set (Supplemental Table S2). Compared with WT mice, a ratio >1.5 in Foxo1+/− mice indicates a significant up-regulation (n = 3 per group). Il24 may be a negative regulator of skin wound healing. Expression of Foxo1 serves as a positive control.
In human fibroblasts, FOXO1 and FOXO3A have been shown to affect Foxo1 gene expression.41 Thus, we examined the protein expression and phosphorylation levels of FOXO1, FOXO3A, and FOXO4 at wound sites 3 days after injury of Foxo1+/− mice (Figure 3B). We find that FOXO1 protein expression and its phosphorylation [pFOXO1 (Thr24)] level at wound sites were markedly decreased in Foxo1+/− mice, but expression levels of FOXO3A, pFOXO3A (Ser318/321), and FOXO4 were not altered in either group. These results, and the pattern of expression of Foxo family genes during skin repair (Figure 1A). indicate that FOXOs at the wound site are predominantly regulated by pFOXO1 (Thr24) and pFOXO3A (Ser318/321) in the early stage of skin wound healing.
Activation of ERK1/2 Is Enhanced in Wound Sites of Foxo1+/− Mice
To determine the molecular mechanisms underlying enhanced skin wound healing when FOXO1 protein expression was reduced, we performed microarray analysis on 3-day skin wound samples from Foxo1+/− versus WT mice. By using a fold-change cutoff of 1.5 (with a P value cutoff of 0.05), we identified 387 and 269 differentially regulated genes that are up- and down-regulated in Foxo1+/− mice, respectively (Supplemental Table S2). We next screened for those molecules/pathways that might be most likely to be promoting healing in Foxo1+/− mice by analyzing the molecular interactions between differentially regulated genes, such as gene expression, activation, post-translational modification, and physical interactions (Supplemental Table S3). Previous in vivo studies have revealed skin wound healing–related genes: Fgf2,36–38 adiponectin (Adipoq),8,9 and Notch1.33 Our results showed that Fgf2, Adipoq, and Notch1 were significantly (P < 0.05) increased by 1.65-, 1.82-, and 1.94-fold, respectively, in 3-day wound sites of Foxo1+/− mice (Table 3).
Two key signaling pathways may contribute to the FOXO1 phenotype we observe. The ERK1/2 signaling pathway is involved in cell migration and proliferation,25 and the AKT signaling pathway is associated with skin wound healing functions upstream of FOXO1.42 The ERK1/2 and AKT pathways are activated by FGF2 and ADIPOQ, contributing to epithelial and fibroblast cell proliferation.8,43 Therefore, we investigated whether activation of ERK and AKT pathways in wound sites of Foxo1+/− mice was altered. The phosphorylation levels of ERK1/2 (Thr202/Tyr204) in wound sites of Foxo1+/− mice 3 days after injury were markedly increased compared with WT (1.5 ± 0.13 and 1.0 ± 0.11, respectively) (Figure 3C). In contrast, the phosphorylation levels of AKT (Ser473) in wound sites of Foxo1+/− mice 7 days after injury were markedly decreased compared with WT (0.56 ± 0.054 and 1.0 ± 0.20, respectively) (Figure 3D).
Our microarray analysis showed that several cell migration– and proliferation-related signals were significantly up-regulated, including myosin heavy chain 10 (Myh10) (1.64-fold). MYH10 generates the nonmuscle myosin IIb isoform,44 which is downstream of the ERK1/2 pathway.45 Myosin IIb is expressed in both epidermis and wound fibroblasts,46 and Myh10 is markedly induced at wound sites, in Foxo1+/− mice (Table 3) in the current study. We also see that protein levels of the myosin IIb isoform are markedly increased in wound sites of Foxo1+/− mice compared with WT (1.5 ± 0.21 and 1.0 ± 0.067, respectively) (Figure 3, C and D). Collectively, these results suggest that expression of myosin IIb may be enhanced via the ERK pathway at an early stage of wound repair in Foxo1+/− mice.
We next tested whether FOXO1 was involved in wound FGF2 signaling, which contributes to the enhancement of cell proliferation and migration in PKs. The in vitro wound scratch assay demonstrated that wound closure in Foxo1 siRNA-treated PKs was not altered compared with control PKs (1.23 ± 0.15 and 1.0 ± 0.04, respectively). Interestingly, the migration of mouse FGF2 (mFGF2)–treated Foxo1 siRNA-treated PKs was significantly enhanced from the wound edge to the center of the wound (Figure 3, E and F). Immunocytochemistry (ICC) was then used to investigate the localization of pERK. In PKs exposed to mFGF2-treated Foxo1 siRNA, pERK activation was mainly observed at the wound edge rather than away from the wound site 24 hours after scratching. This response was significantly higher compared with control wounds (4.95 ± 0.63 and 2.82 ± 0.41, respectively) (Figure 3, G and H). Taken together, these findings indicate that FOXO1 may play a role in re-epithelialization and migration of keratinocytes at wound sites via the ERK and FGF2 pathways.
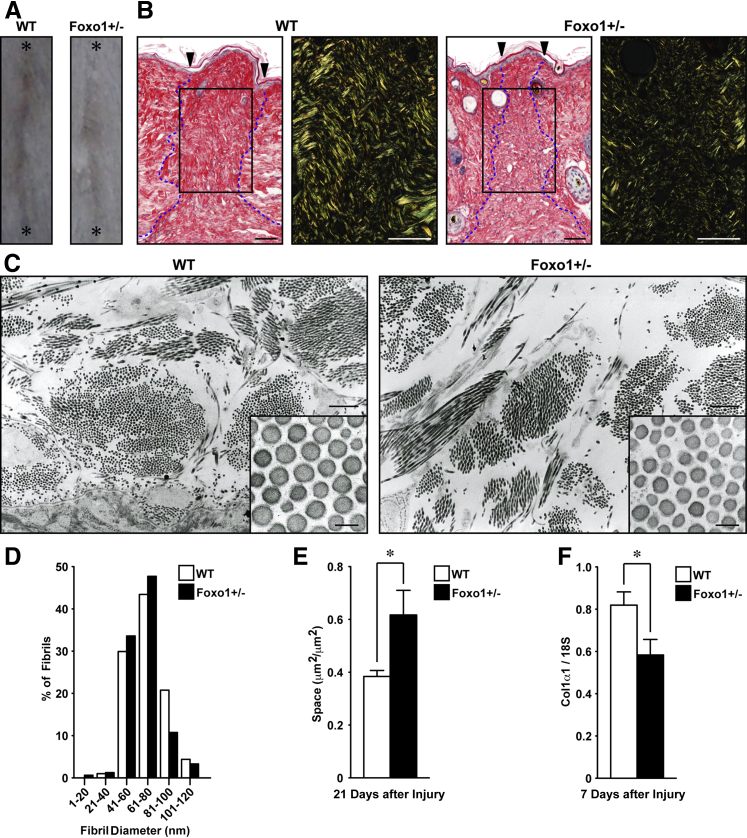
Collagen Organization Is Altered at Wound Sites of Foxo1+/− Mice
Scarring is the final consequence of the wound healing process, and is a measure of wound healing quality.1 To investigate whether altered FOXO1 expression influences the development of scarring at wound sites, we monitored scarring 21 days after making 1-cm incisional wounds to Foxo1+/− and WT mice (Figure 4A). Picrosirius red staining showed type I collagen (red and yellow) and type III collagen (green) bundle organization47 to be markedly reduced in Foxo1+/− mice 21 days after injury (Figure 4B).
Figure 4.
Scarring at wound sites is attenuated in Foxo1+/− mice. A: Gross appearance of scarring at the incisional wound 21 days after injury (asterisk indicates wound edge). Images shown are representative of six independent experiments. B: Picrosirius red–stained sections of incisional wound sites at 21 days after injury for analysis of collagen fibers and alignments [type I collagen (red and yellow), type III collagen (green), and wound edge (arrowheads)]. Granulation tissue was visualized at the midpoint of the wound (dotted lines). Images shown are representative of eight independent experiments. Low magnifications were taken as nonpolarized images. High-magnification details from boxed areas indicated are differential interference contract images using polarized light microscopy. C: TEM images of connective tissue from midwound sites 21 days after injury. High-magnification insets illustrate differing collagen fibril diameters in this tissue. D: Histogram of total range of fibril diameters in the wound site 21 days after injury (n = 1090 fibrils from three WT mice, and n = 1354 fibrils from three Foxo1+/− mice). Fibril diameters at wound sites of Foxo1+/− mice tend to be smaller than in WT mice. E: Quantification of vacant extracellular spaces in wound sites at 21 days (n = 3 to 4). F: Quantification of gene expression of Col1α1 7 days after injury at wound sites (measured by quantitative PCR), relative to 18S ribosomal RNA (n = 8). All values represent means ± SEM. ∗P < 0.05. Scale bars: 50 μm (B); 1 μm (C); 100 nm (inset, C).
To further analyze the development of a scar, we performed TEM to reveal gross collagen bundling patterns, individual collagen fibril diameter, and the density of fibrils at the wound site. The morphological features of collagen within intact skin of Foxo1+/− and WT mice were indistinguishable (Supplemental Figure S4). Interestingly, the fibril diameter at the midregion wound sites of Foxo1+/− mice was markedly (P < 0.001) reduced (61.5 ± 0.49 nm) compared with WT mice (63.3 ± 0.46 nm) (Figure 4, C and D). Furthermore, intracollagen bundle spaces at wound sites of Foxo1+/− mice were significantly (P < 0.05) increased (0.62 ± 0.094 μm2/μm2) compared with WT mice (0.38 ± 0.023 μm2/μm2) (Figure 4E), more closely resembling those of unwounded skin. Gene expression of type I collagen α1 (Col1α1) was significantly decreased at wound sites of Foxo1+/− mice 7 days after injury compared with WT mice (0.58 ± 0.073 and 0.82 ± 0.063, respectively) (Figure 4F). We suggest that these differences of collagen assembly at wound sites play a key role in reducing scar formation during the maturation phase of healing in Foxo1+/− mice.
Acute Knockdown of FOXO1 Using AS ODNs Improves Skin Wound Healing
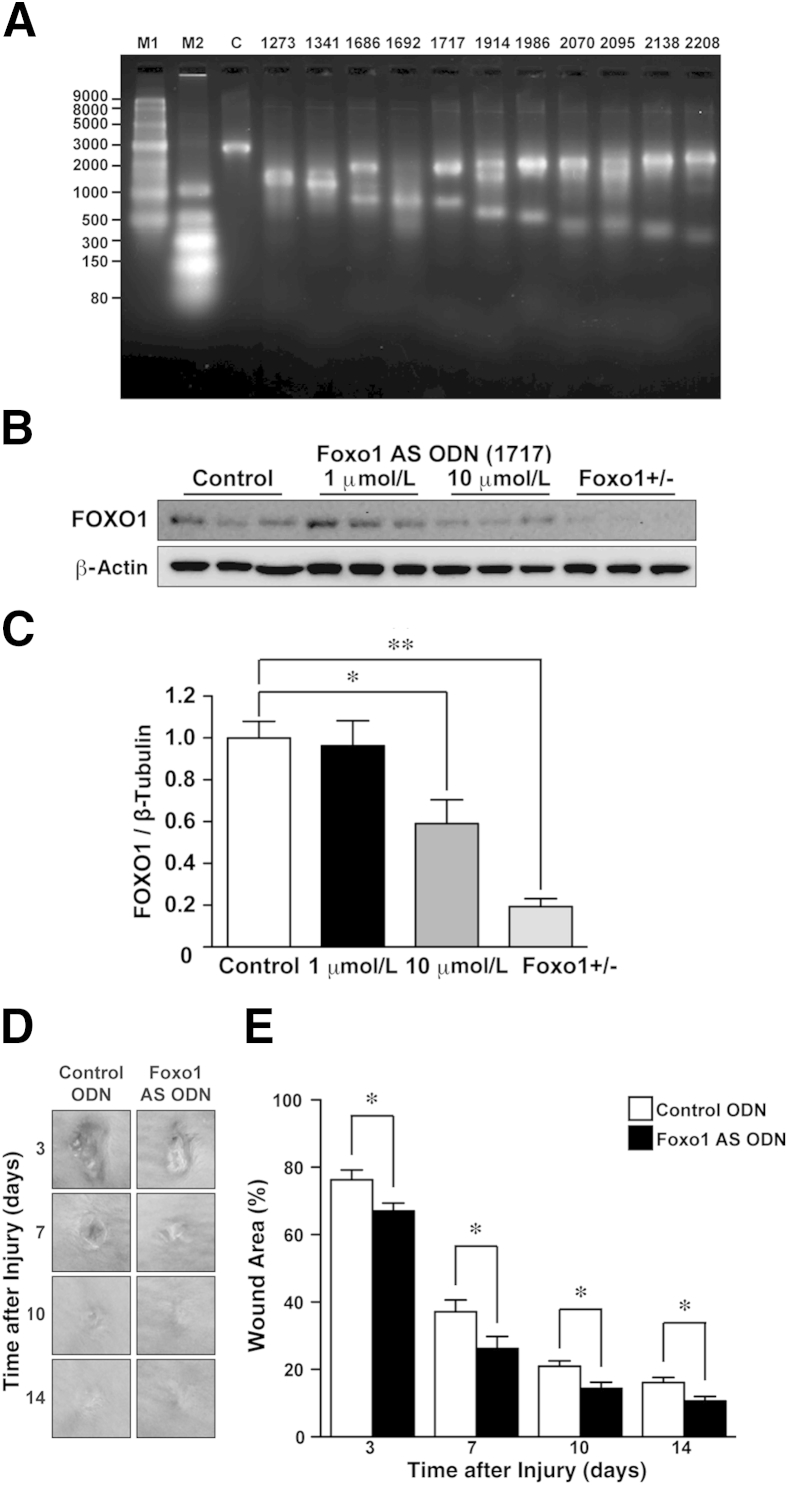
Our Foxo1+/− mouse data provide experimental evidence that attenuation of FOXO1 expression may improve skin wound healing. To further address our hypothesis and test whether reducing levels of this transcription factor during the repair process is a potential therapeutic strategy for improving healing, we designed and optimized Foxo1-specific AS ODNs in vitro (Figure 5A). We applied Foxo1 AS ODN (1717) (10 μmol/L in 30% Pluronic gel for 6 hours at the wound site23) versus control ODN (with sequence predicted to be nonbinding to other mRNAs) to 4-mm-diameter adult skin wound sites (Figure 5, B and C). Macroscopic analysis indicated that wound closure in Foxo1 AS ODN–treated wound sites 3 days after injury was markedly accelerated (67% ± 2.4%) at early time points during the repair process, compared with the control ODN-treated wound (76% ± 3.0%) (Figure 5, D and E). Next, we made a 1-cm incisional wound in the dorsal skin and analyzed scarring (via TEM) with a 21-day treatment of control ODN or Foxo1 AS ODN at the wound sites. The fibril diameter at the midregion was not altered (66.6 ± 0.65 nm, n = 1336 from three mice) at Foxo1 AS ODN–treated wound sites compared with control ODN-treated wound sites (65.1 ± 0.55 nm, n = 1534 from three mice). Vacant extracellular space at Foxo1 AS ODN–treated wound sites was not altered (0.55 ± 0.038 μm2/μm2, n = 3) compared with control ODN-treated wound sites (0.44 ± 0.038 μm2/μm2, n = 3). These results indicated that acute down-regulation of FOXO1 protein at the wound site using Foxo1 AS ODN accelerated skin wound healing, but did not significantly alter scar quality.
Figure 5.
Skin wound healing is accelerated in Foxo1 AS ODN–treated wounds. A: Optimization of Foxo1 AS ODNs in vitro. Cleavage was visualized when transcribed Foxo1 mRNA was incubated with control and AS ODNs in vitro (n = 2 independent experiments) (M1 and M2 = RNA loading markers). Sequences are shown in Table 2. Foxo1 AS ODN (1717) is more efficient than other Foxo1 AS ODNs. B: Western immunoblot analysis from wound sites (6 hours after injury) to detect the effective dose of Foxo1 AS ODN (1717) to decrease FOXO1 protein expression in wounds in vivo. Full scans for Western immunoblots in Supplemental Figure S5D. C: Quantification of FOXO1 protein expression in wound sites exposed to 10 μmol/L Foxo1 AS ODN (1717) (n = 6 per group) reveals that FOXO1 is significantly reduced. Data are presented as the means ± SEM and analyzed by Tukey’s multiple-comparison tests. D: Representative photomicrographs for gross appearances of excisional control and Foxo1 AS ODN–treated wounds at various time points after wounding (n = 12). E: The proportion of the wound remaining open relative to the initial wound area at each time point after the injury in control versus Foxo1 AS ODN–treated wounds (n = 12 per group). All values represent means ± SEM. ∗P < 0.05, ∗∗P < 0.01.
Increased FOXO1 Is Associated with Human Keloid Scars
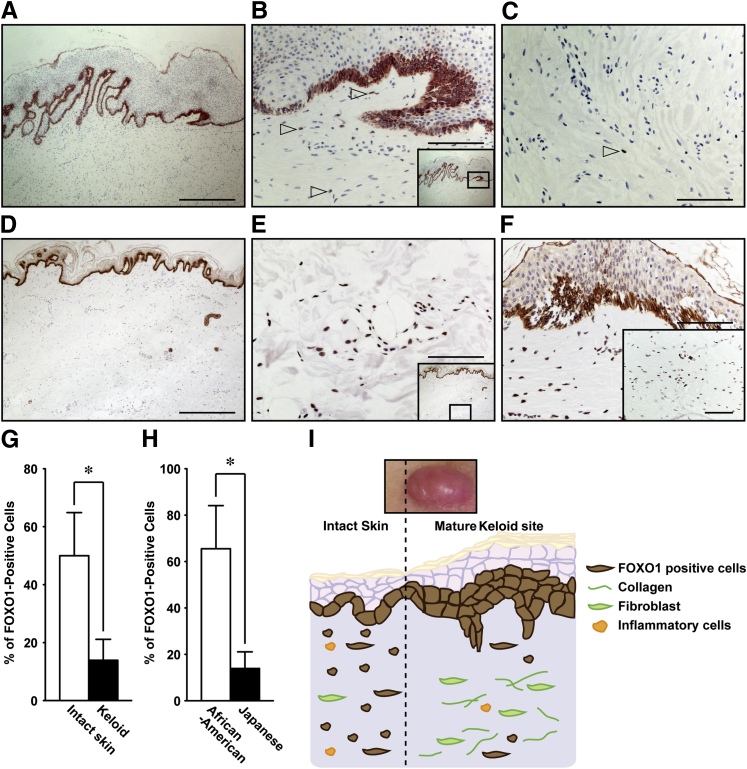
Because of the altered level of scarring in our whole body Foxo1+/− mouse studies, we next chose to investigate whether the expression pattern of FOXO1 was altered in human keloid scars, which are an extreme instance of human skin fibrotic disease, typified by a hypertrophic epidermis, and overgrowth of granulation tissue, which expands in a claw-like way to invade adjacent normal skin.48,49 IHC showed that FOXO1 was prominently present in suprabasal keratinocytes of the hypertrophic epidermal layer of keloids (Figure 6A), in addition to some fibroblasts and inflammatory cells (Figure 6B). Although FOXO1 in fibroblasts was not strongly expressed at deep keloid tissue sites (Figure 6C), it was notably present in numerous fibroblasts and inflammatory cells in the immediate vicinity of keloid sites of the normal dermal layer (Figure 6, D, E, and G). These results suggested that FOXO1 was involved in expanding the growth of keloid into adjacent normal skin sites and might be driving the production of excessive extracellular matrix protein.
Figure 6.
Expression of FOXO1 at human keloids. A–C: IHC for FOXO1 shows that FOXO1 (brown) is highly present in basal lamina keratinocytes at keloid sites in the Japanese population [nuclei are stained with hematoxylin (violet)] [FOXO1-expressing cells (arrowheads)]. D and E: FOXO1 is present in normal epidermis and dermis in the immediate vicinity of keloid sites compared with mature keloid sites. F: IHC for FOXO1 shows highly prominent FOXO1-positive cells at the surface and deep in keloid sites of African Americans compared with Japanese [mature deep keloid sites are shown (inset)]. Micrographs are representative of three and six sections for D–F and A–C, respectively. G: Percentage of FOXO1-positive cells at the intact skin in the immediate vicinity of keloid sites (three cases) and in keloid sites (six cases). H: Percentage of FOXO1-positive cells at mature keloid sites of an African American (three cases) and two Japanese (six cases) patients. I: A proposed model of the exacerbation of keloids by FOXO1. Elevated expression of FOXO1 at the epidermis may cause hyperplasia of keloids. In contrast, numbers of FOXO1-expressing cells at deeply mature keloid sites are markedly reduced. Intact skin in the vicinity of keloids appears normal. However, expression of FOXO1 is markedly increased compared with mature keloid, suggesting that FOXO1-positive cells are associated with keloid expansion. Collectively, many FOXO1-expressing cells produce collagen and enhance the inflammatory response, leading to the exacerbation of keloid scarring. All values represent means ± SEM. ∗P < 0.05. Scale bars: 500 μm (A and D); 100 μm (B, C, E, and F).
The development of a keloid is known to be associated with age, physiological conditions, and genetic backgrounds. Keloids occur most frequently in individuals of African American descent.50 Therefore, we next performed a comparative case report of keloids between African American and Japanese to investigate the expression of FOXO1. Levels of expression of FOXO1 in keratinocytes, fibroblasts, and inflammatory cells in all keloid sites were markedly higher in African Americans compared with Japanese (Figure 6, F and H). Collectively, these results indicate that the development of skin fibrotic diseases may, in part, be regulated by FOXO1 (Figure 6I).
Discussion
In the present study, we report accelerated and improved eventual quality of skin wound healing in Foxo1+/− mice, because of enhanced re-epithelialization and a reduced inflammatory response at sites of tissue damage. Foxo1 AS ODN–treated wounds also exhibit improved skin wound healing. Abnormal and diverse expression of FOXO1 is also associated with the development of keloids in human patients. The present data provide a novel molecular insight into the function of FOXO1 in skin wound healing and suggest its potential as an anti-scarring therapeutic target.
Skin wounding initially leads to clot formation and a significant recruitment of neutrophils, which protect the tissue breach by killing microbes and also release proinflammatory cytokines, some of which act to draw in macrophages to the wound. Thereafter, macrophages infiltrate the wound site and secrete cytokines, chemokines, and growth factors, according to the extent of tissue damage and infection state, thus reflecting the degree of the inflammatory phase.1 The FOXO family has been previously shown to regulate the homeostasis of the immune system and the inflammatory response.51 Conditional Foxo1−/− mice exhibit an altered phenotype, including T-cell homeostasis and tolerance.52–54 FOXO1 may play a role during the infection recognition and clearance process55 because it is known that the bacterial product, N-formylmethionyl-leucyl-phenylalanine, triggers neutrophils to up-regulate myeloid leukemia cell differentiation protein MCL1, which can form a complex with FOXO1. Moreover, chromatin immunoprecipitation sequencing analysis using next-generation sequencing has revealed that FOXO1 significantly enhances Toll-like receptor 4 signaling in macrophages.56 Activation of NF-κB affects the AKT-FOXO1 signaling pathway.57 In the present study, the infiltration of neutrophils and macrophages into wound sites and the phosphorylation of NF-κB and AKT were attenuated in Foxo1+/− mice. Overall, the FOXO1-mediated inflammatory response may link to leukocyte recruitment and activation in the skin wound healing process.
Re-epithelialization, involving keratinocyte migration and proliferation, commences soon after skin damage and is regulated by various factors, including keratinocyte growth factor and others released by infiltrating inflammatory cells, and fibroblasts.2,3 Previous studies suggest that the effect of FOXO1 in guiding cell migration/proliferation may be cell type specific. For example, knockdown of Foxo1 in platelet-derived growth factor–treated fibroblasts has been shown to enhance proliferation, indicating that attenuation of the expression of FOXO1 is sufficient for the enhancement of cell growth.41 In contrast, keratinocyte-specific Foxo1−/− in mice impairs scalp wound healing because of reduced expression of transforming growth factor β1.21 In our current study, enhancement of re-epithelialization in Foxo1+/− mice accelerated skin wound healing, which corresponded with an increase in the expression of Fgf2, Adipoq, Notch1, and Myo10 (Table 3). Each of these, in turn, is known to play key roles in various aspects of the repair process: FGF2 is crucial for re-epithelialization in skin wound healing,37 whereas genetic and pharmacological inhibition of Notch1 in mice markedly impairs skin wound healing.33 Calmodulin-like protein-mediated expression of MYO10 contributes to keratinocyte motility and migration in humans and mice.39 ADIPOQ promotes keratinocyte proliferation and migration via the ERK pathway in vivo.8,9 We are exploring the mechanism underlying FOXO-mediated regulation of cell proliferation and migration in the presence of FGF2 at wound sites.
As well as enhanced rate of wound repair, we observe reduced scarring in Foxo1+/− mice. Scarring appears at the final stage of the skin wound healing process, and the phenotype of scarring is dependent on diverse factors, including inflammation, delayed healing, physiological condition, age, and race.6,50,58 The regulation of collagen expression via FOXO1 may depend on the tissue and cell type, and this may be a consequence of both direct and/or indirect effects. Knockdown of Foxo1 in UV-irradiated human dermal fibroblasts was shown to significantly decrease the expression of Col1α1.59 In contrast, expression of liver Col1α1 was increased in the bile duct ligation-induced experimental liver fibrosis model of Foxo1+/−, resulting in liver fibrosis.60 Collagen organization is controlled by several enzymes and extracellular matrix proteins,61 and is considerably altered by normal aging.62,63 Activation of AKT is one of the main signaling pathways for type I procollagen synthesis.64 In the present study, expression of Col1α1, collagen density, and AKT phosphorylation were all markedly decreased in wound sites of Foxo1+/− mice 7 days after injury. We also found that knockdown of FOXO1 in PKs significantly enhanced the ERK pathway after mFGF2 application. Local administration of FGF2 to the human incisional wound reduces scarring.65 We speculate that the attenuation of FOXO1 in wound fibroblasts contributes to reduced scarring through the FGF2 pathway. We are presently exploring how FOXO1 regulates scarring at wound sites in the presence of several wound growth factors, including FGF2. Further studies using other models, such as keratinocyte-specific Foxo1−/− mice,21 are required to better elucidate the pathophysiological significance of FOXO1 function in skin fibrosis.
In patients, the most extreme scarring phenotype is that of keloid scarring, where scar tissue spills out from the initial site of tissue damage. Our keloid studies showed FOXO1 is highly expressed in fibroblasts and inflammatory cells at the margin of a keloid compared with those fibroblasts in mature keloid sites, and that FOXO1 expression level was altered between African Americans and Japanese. These results indicate that FOXO1 may regulate the expression of collagen and, thus, its expression level may play a key role in scarring and keloids. Current investigations are focusing on the implications of FOXO1 polymorphisms on fibrotic diseases. Disequilibrium of FOXO1 is believed to affect human longevity66,67 and, thus, polymorphism of FOXO1 may also affect homeostasis at the cellular and individual level as well. Therefore, a functional analysis of FOXO1 polymorphisms may further elucidate the differences in keloid morbidity and repair phenotypes by age and race. Determining how the FOXO1 signaling pathway regulates keloid progression for the homeostatic maintenance between proliferation and differentiation will thus be important to explore in future studies.
In conclusion, the age-related gene FOXO1 plays a central role for tissue repair and remodeling. It may be considered a potential therapeutic target for enhancing tissue repair and remodeling as well as for dampening inflammatory diseases and fibrosis.
Acknowledgments
We thank Takashi Suematsu (Department of Electron Microscopy, Nagasaki University) for assistance with the TEM analysis and Kazutaka Hayashida and Shin-ichi Yokota (Nikon Instech, Japan) for assistance with microscopic and imaging analysis.
Footnotes
Supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants-in-Aid for Young Scientists (A) 21689049 and 24689069, Challenging Exploratory Research grants 23650484 and 25560055 (R.M.), Takeda Science Foundation (R.M.), Uehara Memorial Foundation (R.M.), Nakatomi Foundation (R.M.), The Wellcome Trust Senior Investigator Award 097791MA (P.M.), and The Royal Society International Joint Project (R.M. and P.M.).
Disclosures: None declared.
Supplemental Data
Expression of FOXO1 protein at wound sites. IHC for FOXO1 (brown) using diaminobenzidene, showing prominent presence of FOXO1 in the epithelium and hair follicle of WT mice 1 day after injury [nuclei are stained with hematoxylin (violet)]. The arrow and arrowhead indicate the leading edge of epithelia and wound margin, respectively. Micrographs are representative of six sections. Scale bar = 100 μm.
Angiogenesis at wound sites of heterozygous Foxo1+/− and WT mice. A–D: Confocal imaging of three-dimensional blood vessel structures in intact skin and at the wound site. The arrowheads (B and D) indicate the wound margin. E: Vascular density is not altered in Foxo1+/− mice (n = 6 for intact skin and 14 days; n = 15 for 7 days) compared with WT mice (n = 6 for intact skin; n = 12 for 7 days; n = 7 for 14 days). Data are presented as the means ± SEM. Scale bar = 100 μm.
Foxo1+/− mice are resistant to LPS-induced endotoxin shock. Age-matched WT (n = 14) and Foxo1+/− (n = 11) mice were i.p. injected with 1 mg of LPS from E. coli O55. Survival was monitored for 4 days. The survival curve was analyzed using Kaplan-Meier survival analysis and was compared with the log-rank test. P = 0.0326.
Collagen organization in intact skin of Foxo1+/− and WT mice. A: TEM images of intact skin tissue from WT and Foxo1+/− mice. High-magnification insets illustrate collagen fibril diameter in this tissue. B: Histograms of fibril diameters indicate total range of fibril diameters in intact skin from at least three fields of wound site from each mouse of WT (n = 839 fibrils from five mice) and Foxo1+/− (n = 864 fibrils from five mice). Scale bars: 1 μm (A); 100 nm (insets). Original magnification, ×50,000 (insets).
Full scans of Western immunoblots for the indicated figures. Figure 2E (A), Figure 3B (B), Figure 3C (C), and Figure 5B (D). Molecular weights of the target protein are in parentheses.
References
- 1.Shaw T.J., Martin P. Wound repair at a glance. J Cell Sci. 2009;122:3209–3213. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eming S.A., Krieg T., Davidson J.M. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 3.Werner S., Krieg T., Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 4.Martins V.L., Caley M., O’Toole E.A. Matrix metalloproteinases and epidermal wound repair. Cell Tissue Res. 2013;351:255–268. doi: 10.1007/s00441-012-1410-z. [DOI] [PubMed] [Google Scholar]
- 5.Gosain A., DiPietro L.A. Aging and wound healing. World J Surg. 2004;28:321–326. doi: 10.1007/s00268-003-7397-6. [DOI] [PubMed] [Google Scholar]
- 6.Guo S., Dipietro L.A. Factors affecting wound healing. J Dent Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson M.W., O’Kane S. Scar-free healing: from embryonic mechanisms to adult therapeutic intervention. Philos Trans R Soc Lond B Biol Sci. 2004;359:839–850. doi: 10.1098/rstb.2004.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata S., Tada Y., Asano Y., Hau C.S., Kato T., Saeki H., Yamauchi T., Kubota N., Kadowaki T., Sato S. Adiponectin regulates cutaneous wound healing by promoting keratinocyte proliferation and migration via the ERK signaling pathway. J Immunol. 2012;189:3231–3241. doi: 10.4049/jimmunol.1101739. [DOI] [PubMed] [Google Scholar]
- 9.Salathia N.S., Shi J., Zhang J., Glynne R.J. An in vivo screen of secreted proteins identifies adiponectin as a regulator of murine cutaneous wound healing. J Invest Dermatol. 2013;133:812–821. doi: 10.1038/jid.2012.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goren I., Kampfer H., Podda M., Pfeilschifter J., Frank S. Leptin and wound inflammation in diabetic ob/ob mice: differential regulation of neutrophil and macrophage influx and a potential role for the scab as a sink for inflammatory cells and mediators. Diabetes. 2003;52:2821–2832. doi: 10.2337/diabetes.52.11.2821. [DOI] [PubMed] [Google Scholar]
- 11.Squarize C.H., Castilho R.M., Bugge T.H., Gutkind J.S. Accelerated wound healing by mTOR activation in genetically defined mouse models. PLoS One. 2010;5:e10643. doi: 10.1371/journal.pone.0010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serravallo M., Jagdeo J., Glick S.A., Siegel D.M., Brody N.I. Sirtuins in dermatology: applications for future research and therapeutics. Arch Dermatol Res. 2013;305:269–282. doi: 10.1007/s00403-013-1320-2. [DOI] [PubMed] [Google Scholar]
- 13.Huang H., Tindall D.J. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 14.Furuyama T., Kitayama K., Shimoda Y., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Motoyama N., Mori N. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 15.Hosaka T., Biggs W.H., 3rd, Tieu D., Boyer A.D., Varki N.M., Cavenee W.K., Arden K.C. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaza H., Komatsu T., Wakita S., Kijogi C., Park S., Hayashi H., Chiba T., Mori R., Furuyama T., Mori N., Shimokawa I. FoxO1 is involved in the antineoplastic effect of calorie restriction. Aging Cell. 2010;9:372–382. doi: 10.1111/j.1474-9726.2010.00563.x. [DOI] [PubMed] [Google Scholar]
- 17.Snijders T., Verdijk L.B., van Loon L.J. The impact of sarcopenia and exercise training on skeletal muscle satellite cells. Ageing Res Rev. 2009;8:328–338. doi: 10.1016/j.arr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Lara-Pezzi E., Winn N., Paul A., McCullagh K., Slominsky E., Santini M.P., Mourkioti F., Sarathchandra P., Fukushima S., Suzuki K., Rosenthal N. A naturally occurring calcineurin variant inhibits FoxO activity and enhances skeletal muscle regeneration. J Cell Biol. 2007;179:1205–1218. doi: 10.1083/jcb.200704179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roupe K.M., Alberius P., Schmidtchen A., Sorensen O.E. Gene expression demonstrates increased resilience toward harmful inflammatory stimuli in the proliferating epidermis of human skin wounds. Exp Dermatol. 2010;19:e329–e332. doi: 10.1111/j.1600-0625.2009.01038.x. [DOI] [PubMed] [Google Scholar]
- 20.Siqueira M.F., Li J., Chehab L., Desta T., Chino T., Krothpali N., Behl Y., Alikhani M., Yang J., Braasch C., Graves D.T. Impaired wound healing in mouse models of diabetes is mediated by TNF-alpha dysregulation and associated with enhanced activation of forkhead box O1 (FOXO1) Diabetologia. 2010;53:378–388. doi: 10.1007/s00125-009-1529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponugoti B., Xu F., Zhang C., Tian C., Pacios S., Graves D.T. FOXO1 promotes wound healing through the up-regulation of TGF-beta1 and prevention of oxidative stress. J Cell Biol. 2013;203:327–343. doi: 10.1083/jcb.201305074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto K., Araki K.Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M., Chen C., Hosokawa K., Nakauchi H., Nakayama K., Nakayama K.I., Harada M., Motoyama N., Suda T., Hirao A. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Mori R., Shaw T.J., Martin P. Molecular mechanisms linking wound inflammation and fibrosis: knockdown of osteopontin leads to rapid repair and reduced scarring. J Exp Med. 2008;205:43–51. doi: 10.1084/jem.20071412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori R., Ikematsu K., Kitaguchi T., Kim S.E., Okamoto M., Chiba T., Miyawaki A., Shimokawa I., Tsuboi T. Release of TNF-alpha from macrophages is mediated by small GTPase Rab37. Eur J Immunol. 2011;41:3230–3239. doi: 10.1002/eji.201141640. [DOI] [PubMed] [Google Scholar]
- 25.Matsubayashi Y., Ebisuya M., Honjoh S., Nishida E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr Biol. 2004;14:731–735. doi: 10.1016/j.cub.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 26.Newman P.J. The biology of PECAM-1. J Clin Invest. 1997;99:3–8. doi: 10.1172/JCI119129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austyn J.M., Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 28.DiDonato J.A., Mercurio F., Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 29.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 30.Poindexter N.J., Williams R.R., Powis G., Jen E., Caudle A.S., Chada S., Grimm E.A. IL-24 is expressed during wound repair and inhibits TGFalpha-induced migration and proliferation of keratinocytes. Exp Dermatol. 2010;19:714–722. doi: 10.1111/j.1600-0625.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregorio J., Meller S., Conrad C., Di Nardo A., Homey B., Lauerma A., Arai N., Gallo R.L., Digiovanni J., Gilliet M. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J Exp Med. 2010;207:2921–2930. doi: 10.1084/jem.20101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner H., Broberg A., Pozzi A., Laato M., Heino J. Absence of integrin alpha1beta1 in the mouse causes loss of feedback regulation of collagen synthesis in normal and wounded dermis. J Cell Sci. 1999;112(Pt 3):263–272. doi: 10.1242/jcs.112.3.263. [DOI] [PubMed] [Google Scholar]
- 33.Chigurupati S., Arumugam T.V., Son T.G., Lathia J.D., Jameel S., Mughal M.R., Tang S.C., Jo D.G., Camandola S., Giunta M., Rakova I., McDonnell N., Miele L., Mattson M.P., Poosala S. Involvement of notch signaling in wound healing. PLoS One. 2007;2:e1167. doi: 10.1371/journal.pone.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekstrand A.J., Cao R., Bjorndahl M., Nystrom S., Jonsson-Rylander A.C., Hassani H., Hallberg B., Nordlander M., Cao Y. Deletion of neuropeptide Y (NPY) 2 receptor in mice results in blockage of NPY-induced angiogenesis and delayed wound healing. Proc Natl Acad Sci U S A. 2003;100:6033–6038. doi: 10.1073/pnas.1135965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akita S., Daian T., Ishihara H., Fujii T., Akino K. Leukemia inhibitory factor-transfected embryonic fibroblasts and vascular endothelial growth factor successfully improve the skin substitute wound healing by increasing angiogenesis and matrix production. J Dermatol Sci. 2004;36:11–23. doi: 10.1016/j.jdermsci.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Miller D.L., Ortega S., Bashayan O., Basch R., Basilico C. Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol Cell Biol. 2000;20:2260–2268. doi: 10.1128/mcb.20.6.2260-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega S., Ittmann M., Tsang S.H., Ehrlich M., Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc Natl Acad Sci U S A. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer M., Müller A.K., Yang J., Moik D., Ponzio G., Ornitz D.M., Grose R., Werner S. FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. J Cell Sci. 2012;125(Pt 23):5690–5701. doi: 10.1242/jcs.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett R.D., Mauer A.S., Pittelkow M.R., Strehler E.E. Calmodulin-like protein upregulates myosin-10 in human keratinocytes and is regulated during epidermal wound healing in vivo. J Invest Dermatol. 2009;129:765–769. doi: 10.1038/jid.2008.288. [DOI] [PubMed] [Google Scholar]
- 40.Sun L.L., Xu L.L., Nielsen T.B., Rhee P., Burris D. Cyclopentyladenosine improves cell proliferation, wound healing, and hair growth. J Surg Res. 1999;87:14–24. doi: 10.1006/jsre.1999.5716. [DOI] [PubMed] [Google Scholar]
- 41.Essaghir A., Dif N., Marbehant C.Y., Coffer P.J., Demoulin J.B. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284:10334–10342. doi: 10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somanath P.R., Chen J., Byzova T.V. Akt1 is necessary for the vascular maturation and angiogenesis during cutaneous wound healing. Angiogenesis. 2008;11:277–288. doi: 10.1007/s10456-008-9111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chrissouli S., Pratsinis H., Velissariou V., Anastasiou A., Kletsas D. Human amniotic fluid stimulates the proliferation of human fetal and adult skin fibroblasts: the roles of bFGF and PDGF and of the ERK and Akt signaling pathways. Wound Repair Regen. 2010;18:643–654. doi: 10.1111/j.1524-475X.2010.00626.x. [DOI] [PubMed] [Google Scholar]
- 44.Conti M.A., Adelstein R.S. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.H., Wang A., Conti M.A., Adelstein R.S. Nonmuscle myosin II is required for internalization of the epidermal growth factor receptor and modulation of downstream signaling. J Biol Chem. 2012;287:27345–27358. doi: 10.1074/jbc.M111.304824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bond J.E., Ho T.Q., Selim M.A., Hunter C.L., Bowers E.V., Levinson H. Temporal spatial expression and function of non-muscle myosin II isoforms IIA and IIB in scar remodeling. Lab Invest. 2011;91:499–508. doi: 10.1038/labinvest.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T., Feng Y., Sun H., Zhang L., Hao L., Shi C., Wang J., Li R., Ran X., Su Y., Zou Z. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol. 2012;181:1911–1920. doi: 10.1016/j.ajpath.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Hunasgi S., Koneru A., Vanishree M., Shamala R. Keloid: a case report and review of pathophysiology and differences between keloid and hypertrophic scars. J Oral Maxillofac Pathol. 2013;17:116–120. doi: 10.4103/0973-029X.110701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robles D.T., Moore E., Draznin M., Berg D. Keloids: pathophysiology and management. Dermatol Online J. 2007;13:9. [PubMed] [Google Scholar]
- 50.Shih B., Bayat A. Genetics of keloid scarring. Arch Dermatol Res. 2010;302:319–339. doi: 10.1007/s00403-009-1014-y. [DOI] [PubMed] [Google Scholar]
- 51.Peng S.L. Foxo in the immune system. Oncogene. 2008;27:2337–2344. doi: 10.1038/onc.2008.26. [DOI] [PubMed] [Google Scholar]
- 52.Gubbels Bupp M.R., Edwards B., Guo C., Wei D., Chen G., Wong B., Masteller E., Peng S.L. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur J Immunol. 2009;39:2991–2999. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouyang W., Beckett O., Flavell R.A., Li M.O. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang W., Liao W., Luo C.T., Yin N., Huse M., Kim M.V., Peng M., Chan P., Ma Q., Mo Y., Meijer D., Zhao K., Rudensky A.Y., Atwal G., Zhang M.Q., Li M.O. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crossley L.J. Neutrophil activation by fMLP regulates FOXO (forkhead) transcription factors by multiple pathways, one of which includes the binding of FOXO to the survival factor Mcl-1. J Leukoc Biol. 2003;74:583–592. doi: 10.1189/jlb.0103020. [DOI] [PubMed] [Google Scholar]
- 56.Fan W., Morinaga H., Kim J.J., Bae E., Spann N.J., Heinz S., Glass C.K., Olefsky J.M. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D.H., Kim J.Y., Yu B.P., Chung H.Y. The activation of NF-kappaB through Akt-induced FOXO1 phosphorylation during aging and its modulation by calorie restriction. Biogerontology. 2008;9:33–47. doi: 10.1007/s10522-007-9114-6. [DOI] [PubMed] [Google Scholar]
- 58.Martin P., Leibovich S.J. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15:599–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka H., Murakami Y., Ishii I., Nakata S. Involvement of a forkhead transcription factor, FOXO1A, in UV-induced changes of collagen metabolism. J Investig Dermatol Symp Proc. 2009;14:60–62. doi: 10.1038/jidsymp.2009.2. [DOI] [PubMed] [Google Scholar]
- 60.Adachi M., Osawa Y., Uchinami H., Kitamura T., Accili D., Brenner D.A. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology. 2007;132:1434–1446. doi: 10.1053/j.gastro.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 61.Canty E.G., Kadler K.E. Procollagen trafficking, processing and fibrillogenesis. J Cell Sci. 2005;118:1341–1353. doi: 10.1242/jcs.01731. [DOI] [PubMed] [Google Scholar]
- 62.Puschmann S., Rahn C.D., Wenck H., Gallinat S., Fischer F. Approach to quantify human dermal skin aging using multiphoton laser scanning microscopy. J Biomed Opt. 2012;17:036005. doi: 10.1117/1.JBO.17.3.036005. [DOI] [PubMed] [Google Scholar]
- 63.Wu S., Li H., Yang H., Zhang X., Li Z., Xu S. Quantitative analysis on collagen morphology in aging skin based on multiphoton microscopy. J Biomed Opt. 2011;16:040502. doi: 10.1117/1.3565439. [DOI] [PubMed] [Google Scholar]
- 64.Park J.H., Kim S.R., An H.J., Kim W.J., Choe M., Han J.A. Esculetin promotes type I procollagen expression in human dermal fibroblasts through MAPK and PI3K/Akt pathways. Mol Cell Biochem. 2012;368:61–67. doi: 10.1007/s11010-012-1342-7. [DOI] [PubMed] [Google Scholar]
- 65.Ono I., Akasaka Y., Kikuchi R., Sakemoto A., Kamiya T., Yamashita T., Jimbow K. Basic fibroblast growth factor reduces scar formation in acute incisional wounds. Wound Repair Regen. 2007;15:617–623. doi: 10.1111/j.1524-475X.2007.00293.x. [DOI] [PubMed] [Google Scholar]
- 66.Kleindorp R., Flachsbart F., Puca A.A., Malovini A., Schreiber S., Nebel A. Candidate gene study of FOXO1, FOXO4, and FOXO6 reveals no association with human longevity in Germans. Aging Cell. 2011;10:622–628. doi: 10.1111/j.1474-9726.2011.00698.x. [DOI] [PubMed] [Google Scholar]
- 67.Li Y., Wang W.J., Cao H., Lu J., Wu C., Hu F.Y., Guo J., Zhao L., Yang F., Zhang Y.X., Li W., Zheng G.Y., Cui H., Chen X., Zhu Z., He H., Dong B., Mo X., Zeng Y., Tian X.L. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of FOXO1 protein at wound sites. IHC for FOXO1 (brown) using diaminobenzidene, showing prominent presence of FOXO1 in the epithelium and hair follicle of WT mice 1 day after injury [nuclei are stained with hematoxylin (violet)]. The arrow and arrowhead indicate the leading edge of epithelia and wound margin, respectively. Micrographs are representative of six sections. Scale bar = 100 μm.
Angiogenesis at wound sites of heterozygous Foxo1+/− and WT mice. A–D: Confocal imaging of three-dimensional blood vessel structures in intact skin and at the wound site. The arrowheads (B and D) indicate the wound margin. E: Vascular density is not altered in Foxo1+/− mice (n = 6 for intact skin and 14 days; n = 15 for 7 days) compared with WT mice (n = 6 for intact skin; n = 12 for 7 days; n = 7 for 14 days). Data are presented as the means ± SEM. Scale bar = 100 μm.
Foxo1+/− mice are resistant to LPS-induced endotoxin shock. Age-matched WT (n = 14) and Foxo1+/− (n = 11) mice were i.p. injected with 1 mg of LPS from E. coli O55. Survival was monitored for 4 days. The survival curve was analyzed using Kaplan-Meier survival analysis and was compared with the log-rank test. P = 0.0326.
Collagen organization in intact skin of Foxo1+/− and WT mice. A: TEM images of intact skin tissue from WT and Foxo1+/− mice. High-magnification insets illustrate collagen fibril diameter in this tissue. B: Histograms of fibril diameters indicate total range of fibril diameters in intact skin from at least three fields of wound site from each mouse of WT (n = 839 fibrils from five mice) and Foxo1+/− (n = 864 fibrils from five mice). Scale bars: 1 μm (A); 100 nm (insets). Original magnification, ×50,000 (insets).
Full scans of Western immunoblots for the indicated figures. Figure 2E (A), Figure 3B (B), Figure 3C (C), and Figure 5B (D). Molecular weights of the target protein are in parentheses.