Abstract
Hypoxia inducible factor 1alpha (HIF-1α) plays a central role in regulating tumor angiogenesis via its effects on vascular endothelial growth factor (VEGF) transcription, and its expression is regulated through proteasome-mediated degradation. Paradoxically, previous studies have shown that proteasome inhibitors (PIs) block tumor angiogensis by reducing VEGF expression, but the mechanisms have not been identified. Here we report that PIs downregulated HIF-1α protein levels and blocked HIF-1α transcriptional activity in human prostate cancer cells. PIs induced phosphorylation of the translation intiation factor 2α (eIF2α) which inhibited caused general translational repression to inhibit HIF-1α expression. Furthermore, PIs induced HIF-1α accumulation in LNCaP-Pro5 cells depleted of eIF2α via siRNA transfection and in MEFs expressing a phosphorylation-deficient mutant form of eIF2α. Finally, PIs failed to induce eIF2α phosphorylation or translational attenuation in DU145 or 253JB-V cells and in these cells PIs promoted HIF-1α accumulation. Our data established that PIs downregulated HIF-1α expression in cells that display activation of the unfolded protein response (UPR) by stimulating phosphorylation of eIF2α and inhibiting HIF-1α translation.
Keywords: Bortezomib, NPI-0052, HIF-1α, VEGF, ER stress, prostate cancer
Introduction
The 26S proteasome is a large, multicatalytic enzyme that functions as a major route of intracellular protein degradation (1–3). Bortezomib (PS-341/Velcade®, Millennium Pharmaceuticals, Inc.) is a peptide boronate inhibitor of the chymotryptic activity of the proteasome that received FDA approval for the treatment of multiple myeloma and mantle cell lymphoma (1, 2, 4). Its clinical success has prompted other companies to develop chemically distinct PIs that might be even more active. One such compound is NPI-0052 (salinosporamide A, Nereus Pharmaceuticals), a structural analog of the proteasome inhibitor lactacystin that is currently being evaluated in phase I clinical trials. NPI-0052 is orally bioactive, irreversible, and has broader proteasome inhibitory activity than bortezomib (5, 6).
Analyses of the direct cytotoxic effects of PIs in tumor cells have identified a number of different biochemical mechanisms, including inhibition of pro-survival transcription factor nuclear factor kappaB (NFκB), accumulation of pro-apopototic proteins, like p53, Bax, and NOXA, and induction of endoplasmic reticular (ER) stress (2, 3, 7, 8). PIs also suppress angiogenesis by downregulating VEGF expression (5, 9). Tumor VEGF expression is controlled in large part by the transcription factor HIF-1 (10, 11), a heterodimer composed of an O2 sensitive alpha subunit (HIF-1α) and a constitutively expressed beta subunit (HIF-1β/ARNT) (10). Under normoxic conditions, HIF-1α is hydroxylated at two proline residues (P402 and P564) by prolyl hydroxylase-domain proteins and is subsequently recongnized by Von Hippel-Lindau (VHL), a component of an E3 ubiquitin-protein ligase that targets HIF-1α for degradation by the proteasome (10). Under hypoxic conditions HIF-1α is not hydroxylated and it accumulates. Loss of VHL expression is a common feature of renal cell carcinoma that results in overexpression of VEGF and increased angiogenesis (12). Overexpression of HIF-1α has been observed in many other solid tumors, including prostate, breast, lung and head and neck cancers, and chemical inhibitors of HIF-1α are being developed for cancer therapy (10, 13–15).
It seemed paradoxical to us that HIF-1α expression is controlled primarily by the proteasome yet PIs downregulate VEGF expression. We therefore initiated the present study to characterize the effects of PIs on HIF-1α function. Here we report that bortezomib and NPI-0052 selectively blocked HIF-1α’s expression and activity in a subset of human prostate cancer cells. Analyses of the biochemical mechanisms implicated processes observed during ER stress, including phosphorylation of eIF2α leading to inhibition of HIF-1α translation. Similar effects were observed in cells exposed to other stimuli known to induce eIF2α phosphorylation, indicating that the response may have broader biological significance.
Materials and Methods
Cell lines and culture
Human LNCaP-Pro5 prostate cancer cells and 253JB-V bladder cancer cells were provided by Dr. Curtis Pettaway and Dr. Colin Dinney, respectively (Department of Urology, University of Texas M.D. Anderson Cancer Center, TX). Human PC3 and DU145 prostate cancer cells were obtained from American Type Culture Collection (Rockville, MD). eIF2α51SS wild type and eIF2α51AA knock-in mutant mouse embryonic fibroblast cells (MEFs) were kindly provided by Dr. David Ron (New York University School of Medicine, NY). The prostate cancer cells were grown in RPMI 1640 media (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (Life Technologies), 1% vitamins (Life Technologies), sodium pyruvate (Bio Whittaker, Rockland, ME), L-glutamine (Bio Whittaker), penicillin/streptomycin solution (Bio Whittaker), and non-essential amino acids (Life Technologies) under conditions of 5% CO2 at 37°C in an incubator. 253JB-V cells were cultured in MEM media containing the same supplements. The MEFs were grown in dMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin solution, L-glutamine, non-essential amino acids, 55mM β-mercaptoethanol. Leucine free media was purchased from MP Biomedicals (Solon, OH) for measuring protein synthesis. For hypoxic exposure, cells were placed in a NAPCO Water-Jacketed CO2 incubator (Thermo Fisher Scientific, Inc., Waltham, MA) flushed with 0.2% oxygen, 5% CO2 and 95% nitrogen. Hypoxia was also mimicked by incubating cells with 100μM cobalt chloride (CoCl2) in a regular atmosphere.
Reagents, antibodies and plasmids
The proteasome inhibitor bortezomib and the IkappaB kinase (IKK)/NFκB inhibitor PS-1145 were provided by Millenium Pharmaceuticals (Cambridge, MA). The proteasome inhibitor NPI-0052 was provided by Nereus Pharmaceuticals (San Diego, CA). Cycloheximde (CHX), thapsigargin (TG), tunicamycin (TM), heparin and CoCl2 were purchased from Sigma Chemical Co. (St. Louis, MO). Antibodies were obtained from the following commercial sources: human HIF-1α and p21WAF1 (BD Biosciences Transduction Laboratories™); HIF-2α, HIF-1β and mouse HIF-1α (Novus Biologicals Inc., Littleton, CO); phosphorylated eIF2α(Ser52) and total eIF2α (Invitrogen Biosource™, Carlsbad, CA); phosphorylated eIF2α(Ser51) (Cell Signaling Technology, Inc., Danvers, MA); phosphorylated PERK(Thr851), P300 and Ref-1 (Santa Cruz Biotechnology, Santa Cruz, CA); and actin (Sigma Chemical Co., St. Louis, MO). Horseradish peroxidase-conjugated secondary antibodies were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Hypoxia response element (HRE)-driven firefly luciferase plasmid was purchased from Panomics Inc. (Fremont, CA).
Immunoblotting
Total cell lysates were prepared using a 1% Triton X-100 buffer as described previously (16). Densitometric quantification of proteins levels was performed using ImageJ software [National Institutes of Health (NIH), Bethesda, MD].
Quantification of VEGF by enzyme-linked immunosorbent assays (ELISA)
LNCaP-Pro5 cells (5×104 cells/well) were plated in 24-well plates and were incubated with indicated chemicals for 24h under normoxic or hypoxic conditions (0.2% O2). Media were then collected and VEGF levels were determined using Quantikine ELISA kits (R&D Systems, Inc., Minneapolis, MN). The results were expressed as concentrations of VEGF (pg/ml) per 5×104 cells/well. At these time points the drugs did not produce significant toxicity in LNCaP-Pro5 cells.
Luciferase reporter assays
To examine the transcriptional activity of HIF-1α, LNCaP-Pro5 cells (5×104 cells/well) were cotransfected with plasmids encoding a firefly luciferase reporter driven by a promoter containing an HRE and renilla luciferase under the control of an autologous promoter (pRL-CMV) (internal control for transfection efficiency) using TransFast (Promega Co., Madison, WI) following the manufacturer’s instructions. Luciferase activity was measured using the Dual-luciferase assay system (Promega Co, Madison, WI). Firefly luciferase activity was normalized by renilla luciferase activity and the indicated promoter activities were expressed as the average ratios (±SEM) from three independent experiments.
Small interfering RNA (siRNA)-mediated silencing of HIF-1α and eIF2α
siRNA mediated silencing of HIF-1α and eIF2α (Dharmacon RNA Technologies, Lafayette, CO) were performed as previously described (16). Following silencing, cells were incubated as indicated, VEGF expression was measured by ELISA, and HIF-1α expression was examined by immunoblotting. The efficiency of gene silencing was verified in each experiment by immunoblotting.
Quantitative real time-PCR
Total cellular mRNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. One microgram of total mRNA was reverse transcribed using SuperArray First-Strand cDNA Kit (Superarray Bioscience Co., Frederick, MD) and qPCR for HIF-1α and GAPDH (SuperArray Bioscience Co., Frederick, MD) were performed in triplicate using a Bio-Rad iCycler real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA). The amplification protocol consisted of one cycle at 95°C for 3 min, followed by 40 cycles at 95°C for 30 sec, 58°C for 30 sec, then 72°C for 30 sec. The melt-curve protocol, performed at the end of the amplification, consisted of 80 cycles beginning at 55°C for 10 sec, and then the temperature was increased by 0.5°C/cycle. A standard curve for each target gene was generated to determine the linear range and amplification efficiency. PCR efficiency greater than 80% was considered sufficient. The threshold cycle for each sample was fitted to the standard curve to calculate the expression level of the target gene relative to the input mRNA. The resulting data were analyzed with the iCycler iQ Real-Time Detection System software and expressed as the mean of ratios (relative expression to control) ±SEM and GAPDH served as internal loading control.
Polysome profile analysis
Polysome fractionation was performed as previously described (17). Briefly, cells were lysed with polysome lysis buffer (60mM NaCl, 15mM Tris-HCl pH7.5, 15mM MgCl2, 0.5% Triton X-100, 100μg/ml cycloheximide, and 1mM DTT) on ice for 10min. After centrifugation, supernatants were layered onto 10ml, 10–50% sucrose gradients composed of polysome extraction buffer (60mM NaCl, 15mM Tris-HCl pH7.5, 15mM MgCl2, 100μg/ml cycloheximide and 1mg/ml heparin). The gradients were centrifuged at 36,000rpm for 2h and 15min at 4°C in a SW41 rotor (Beckman L8–70M Ultra-centrifuge). Gradients were then collected as 0.5 ml fractions by pumping 60% sucrose into the bottom of the gradients and collecting them from the top using an ISCO fraction collection system (Density Gradient Fractionation System, TELEDYNE, ISCO Inc.) with concomitant measurement of the absorbance at 254nm (UA-6, UV.VIS Detector, BRABDEL). Total mRNA from each fraction was extracted using RNeasy kits (Qiagen, Valencia, CA) and One-step RT-PCR was performed in triplicate using the AgPath-ID One-Step RT-PCR Kit (Ambion, Inc., Autsin, TX) using primer pairs for HIF-1α and cyclophilin A (Applied Biosystems, Foster City, CA). The expression of each target gene was quantified using the StepOne™ Real-time PCR systems (Applied Biosystems, Foster City, CA).
Quantification of protein synthesis by [L-4,5-3H]Leucine incorporation
Equal numbers of cells were plated in 6-well plates and were incubated with 100nM bortezomib, 100nM NPI-0052, 40μM cycloheximide or 10μM thapsigargin for 4h under normoxic conditions. At the end of the experiments no significant differences in viable cell numbers were observed between untreated and treated samples. Cells were then trans-labeled with leucine-free medium (MP Biomedicals, Solon, OH) containing [L-4,5-3H]Leucine [2μCi/ml, GE Healthcare Bio-Sciences Corp., (Piscataway, NJ)] for 2h at 37°C under normoxic conditions. Excess unincorporated [L-4,5-3H]Leucine was removed by washing cells with ice-cold PBS. Cells were collected and lysed as previously described. Equal volumes of cellular protein (~20 μl) from each sample were precipitated by ice cold trichloroacetic acid (TCA, 10%, w/v) at 4°C for 30min. Precipitated proteins were then dissolved in 0.1M KOH and transferred to vials for scintillation counting.
Data analysis
Experiments presented in the figures are derived from or are representative of at least three independent repetitions. Statistical analyses were performed using GraphPad3.05 statistical software (GraphPad Software, San Diego, CA) using the Student’s t test, or one-way ANOVA where appropriate (P<0.05 was considered statistically significant).
Results
Bortezomib and NPI-0052 downregulate HIF-1α, HIF-2α and VEGF expression
Bortezomib and NPI-0052 inhibit angiogenesis but the molecular mechanisms involved remain unclear (4, 5, 9). HIF-1α is considered one of the most important pro-angiogenic transcription factors and its expression is tightly regulated by the ubiquitin/proteasome degradation pathway (10). We therefore assessed the effects of PIs on HIF-1α protein accumulation and its transcriptional activity in LNCaP-Pro5 cells. HIF-1α was almost undetectable under normoxic conditions but was strongly upregulated by exposure to hypoxia or CoCl2 (Figure 1A). Paradoxically, both PIs caused concentration dependent downregulation of HIF-1α protein (Figure 1A). Both PIs also downregulated HIF-2α (Figure 1A), which is regulated by the proteasome in a similar manner (18). These effects were associated with reduced HIF-1 transcriptional activity (Figure 1B) and VEGF expression (Figure 1C). The effects of the PIs were very similar to those caused by HIF-1α silencing (Figure 1D).
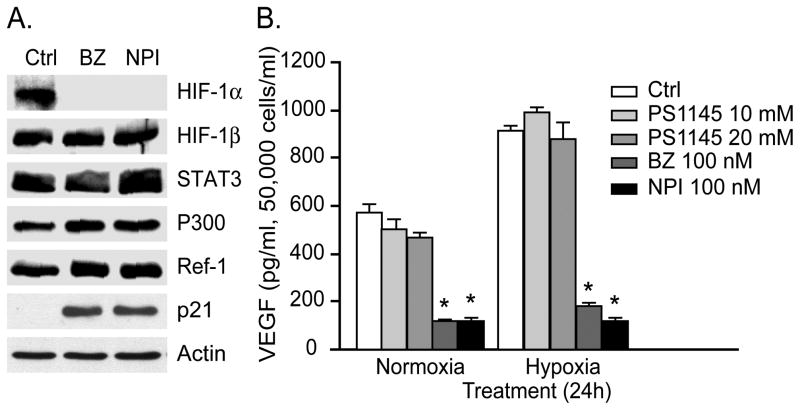
Figure 1. Effects of PIs on HIF-1α, HIF-2α and VEGF expression.
A. PIs downregulate HIF-1α and HIF-2α protein levels. LNCaP-Pro5 cells were exposed to increasing concentrations of bortezomib (BZ) or NPI-0052 (NPI) for 12h under normoxic or hypoxic conditions and HIF-1α was measured by immunoblotting. Actin served as a loading control. B. PIs block HIF-1α transcriptional activity. LNCaP-Pro5 cells were treated as above for 8h under normoxic or hypoxic conditions. HIF-1α transcriptional activities were measured using an HRE-driven firefly luciferase expression construct and a renilla luciferase construct as an internal normalization control. Data shown are mean±S.E., *P<0.001, compared to controls. C. PIs inhibit VEGF expression. LNCaP-Pro5 cells were treated as above for 12h or 24h under normoxic or hypoxic conditions. VEGF expression was measured by VEGF ELISA (R&D System) in conditioned media. Data shown are mean±S.E., *P<0.001, compared to control (Ctrl). D. Knockdown of HIF-1α inhibits VEGF expression. siRNA mediated knockdown of HIF-1α was carried out as described in Materials and Methods and VEGF expression was measured by VEGF ELISA (R&D System). Data shown are mean ±S.E., *P<0.001, compared to control. In parallel, HIF-1α expression was examined to confirm the silencing efficiency and actin served as a loading control.
The VEGF promoter contains several distinct cis-acting elements, including the HIF-1 binding sites (HREs) as well as binding sites for signal transducer and activator of transcription 3 (STAT3), activating protein-1 (AP-1), AP-2, NFκB, and SP-1 (11, 19). We therefore examined the effects of the PIs on several other VEGF regulators in LNCaP-Pro5 cells. We focused on HIF-1β/ARNT, STAT3, P300 and Ref-1/APE (Redox effector factor-1/apurinic/apyrimidinic endonuclease) because these factors form a hypoxia-inducible transcriptional complex with HIF-1α on the VEGF promoter’s HRE region (11, 19, 20). Strikingly, none of these molecules was downregulated by PIs (Figure 2A). We also examined the potential role of NFκB in the PIs-mediated suppression of VEGF expression since PIs inhibit NFκB activity via stabilization of IκBα. The IKK inhibitor, PS-1145, had no effect on VEGF expression (Figure 2B), even though NFκB activity was inhibited at these doses as measured by NFκB EMSA (electrophoretic mobility shift assay) (21).
Figure 2. Effects of PIs on other candidate VEGF promoter regulators.
A. LNCaP-Pro5 cells were exposed to 100nM bortezomib or 100nM NPI-0052 for 12h under hypoxic conditions. Total lysates were probed for expression of HIF-1α, HIF-1β, STAT3, p300, Ref-1 or p21WAF1 by immunoblotting and actin served as a loading control. B. Effects of PS-1145 on VEGF expression. LNCaP-Pro5 cells were exposed to 10/20μM PS-1145, 100nM bortezomib, or 100nM NPI-0052 for 24h under normoxic or hypoxic conditions. VEGF levels were measured by ELISA (R&D System) in conditioned media. Data shown are mean ±S.E., *P<0.001, compared to control.
Biphasic effects of PIs on HIF-1α’s stability
We next examined the effects of PIs on HIF-1α stability by immunoblotting using cellular extracts prepared from cells incubated with the protein translation inhibitor cycloheximide. LNCaP-Pro5 cells were preincubated with CoCl2 for 16h to stimulate HIF-1α accumulation and then exposed to PIs with or without CHX (Figure 3 and Table 1). Levels of HIF-1α were detectably lower in CHX-exposed cells as compared to controls at 30min and were almost undetectable by 2h. In cells exposed to CHX plus PIs, levels of HIF-1α were about 60% higher than the levels observed in cells exposed to CHX alone at 30min, although HIF-1α levels were significantly reduced by 2h. In cells exposed to PIs alone, HIF-1α accumulation was observed at 30min (~50% induction compared to control), but HIF-1α protein levels returned to control levels by 1h and then decreased. PIs downregulated HIF-1α so rapidly that there was almost no detectable HIF-1α in cells exposed to the PIs with or without CHX by 4h. Based on these observations, we conclude that PIs have biphasic effects on HIF-1α, causing an early accumulation of HIF-1α followed by profound downregulation of protein expression. Importantly, both PIs cause sustained proteasome inhibition in the same cells (>24h) and other labile proteins do in fact accumulate in the cells at 4h and later [Figure 2A and (6, 16)].
Figure 3. Effects of PIs on HIF-1α protein stability and HIF-1α mRNA translation.
A. Biphasic effects of PIs on HIF-1α stability. LNCaP-Pro5 cells were preincubated with 50μM CoCl2 for 16h, then exposed to 100nM bortezomib or 100nM NPI-0052 with or without 20μM cycloheximde for the time indicated in the presence of CoCl2. Total lysates were probed for HIF-1α expression by immunoblotting. B. PIs have no effect on total HIF-1α mRNA. LNCaP-Pro5 cells were exposed to 100nM bortezomib or 100nM NPI-0052 for 12h under normoxic or hypoxic conditions. Real time RT-PCR for HIF-1α was performed using the BioRad iCycler. Data shown are mean±S.E. C. PIs induce polysome dissociation. Representative polysome profiles isolated from total cellular lysates fractionated by a continuous sucrose gradient. LNCaP-Pro5 cells were exposed to 100nM bortezomib or 100nM NPI-0052 for 4h. The positions of the polysomes and non-polysomes (monosomes and mRNPs) are indicated. Note the accumulation of non-polysomes in the treated cells is indicative of decreased protein translation. D. PIs inhibit HIF-1α translation efficiency. Real-time RT-PCR analysis (RQ) of HIF-1α mRNA levels in each fraction was performed and the relative mRNA level of the first fraction was arbitrarily set at the value of 1. That HIF-1α mRNA shifted to lighter fractions in the treated cells is indicative of its translation being repressed. Bars, standard deviation.
Table 1.
Densitometric quantification of the results shown in Figure 3A.
| 15min | 30min | 1h | 2h | 4h | |
|---|---|---|---|---|---|
| Ctrl | 1 | 1 | 1 | 1 | 1 |
| CHX | 0.85 ± 0.05 | 0.44 ± 0.11 | 0.37 ± 0.09 | 0.14 ± 0.08 | 0.11 ± 0.04 |
| CHX+BZ | 0.81 ± 0.04 | 0.73 ± 0.25 | 0.16 ± 0.01 | 0.17 ± 0.01 | 0.07 ± 0.02 |
| CHX+NPI | 0.78 ± 0.11 | 0.67 ± 0.28 | 0.28 ± 0.11 | 0.20 ± 0.04 | 0.11 ± 0.01 |
| BZ | 0.91 ± 0.08 | 1.39 ± 0.21 | 0.93 ± 0.18 | 0.70 ± 0.19 | 0.11 ± 0.02 |
| NPI | 0.95 ± 0.05 | 1.76 ± 0.3 | 0.91 ± 0.12 | 0.77 ± 0.21 | 0.10 ± 0.03 |
HIF-1α/actin levels from untreated cells were arbitrarily set at the value of 1. HIF-1α signals following normalization to actin levels were expressed as relative densitometry units of the mean of the three repetitions. Data shown are mean±SEM.
Effects of PIs on polysome profiles and HIF-1α translation efficiency
PIs had no effects on HIF-1α total mRNA levels (Figure 3B), which excluded the possibility that PIs inhibit HIF-1α transcription. Considering that short-lived proteins are particularly sensitive to translational regulation, we performed polysome profiling analysis to directly determine whether or not PIs depress HIF-1α translation. The results revealed that PIs and thapsigargin (which inhibits the sarcoplasmic/endoplasmic Ca2+-ATPase to induce ER stress) induced substantial polysome dissociation and accumulation of monosomes and messenger ribonucleoprotein (mRNPs) at 4h (Figure 3C and data not shown). Longer periods of drug exposure triggered almost complete polysome dissociation (data not shown). We next quantified the relative expression of HIF-1α and cyclophilin A (internal control) in each fraction using real time RT-PCR. In untreated cells, the HIF-1α mRNA peak coincided with heavy polysome fractions, whereas PIs induce the HIF-1α mRNA to shift to the lighter fractions, confirming that they disrupt HIF-1α translation. The normally well-translated cyclophilin A mRNA was modestly affected, suggesting that the effects were somewhat selective for HIF-1α mRNA (Figure 3D and data not shown).
Effects of PIs on the UPR and eIF2α-dependent protein translation
Recent studies have demonstrated that PIs induce ER stress in cancer cells (7, 8). Cells have evolved the UPR to alleviate ER stress or to induce apoptosis if the stress is excessive. Protein kinase R (PKR)-like ER kinase (PERK) plays an important role in the UPR by phosphorylating eIF2α and downregulating global protein synthesis (22, 23). Bortezomib, NPI-0052, and thapsigargin all induced phosphorylation of PERK and eIF2α in LNCaP-Pro5 cells (Figure 4A and 4B). Hypoxia itself slightly induced eIF2α phosphorylation, consistent with other studies implicating PERK/eIF2α in the adaptive response to hypoxia (24, 25). The effects of PIs- or TG- were associated with inhibition of global protein synthesis as measured by [L-4,5-3H]Leucine incorporation (Figure 4C) and the inhibitory effects of PIs on protein synthesis lasted at least 12h (data not shown). These data are consistent with the observation that PIs induced polysome dissociation (Figure 3C). Finally, thapsigargin and another classic ER stress inducer tunicamycin (which inhibits N-linked protein glycosylation) also induced eIF2α phosphorylation, attenuated protein synthesis and downregulated HIF-1α protein expression (Figure 4D).
Figure 4. Effects of PIs on the unfolded protein response.
A. PIs induce PERK phosphorylation. LNCaP-Pro5 cells were exposed to 100nM bortezomib, 100nM NPI-0052 or 10μM thapsigargin for 4h and phosphorylated PERK and actin levels were measured by immunoblotting. B. PIs induce eIF2α phosphorylation. LNCaP-Pro5 cells were treated as above under normoxic (Nor) and hypoxic conditions and phosphorylated and total eIF2α levels were measured by immunoblotting. The numbers located below each lane correspond to the quantification of the phosphorylated eIF2α signals by densitometry adjusted to the total eIF2α protein levels. The phosphorylation of eIF2α in the untreated group is arbitrarily set at the value of 1. C. PIs attenuate general protein synthesis. LNCaP-Pro5 cells were exposed to 100nM bortezomib, 100nM NPI-0052, 10μM thapsigargin or 40μM cycloheximide for 4h under normoxic conditions and protein synthesis was measured by [L-4,5-3H]Leucine incorporation. Data shown are mean±S.E., *P<0.001. D. Thapsigargin, tunicamycin and cycloheximide downregulated HIF-1α protein levels. LNCaP-Pro5 cells were incubated with 100nM bortezomib, 100nM NPI-0052, 10μM thapsigargin, 5μg/ml tunicamycin or 40μM cycloheximide for 12h under hypoxic conditions and HIF-1α levels were determined by immunoblotting.
Direct role of eIF2α phosphorylation in HIF-1α downregulation
To determine whether downregulation of HIF-1α was phospho-eIF2α dependent, we first examined the levels of HIF-1α in LNCaP-Pro5 cells transfected with siRNA specific for eIF2α or an off-target control construct. PIs induced the accumulation of HIF-1α in cells depleted eIF2α, and similar effects were observed in cells exposed to thapsigargin (Figure 5A). Knockdown of eIF2α attenuated basal protein synthesis (about 35% reduction compared to the off-target control transfected cells) and partially reversed the effects of PIs on protein translation repression (about 60% rescue compared to the off-target control transfected cells in treated groups, Supplemental Figure 1). We next compared the effects of PIs on HIF-1α in MEF cells expressing wild type (eIF2α51SS) or a phosphorylation-deficient knock-in mutant form of eIF2α (eIF2α51AA). PIs induced phosphorylation of eIF2α and translational repression in wild-type MEFs but not in the mutant MEFs. Consistent with the results obtained with LNCaP-Pro5 cells, PIs induced strong HIF-1α accumulation in the mutant MEFs while only have modest effects on HIF-1α accumulation in the wild-type MEFs (Figure 5B and 5C). Thus, PI-induced downregulation of HIF-1α is largely eIF2α phosphorylation dependent.
Figure 5. Role of eIF2α in the effects of PIs on HIF-1α expression and protein synthesis.
A. Knockdown of eIF2α rescued HIF-1α expression. LNCaP-Pro5 cells were transfected with a siRNA construct specific for eIF2α or a non-targeted control siRNA. Transfected cells were incubated with 100nM bortezomib, 10nM NPI-0052 or 5μM thapsigargin for 8h under hypoxic conditions. Levels of HIF-1α and eIF2α were examined by immunoblotting and actin served as a loading control. B. Effects of PIs on eIF2α and protein translation in MEFs. MEFs were incubated with 100nM bortezomib, 100nM NPI-0052 or 10μM thapsigargin for 4h. Phosphorylated eIF2α(Ser51) and total eIF2α levels were measured by immunoblotting. Protein synthesis was measured by [L-4,5-3H]Leucine incorporation. Data shown are mean±S.E. C. Effects of PIs on HIF-1α in MEFs. MEFs were treated as above for 12h under hypoxic conditions. Levels of HIF-1α and total eIF2α were measured by immunoblotting.
Cell line-dependent effects of PIs on eIF2α phosphorylation and HIF-1α accumulation
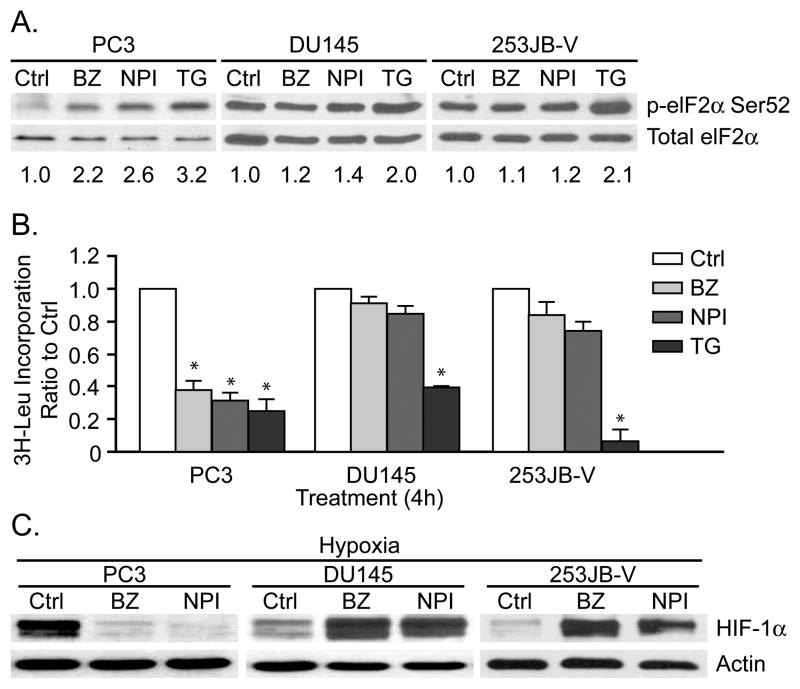
PIs induce phosphorylation of eIF2α in some cancer cell lines but not in others (26–28). Therefore, we characterized the effects of PIs on eIF2α phosphorylation and HIF-1α protein levels in three additional genitourinary cancer cell lines (Figure 6). In PC3 prostate cancer cells, both PIs induced eIF2α phosphorylation and global translation repression, and these effects were associated with HIF-1α downregulation. In contrast, both drugs failed to induce phosphorylation of eIF2α or inhibit protein synthesis in DU145 or 253JB-V cells, and they actually promoted the accumulation of HIF-1α in both cell lines. Both PIs inhibited the 20S proteasome and caused accumulation of p21WAF1 in all four cell lines (Figure 2A and data not shown), demonstrating that the observed differences were not caused by differential drug-target interactions. Therefore, the effects of PIs are heterogeneous, and downregulation of HIF-1α only occurs in cells that display PIs induced phosphorylation of eIF2α and protein translation repression.
Figure 6. Differential effects of PIs on UPR and HIF-1α protein levels in different cancer cells.
A. Differential effects of PIs on eIF2α phosphorylation. PC3, DU145 and 253JB-V cells were incubated with 100nM bortezomib, 100nM NPI-0052, or 10μM thapsigargin for 4h. Phosphorylated eIF2α(Ser52) and total eIF2α levels were measured by immunoblotting. The numbers located below each lane corresponded to the levels of phospho-eIF2α which were determined by densitometry analysis and adjusted to total eIF2α protein levels. The phosphorylation of eIF2α in the untreated group was arbitrarily set at the value of 1. B. Differential effects of PIs on protein synthesis. PC3, DU145 and 253JB-V cells were treated as above for 4h and protein synthesis was measured by [L-4,5-3H]Leucine incorporation. Data shown are mean±S.E., *P<0.001, compared to control. C. Differential effects of PIs on HIF-1α. PC3, DU145 and 253JB-V cells were incubated with 100nM bortezomib or 100nM NPI-0052 for 12h under hypoxic conditions and HIF-1α levels were measured by immunoblotting.
Discussion
Proteasome inhibition is an effective therapeutic strategy for certain cancers, and it also appears to underlie the tissue damage that occurs in neurodegenerative diseases (1, 2, 29). We previously showed that the anti-tumor effects of bortezomib are associated with suppression of VEGF expression (9). The effects of PIs on VEGF expression are paradoxical given the well established role VHL/proteasome- mediated degradation plays in the control of HIF-1α protein expression. Here we report that PIs induce an unexpected downregulation of HIF-1α at translational level via a PERK/eIF2α-mediated mechanism. Several other anti-tumor agents, including gefitinib, cetuximab, nelfinavir and rapamycin, also suppress HIF-1α expression by blocking its translation. However, these effects involve blockade of the phosphatidylinositol-3-kinase (PI3K)/AKT/glycogen synthesis kinase 3 beta (GSK3β)/mammalian target of rapamycin (mTOR) pathway (11, 30–32). The PI3K/AKT/mTOR signaling pathway profoundly affects 7-methyl guanosine cap-dependent mRNA translation through phosphorylation of downstream targets such as 4E-BP and S6K and inactivation of the eIF4F translation complex rather than inducing eIF2α phosphorylation (33). Therefore, it is possible that PIs could be combined with these other agents to produce even stronger inhibition of HIF-1α-dependent angiogenesis.
We showed here that two cell lines displayed efficient PIs-mediated eIF2α phosphorylation and downregulation of HIF-1α (LNCaP-Pro5 and PC3) and two did not (DU145 and 253 JB-V). The basal levels of eIF2α phosphorylation appeared to correlate with these differences as DU145 and 253 JB-V cells contained high basal phosphorylated eIF2α and LNCaP-Pro5 and PC3 cells did not. However, there were no measurable differences in global translation rates among the cell lines as measured by [L-4,5-3H]Leucine incorporation (Supplemental figure 2), indicating that basal eIF2α phosphorylation observed in DU145 and 253JB-V did not activate the protein synthesis checkpoint. We are actively seeking mechanistic explanation(s) for the observed heterogeneity that eIF2α is phosphorylated at baseline in some tumor cell lines but not in others.
The proteasome contains three catalytic sites in its inner β-rings (1, 2, 4). We knocked down one (β5) or all three (β1, β2 and β5) of the active sites of the proteasome using siRNA and measured the effects on protein translation and HIF-1α protein level. Partial knockdown of proteasome active site(s) attenuated protein translation, but the effects were much less dramatic than those obtained with the chemical inhibitors and caused no obvious HIF-1α downregulation (Supplemental figure 3). Therefore, we speculate that quantitative differences between the effects of the PIs and proteasome subunits knockdown on translation inhibition and downstream cellular stress responses probably account for the differential effects observed.
The selective effects of PIs on HIF-1α expression also raise an additional question. Global translational repression would be expected to affect all short-lived proteins, yet p53 and p21 accumulate in LNCaP-Pro5 cells exposed to PIs [Figure 2A and (34)]. Furthermore, previous studies reported that phospho-eIF2α mediated translational repression were associated with downregulation of IκBα and cyclin D in MEFs (35, 36), but PIs failed to downregulate cyclin D in LNCaP-Pro5 cells while thapsigargin did (data not shown). Our explanation for these discrepancies is that the translation of all of these proteins is probably inhibited in a phospho-eIF2α-dependent manner in cells exposed to PIs, but what distinguishes the outcome of the protein in question largely depends on whether it is efficiently degraded by a proteasome-independent pathway or not. Interestingly, other studies have shown that bortezomib has inhibitory effects on HIF-1α transcriptional activity even when HIF-1α accumulates (37, 38). Consistent with this conclusion, VEGF expression was blocked by PIs in DU145 and 253JB-V cells, even though the PIs promoted HIF-1α accumulation in these two cell lines (Figure 6 and data not shown). Therefore, PIs might function as HIF-1α inhibitors regardless of whether they deplete cells of HIF-1α or not. In future studies it will be important to determine whether these mechanisms contribute to the anti-angiogenic effects of proteasome inhibitors in preclinical models and patient tumors in vivo.
Supplementary Material
References
- 1.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–21. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 2.Voorhees PM, Dees EC, O’Neil B, Orlowski RZ. The proteasome as a target for cancer therapy. Clin Cancer Res. 2003;9:6316–25. [PubMed] [Google Scholar]
- 3.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Proteasome inhibition in the treatment of cancer. Cell cycle. 2005;4:290–6. [PubMed] [Google Scholar]
- 4.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer Invest. 2004;22:304–11. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 5.Chauhan D, Catley L, Li G, et al. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8:407–19. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz S, Krupnik Y, Keating M, Chandra J, Palladino M, McConkey D. The proteasome inhibitor NPI-0052 is a more effective inducer of apoptosis than bortezomib in lymphocytes from patients with chronic lymphocytic leukemia. Mol Cancer Ther. 2006;5:1836–43. doi: 10.1158/1535-7163.MCT-06-0066. [DOI] [PubMed] [Google Scholar]
- 7.McConkey DJ, Zhu K. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist Updat. 2008;11:164–79. doi: 10.1016/j.drup.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Egger L, Madden DT, Rheme C, Rao RV, Bredesen DE. Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell death and differentiation. 2007;14:1172–80. doi: 10.1038/sj.cdd.4402125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams S, Pettaway C, Song R, Papandreou C, Logothetis C, McConkey DJ. Differential effects of the proteasome inhibitor bortezomib on apoptosis and angiogenesis in human prostate tumor xenografts. Mol Cancer Ther. 2003;2:835–43. [PubMed] [Google Scholar]
- 10.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 11.Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanisms of anti-tumour activity. Nat Rev Cancer. 2008;8:579–91. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 12.Kaelin WG. Von hippel-lindau disease. Annu Rev Pathol. 2007;2:145–73. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 13.Alqawi O, Moghaddas M, Singh G. Effects of geldanamycin on HIF-1alpha mediated angiogenesis and invasion in prostate cancer cells. Prostate cancer and prostatic diseases. 2006;9:126–35. doi: 10.1038/sj.pcan.4500852. [DOI] [PubMed] [Google Scholar]
- 14.Powis G, Kirkpatrick L. Hypoxia inducible factor-1alpha as a cancer drug target. Mol Cancer Ther. 2004;3:647–54. [PubMed] [Google Scholar]
- 15.Trastour C, Benizri E, Ettore F, et al. HIF-1alpha and CA IX staining in invasive breast carcinomas: prognosis and treatment outcome. Int J Cancer. 2007;120:1451–8. doi: 10.1002/ijc.22436. [DOI] [PubMed] [Google Scholar]
- 16.Lashinger LM, Zhu K, Williams SA, Shrader M, Dinney CP, McConkey DJ. Bortezomib abolishes tumor necrosis factor-related apoptosis-inducing ligand resistance via a p21-dependent mechanism in human bladder and prostate cancer cells. Cancer research. 2005;65:4902–8. doi: 10.1158/0008-5472.CAN-04-3701. [DOI] [PubMed] [Google Scholar]
- 17.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. The Journal of cell biology. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Uchida K, Endler A, Shibasaki F. Mammalian Tumor Suppressor Int6 Specifically Targets Hypoxia Inducible Factor 2{alpha} for Degradation by Hypoxia- and pVHL-independent Regulation. The Journal of biological chemistry. 2007;282:12707–16. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
- 19.Semenza G. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002;64:993–8. doi: 10.1016/s0006-2952(02)01168-1. [DOI] [PubMed] [Google Scholar]
- 20.Gray MJ, Zhang J, Ellis LM, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–20. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 21.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. The Journal of biological chemistry. 2002;277:16639–47. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 22.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nature reviews. 2008;8:663–74. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 23.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 24.Bi M, Naczki C, Koritzinsky M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. Embo J. 2005;24:3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koumenis C, Naczki C, Koritzinsky M, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Molecular and cellular biology. 2002;22:7405–16. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawrocki ST, Carew JS, Dunner K, Jr, et al. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer research. 2005;65:11510–9. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- 27.Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. The Journal of biological chemistry. 2005;280:14189–202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 28.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 30.Semenza GL. Development of novel therapeutic strategies that target HIF-1. Expert Opin Ther Targets. 2006;10:267–80. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]
- 31.Land SC, Tee AR. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. The Journal of biological chemistry. 2007;282:20534–43. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 32.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Mol Cancer Res. 2006;4:471–9. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 33.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–46. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 34.Williams SA, McConkey DJ. The proteasome inhibitor bortezomib stabilizes a novel active form of p53 in human LNCaP-Pro5 prostate cancer cells. Cancer research. 2003;63:7338–44. [PubMed] [Google Scholar]
- 35.Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng J, Lu PD, Zhang Y, et al. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Molecular and cellular biology. 2004;24:10161–8. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaluz S, Kaluzova M, Stanbridge EJ. Proteasomal inhibition attenuates transcriptional activity of hypoxia-inducible factor 1 (HIF-1) via specific effect on the HIF-1alpha C-terminal activation domain. Molecular and cellular biology. 2006;26:5895–907. doi: 10.1128/MCB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birle DC, Hedley DW. Suppression of the hypoxia-inducible factor-1 response in cervical carcinoma xenografts by proteasome inhibitors. Cancer research. 2007;67:1735–43. doi: 10.1158/0008-5472.CAN-06-2722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.