SUMMARY
Facioscapulohumeral muscular dystrophy (FSHD) is an enigmatic disease associated with epigenetic alterations in the subtelomeric heterochromatin of the D4Z4 macrosatellite repeat. Each repeat unit encodes DUX4, a gene that is normally silent in most tissues. Besides muscular loss, most patients suffer retinal vascular telangiectasias. To generate an animal model, we introduced a doxycycline-inducible transgene encoding DUX4and 3′genomic DNA into a euchromatic region of the mouse X chromosome. Without induction, DUX4 RNA was expressed at low levels in many tissues and animals displayed a variety of unexpected dominant leaky phenotypes, including male-specific lethality. Remarkably, rare live-born males expressed DUX4 RNA in the retina and presented a retinal vascular telangiectasia. By using doxycycline to induce DUX4 expression in satellite cells, we observed impaired myogenesis in vitro and in vivo. This mouse model, which shows pathologies due to FSHD-related D4Z4 sequences, is likely to be useful for testing anti-DUX4 therapies in FSHD.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is a common degenerative myopathy caused by illicit recombination within D4Z4, a subtelomeric macrosatellite repeat on chromosome 4 (van Deutekom et al., 1993; Wijmenga et al., 1992). Array contractions cause chromatin changes (de Greef et al., 2009; Gabellini et al., 2002; van Overveld et al., 2003; Zeng et al., 2009) but cause disease only on a specific allele of chromosome 4, termed 4qA161 (Lemmers et al., 2004; Lemmers et al., 2007). FSHD non-permissive alleles, and related D4Z4 repeats on chromosome 10 (Bakker et al., 1995; Deidda et al., 1996), lack an ATTAAA polyadenylation sequence downstream of the terminal repeat (Dixit et al., 2007; Lemmers et al., 2010). These data suggest that repeat contractions result in an mRNA transcript produced from D4Z4, which must be polyadenylated to cause disease.
The D4Z4 transcript bears an open reading frame encoding a double homeodomain protein named DUX4 (Gabriels et al., 1999) and this protein is specifically expressed, albeit extremely weakly, in FSHD (Dixit et al., 2007; Snider et al., 2010). Expression of DUX4 at very low levels interferes with myogenesis and sensitizes cells to oxidative stress (Bosnakovski et al., 2008b). These low level effects are intriguing as defects in myogenic gene expression (Celegato et al., 2006; Winokur et al., 2003b), and sensitivity to oxidative stress (Turki et al., 2012; Winokur et al., 2003a) have been detected in FSHD muscle and primary cell cultures. High levels of DUX4 expression cause rapid cell death in vitro (Bosnakovski et al., 2008b; Kowaljow et al., 2007).
In addition to muscle wasting, most individuals afflicted with FSHD have subclinical retinal vascular pathologies involving vascular tortuosity, microaneurysms, occlusions, and occasionally small exudates (Fitzsimons et al., 1987; Padberg et al., 1995).
A transgenic mouse has recently been described that carries an insertion of tandem arrays of a 2.5-unit FSHD allele. Although transcription could be detected in several tissues, and very rare DUX4+ nuclei were detected in cultured cells from this animal (Krom et al., 2013), the animal was normal except for an eye keratitis that developed with age. With the aim of studying the effect of DUX4 expression in vivo, we generated mice with a doxycycline (dox)-inducible DUX4 transgene upstream of HPRT. Unexpectedly, in the absence of dox, these animals display a variety of pathologies. We characterize these pathologies, and use this model to investigate effects of D4Z4/DUX4 expression on myogenic progenitor cell activity.
RESULTS
To generate a dox-inducible DUX4-expressing mouse, we used inducible cassette exchange recombination (Bosnakovski et al., 2008b; Iacovino et al., 2011) to insert a genomic DNA fragment encoding the DUX4 ORF and 3′ sequences from the terminal D4Z4 repeat up to the EcoRI site into a euchromatic site on the X-chromosome. This site, upstream of a functional HPRT gene, was chosen for its ability to give reliable transgene expression (Bronson et al., 1996; Cvetkovic et al., 2000; Portales-Casamar et al., 2010; Touw et al., 2007). The integration places this gDNA into a dox-inducible locus regulated by a second generation, tight, tet-response element (Agha-Mohammadi et al., 2004). The rtTA for the Tet-On system is expressed ubiquitously from Rosa26 (Hochedlinger et al., 2005). The integration vector provides an SV40 poly A signal downstream of the inserted DNA. We named this transgene iDUX4(2.7), because DUX4 is on a 2.7 kb genomic fragment (Fig. 1A).
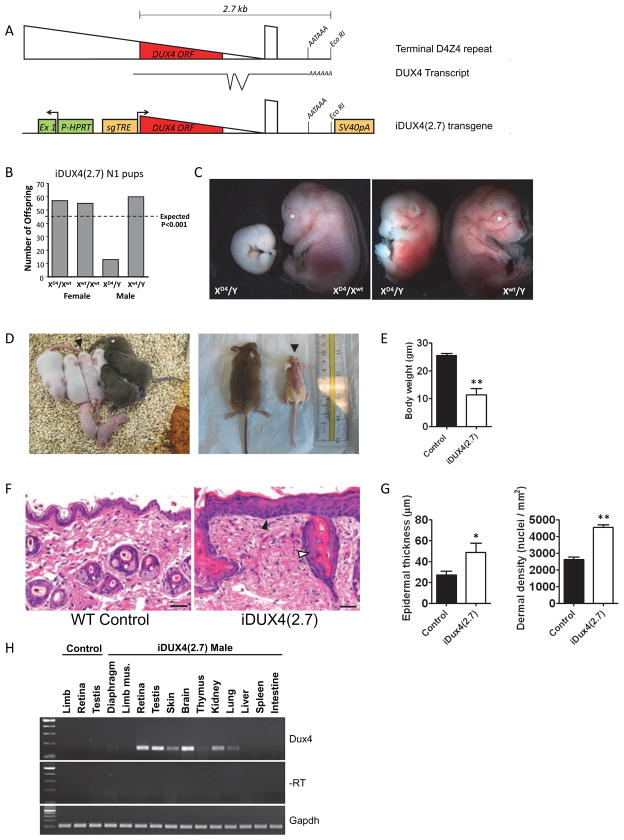
Figure 1. The iDUX4(2.7) transgene is male-specific dominant lethal.
(A) Schematics of the terminal D4Z4 repeat (above), the DUX4 transcript with introns and proposed AATAAA polyadenylation signal (below) and the construct integrated into the X chromosome (bottom). HPRT, hypoxanthine phosphor-ribosyl transferase; Ex1, exon 1, sgTRE, second generation tet-response element.
(B) Genotypes of live pups from iDUX4(2.7) female carriers bred to WT males and chi-square P-values.
(C) iDUX(2.7) embryos at E14.5. XD4 indicates the transgenic iDUX4(2.7)-bearing chromosome. Most male carrier embryos display severe growth delay or resorption.
(D) Examples of litters with runted iDUX4(2.7) males. Left: a P5 litter with two obvious runts, and one animal, identified by arrowhead, with some hair loss that later became runted. Right: littermate WT and iDUX4(2.7) males at P28.
(E) Weight at 6 weeks, p<0.05.
(F) H&E staining of skin. Note the hypertrophic epithelium (filled arrowhead) and the abnormal glands (open arrowhead). Scale bar = 20 μm.
(G) The epidermal thickness and nuclear density in the dermis were measured at 6 locations per section and averaged (* p<0.04; ** p<0.001) and the experiment was repeated with three mice.
(H) RT-PCR-detection of the DUX4 transcript in tissues of iDUX4(2.7) transgenic male mice. Robust and repeatable expression were always seen in the brain, retina and testis. Other tissues displayed sporadic and weaker expression.
The iDUX4(2.7) transgene is male-specific dominant lethal
Blastocyst injection generated chimeric males with poor transmission of the transgene: only two F1 progeny were obtained after a year of mating chimeras to C57BL/6 females. These carrier females displayed a striped mosaic scaly skin, mild alopecia phenotype. They transmitted the transgene to female progeny, but most litters lacked male carriers (Fig. 1B) demonstrating that the DUX4(2.7) transgene behaved as a male-specific dominant embryonic lethal. Evaluation of embryonic litters at E14.5 revealed male carrier fetuses with various degrees of developmental delay (Fig. 1C). Rarely, carrier males survived to birth, and these animals were runted, displayed the skin phenotype homogeneously, and invariably died before two months of age. Notably, when carrier males survived to term, they were usually of similar size to littermate controls at birth. The runting and the skin phenotypes usually became apparent within a few days, but were sometimes not initiated until the second week (Fig. 1D) and by 6 weeks, iDUX4(2.7) males were significantly underweight (Fig. 1E). On histological examination, the skin was found to have excessive numbers of cells, in both the epidermis and dermis (Fig. 1F, G), features that may explain the flaky scaly skin, and alopecia. We evaluated various tissues from affected males for leaky expression of the transgene (Fig. 1H). Unexpectedly, expression in skin, as in most other tissues was very low and inconsistently detected at 38 cycles of RT-PCR. Nor did we detect the DUX4 protein in any tissues in the absence of dox. This situation is very reminiscent of patients with FSHD, in which spliced RNA transcripts from the terminal D4Z4 element can be detected by RT-PCR in most samples, but protein is exceedingly difficult to detect.
However, DUX4 mRNA was consistently detected in testis, retina, and brain. DUX4 has been detected in normal human testis (Snider et al., 2010), and FSHD patients present retinal vascular defects, therefore, we evaluated these tissues in greater detail. Compared to sibling controls, testes of 42 day-old iDUX4(2.7) males displayed a marked defect in gametogenesis. Seminiferous tubule cross sections showed almost complete loss of spermatocytes during late prophase in most tubules. This resulted in relatively few tubules that contained round haploid spermatids, and a lack of elongating spermatids compared to wild-type (WT) siblings (Fig. 2A, B).
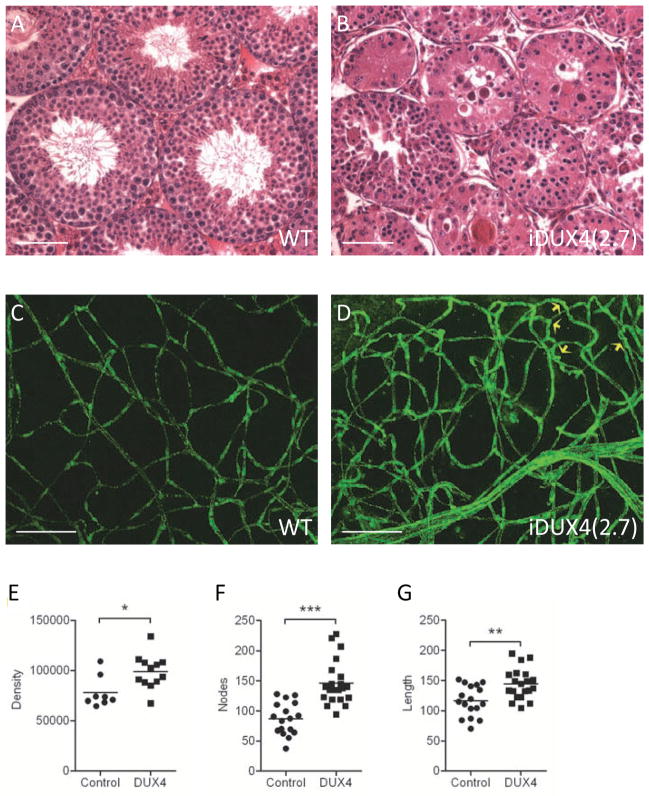
Figure 2. Phenotypes in testis and retina.
(A) Cross section through WT seminiferous tubules. Scale bar = 50 μm.
(B) Cross section through iDUX4(2.7) seminiferous tubules.
(C) Vasculature in WT retina: montage of z-stack laser scanning confocal microscopy images of retina flat mounts stained with anti-CD31/PECAM showing, normal presentation of vasculature. Scale bar = 50 μm.
(D) Vasculature in iDUX4(2.7) retina showing dense network of disorganized and twisted/looping vessels. Yellow arrows indicate twisted vessels.
(E) Morphometric analysis of retinal images following skeletonization showing vascular density measured as immunoreactive pixels (p<0.05).
(F) Morphometric quantification of vessel branching measured by nodes (p<0.001).
(G) Morphometric quantification of total vessel length. For E–G, each point represents values/field, averaged from 3–4 different fields per retina; n=7 iDUX4, n=8 WT (p< 0.01).
To evaluate retinae, we generated montages of z-stack LSCM images of retinal whole mounts stained with PECAM antibody. The iDUX4(2.7) males displayed features associated with telangiectasia: excessive branching with looping and twisted retinal vessels, which appear more dilated than control retinae (Fig. 2C, D). Morphometric analysis of these images revealed a dramatic increase in vessel branching (nodes), as well as significant increases in vessel length and overall density of vessels in iDUX4(2.7) males compared to controls (Fig. 2E–G), suggestive of increased neovascularization.
iDUX4(2.7) males have proportionally weaker muscles but show no evidence of muscular dystrophy prior to 6 weeks of age
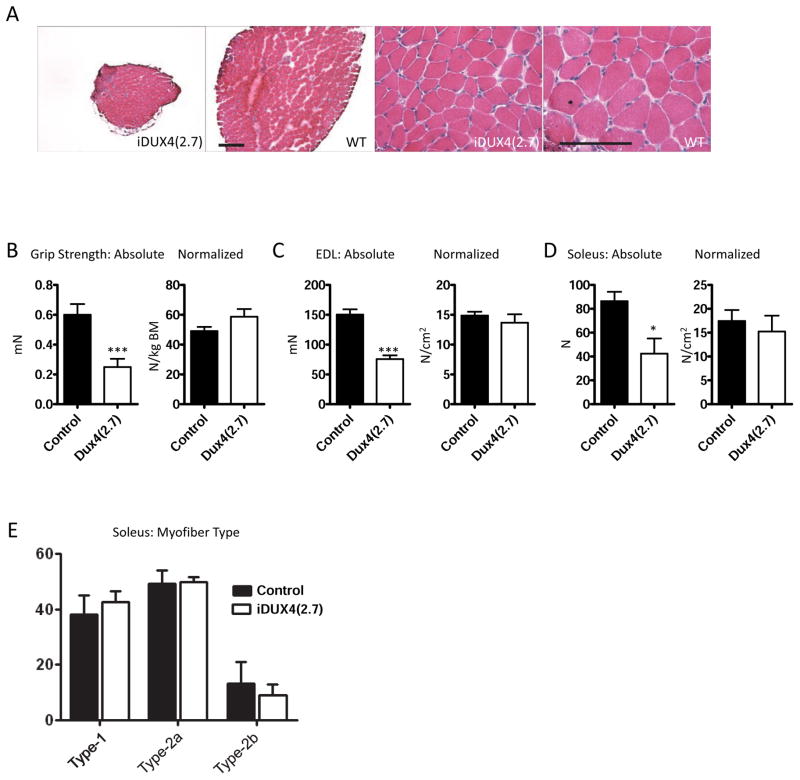
Muscles of the iDUX4(2.7) males were smaller than those of their matched sibling controls, and fibers were fewer and smaller, however muscles did not become overtly dystrophic within the short lifespan of these mice (Fig. 3A). Grip-strength measurements revealed that the iDUX4(2.7) males were much weaker than sibling controls, but strength was proportional to body size (Fig. 3B). We measured the force generation capacity of isolated extensor digitorum longus (EDL) and soleus muscles. Both muscles produced much lower absolute force, however when normalized to muscle cross sectional area, the specific force generating capacity was not significantly different from WT (Fig. 3C–D). Soleus muscle fiber type distribution, based on myosin heavy chain isoform expression (Types 1, 2a, and 2x), was not different between iDUX4(2.7) and wild-type mice (Fig. 3E).
Figure 3. Muscle in iDUX4(2.7) animals.
(A) H+E staining of cross sections through soleus muscles of iDUX4(2.7) males (left) and male littermate controls (right). Scale bar is 100 μm (low magnification images) or 200 μm (high magnification images).
(B) Grip strength measurements at 6 weeks, absolute at left (p<0.001), normalized to body mass at right (N=4).
(C) Maximal isometric force generated by the extensor digitorum longus muscle at 6 weeks, absolute at left (p<0.001), normalized to cross sectional area at right (N=4).
(D) Maximal isometric force generated by the soleus muscle at 6 weeks, absolute at left (p=0.026), normalized to cross sectional area at right (N=4).
(E) Fiber-type analysis based on myosin heavy chain isoform expression.
Demethylation of DUX4 in myogenic and fibroadipogenic (FAP) progenitors from muscle
To determine whether the integrated transgene more resembled an FSHD-associated or a WT allele, we evaluated methylation of the transgene in rare liveborn iDUX4(2.7) males by bisulfite sequencing. We initially evaluated testis and hind limb muscle, tissues with visualizable vs. barely detectable DUX4 mRNA. Remarkably, in both tissues, the transgene was completely demethylated (Fig. 4A). We then initiated cultures of total muscle mononuclear cells and subsequently FACS-purified and subcultured the α7-integrin− PDGFRα+ FAP (Joe et al., 2010; Uezumi et al., 2010) and the α7-integrin+ PDGFRα− myoblast fractions. The transgene was likewise completely demethylated in both progenitor cell types (Fig. 4A and Fig. S1).
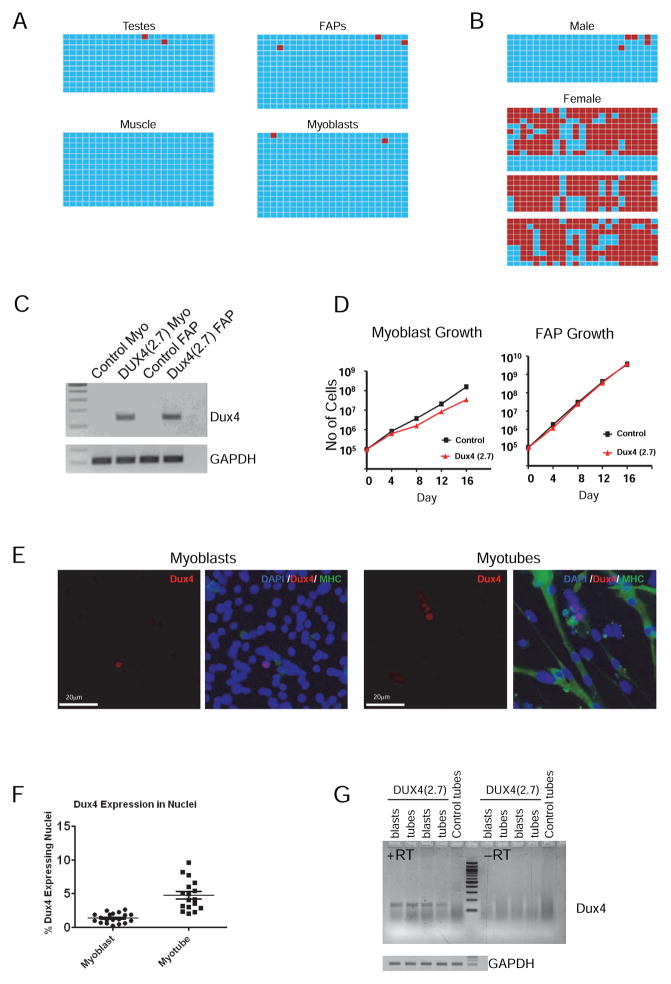
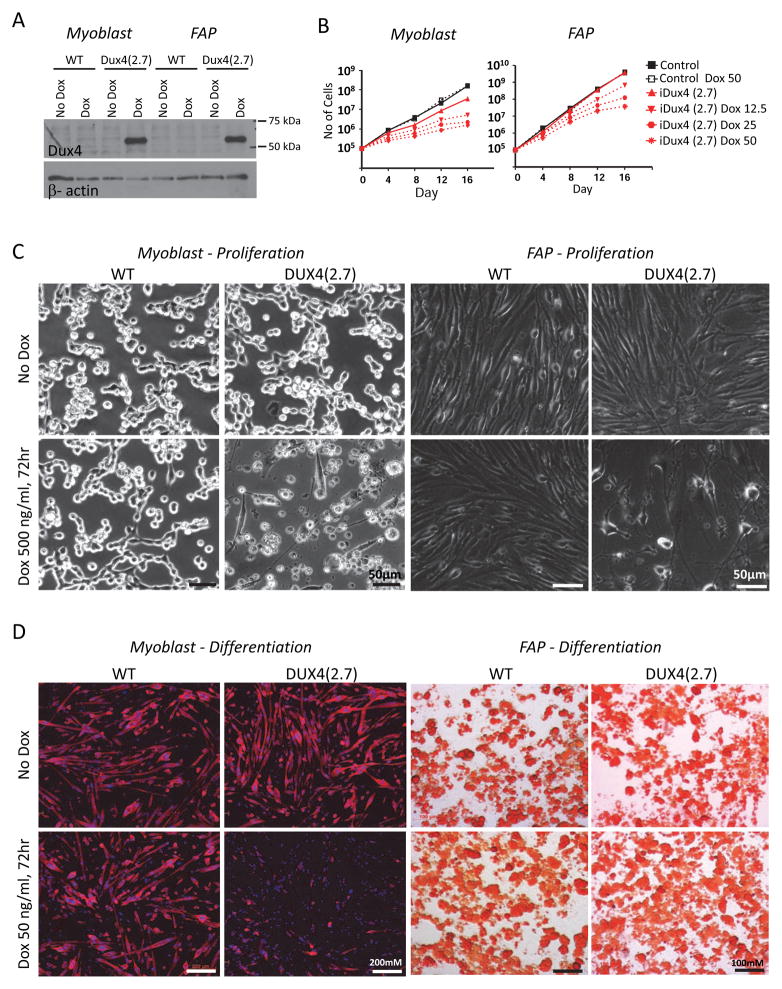
Figure 4. Methylation and DUX4 expression in primary cells.
(A) Methylation of the iDUX4(2.7) transgene in various tissues, and primary muscle cells. Bisulfite-treated DNA was amplified by PCR, cloned and sequenced. Blue boxes indicate unmethylated cytosines of CpG dinucleotides and red boxes indicate methylated cytocines along the PCR product.
(B) Methylation of the transgene in male and female peripheral blood. One male aged 6 weeks, and three independent females aged 6, 9, and 36 weeks are shown. Methylated sequences in females were greatly overrepresented compared to the 1:1 ratio predicted by X-inactivation for a heterozygous female (p=0.0).
(C) RT-PCR detection of the DUX4 transcript in proliferating myoblasts and FAPs.
(D) Growth rate of iDUX4(2.7) myoblasts and FAPs compared to cells from WT littermate controls. Myoblasts displayed a significant growth disadvantage. No difference was seen in FAP growth rate. One experiment of 3 similar replicates is shown.
(E) Immunofluorescent detection of DUX4 protein in myoblasts and myotubes. Sections are also stained with DAPI (blue) and antibody to myosin heavy chain (MHC, in green).
(F) Quantification of DUX4+ nuclei, expressed as average percentage and SD of total per microscopic field, over 19 separate fields. One of 3 similar replicates is shown.
(G) Comparison of levels of DUX4 expression by RT-PCR in myoblasts vs. myotubes. See also Figure S1.
Biased X-inactivation in females
We then investigated methylation in females, using peripheral blood samples of female iDUX4(2.7) heterozygous breeders. In stark contrast to males, sequences from females indicated that the gene could be in one of two states: either similar to that seen in males, or heavily methylated. This is consistent with X-inactivation, which, when it occurred on the DUX4-bearing X, would result in high levels of methylation. Because X-inactivation is random, an equal proportion of the two states is expected. Remarkably, the methylated sequences greatly outnumbered the demethylated sequences in females (Fig. 4B). This X-inactivation bias strongly suggests that there is selection against cells that inactivate the WT X, leading female heterozygotes to be composed mainly of cells in which the DUX4-bearing X was inactivated.
Expression of DUX4 in muscle-derived progenitors
In contrast to the difficulty of detecting DUX4 transcript in primary muscle tissue, the DUX4 transcript was detectable at low levels in both myoblasts and FAPs (Fig. 4C). Notably, in iDUX4(2.7) cultures, the FAP population predominated prior to FACS and isolated iDUX4(2.7) myoblasts had a clear growth disadvantage compared to their WT controls, while iDUX4(2.7) FAPs showed no growth disadvantage (Fig. 4D). We next sought to detect the DUX4 protein in isolated myoblasts and FAPs. We found no detectable protein expression in FAPs (not shown), however myoblast cultures consistently showed scattered cells with nuclear DUX4 protein (Fig. 4E), similar to what was seen with the D4Z4 mouse (Krom et al., 2013). The protein could also be detected, and at greater frequency, in nuclei from differentiated myotubes, often in groups of adjacent nuclei. Quantification revealed that about 5% of myonuclei from differentiated cultures stained DUX4+, while the in myoblast cultures, DUX4 protein was present in about 1.5% of nuclei (Fig. 4F). This result is not necessarily suggestive of greater expression in differentiated cultures: myotubes are syncitia, therefore active DUX4 transcription in one nucleus can lead to uptake and positive staining of nuclear proteins in nearby myonuclei (Block et al., 2013; Tassin et al., 2013), so at the same frequency of expression, myotubes are predicted to have greater numbers of positive nuclei than myoblasts. Indeed, a careful comparison of two independent myoblast and myotube cultures revealed very similar levels of expression at the RNA level before vs. after differentiation (Fig. 4G), although it is important to point out that these results are non-quantitative. The lack of detectable protein in most nuclei by immunostaining, as well as the lack of detectable protein in FAPs, may indicate the sensitivity of the assay is limiting. Indeed, between laboratories, the immunohistochemical method has produced widely varying estimates of frequencies of nuclei positive for DUX4 in FSHD (and control) myoblasts (Jones et al., 2012; Snider et al., 2010).
Induced DUX4 expression is selectively deleterious to myogenic progenitors
The studies described above were undertaken in the absence of dox, therefore phenotypes are due to extremely low level, leaky expression. To evaluate the utility of dox-mediated DUX4 expression, we tested whether the inducible locus was functional by treating myoblast and FAP cultures with a high dose of dox (500 ng/mL) for 24 hours. This confirmed that the inducible system was indeed working as expected: DUX4 protein was expressed in response to dox (Fig. 5A). We then subjected ex vivo cultures of myoblasts and FAPs to various concentrations of dox. In both myoblasts and FAPs, expression of DUX4 was deleterious, however growth inhibition was more severe in myoblasts (Fig. 5B, C).
Figure 5. Effects of induced DUX4 expression.
(A) Western blot for DUX4 expression in proliferating myoblasts and FAPs exposed to a high dose (500 ng/mL) of dox. One representative blot of 3 independent experiments is shown.
(B) Dox dose-response growth curves for myoblasts (left) and FAPs (right) exposed to very low doses of dox to induce DUX4 expression.Myoblasts displayed a more severe growth inhibition and greater sensitivity to dox.
(C) Cellular morphology of proliferating myoblast (left) and FAP (right) cultures exposed to a high dose (500 ng/mL) of dox.
(D) Myoblasts and FAPs exposed to a low dose (50 ng/mL) of dox and cultured under myogenic or adipogenic differentiation conditions, respectively. Myotubes (left) were stained for myosin heavy chain; adipocytes (right) were stained with Oil Red O. Note that low dose induction of DUX4 inhibits myogenic but not adipogenic differentiation.
Low, non-toxic, levels of DUX4 expression were shown to interfere with the differentiation of C2C12 myoblasts (Bosnakovski et al., 2008b). To determine DUX4 effects on primary cells, we induced differentiation of myoblasts (into myotubes) and FAPs (into adipocytes) in the presence of low levels of dox (50 ng/mL). Myotube formation was clearly impaired by DUX4 expression, while adipocyte differentiation was unaffected (Fig. 5D).
DUX4 impairs myogenic regeneration in vivo
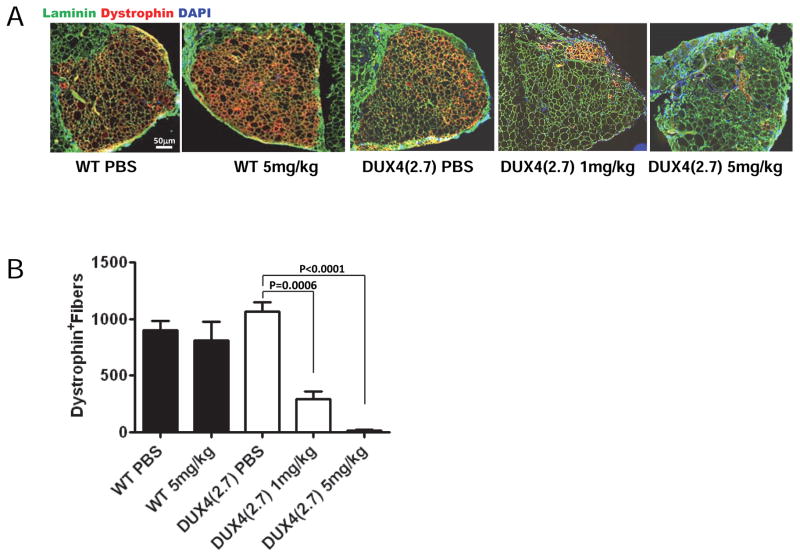
To test the effect of DUX4 expression during myogenic regeneration in vivo, we transplanted 1,800 FACS-isolated satellite cells from hind limb muscle of iDUX4(2.7) mice into pre-injured, irradiated tibialis anterior muscles of NSG-mdx4Cv mice (Arpke et al., 2013). The recipients lack dystrophin, thus the amount of donor muscle tissue produced by the transplanted cells can be quantified by immunostaining for dystrophin. We found that treatment of mice with 5 mg/kg doxycycline severely impaired the ability of donor satellite cells to produce new muscle (Fig. 6A, B). This indicates that DUX4 expression is deleterious to muscle regeneration in vivo.
Figure 6. DUX4 expression impairs myogenic regeneration in vivo.
(A) Representative examples of TA muscles one month after transplantation of 1,800 iDUX4(2.7) satellite cells in the presence or absence of daily doxycycline injection (either 1 mg/kg or 5 mg/kg) to induce DUX4 or control carrier (PBS) injection. The overall muscle architecture is indicated by laminin staining (green). Donor cell contribution to new fibers is evaluated by counting dystrophin+ (red) fibers. Doxycycline treatment impaired contribution to myofiber regeneration in a dose-dependent manner.
(B) Quantification of myofiber engraftment. For each group, N=6. Bars indicate SEM.
DISCUSSION
FSHD is one of the most enigmatic of the muscular dystrophies, and despite many efforts, there is no genetic model that displays any phenotype for this disease. In humans, reduction in repeat number (or second site mutation in rare non-contraction cases i.e. FSHD2, Lemmers et al., 2012), clearly leads to an epigenetic alteration at D4Z4 (van Overveld et al., 2003) that impairs repeat-induced silencing allowing D4Z4 transcription (Block et al., 2013; Jones et al., 2012; Snider et al., 2009). Although a recently described mouse bearing randomly integrated tandem D4Z4 repeats from an FSHD allele showed some expression of D4Z4 transcript (Krom et al., 2013), this animal did not present profound pathologies. With the recognition that DUX4 has potent myogenesis-relevant phenotypes even at very low levels of expression in C2C12 cells (Bosnakovski et al., 2008b), we sought to generate an animal model based on low level, regulated and systemic expression of DUX4. Systemic expression is relevant because D4Z4 misexpression in FSHD has not been shown to be restricted to myoblasts or muscle fibers. Indeed, the presence of non-muscle phenotypes such as retinal vascular pathology (Fitzsimons et al., 1987; Gieron et al., 1985; Small, 1968) and sensorineural hearing loss (Brouwer et al., 1991; Lutz et al., 2013)in FSHD patients, the syndrome of phenotypes associated with very severe infantile FSHD cases with extremely short D4Z4 arrays (Chen et al., 2013), together with the demonstration of epigenetic changes also in the blood cells of FSHD patients (Hartweck et al., 2013; van Overveld et al., 2003), suggests that misexpression is not restricted to muscle and might be a global feature. Remarkably, in our novel allele, we found that low-level leaky and variable expression led to several severe phenotypes, making the iDUX4(2.7) mouse the first animal model in which FSHD allele-specific DNA has been integrated into the genome and caused a serious pathology.
This pathology is dominant, like FSHD, but dramatically more potent, resulting in male-specific lethality. In humans the D4Z4 repeats are embedded in subtelomeric heterochromatin and subject to repeat-induced silencing, whereas in this mouse model, the transgene has been inserted into euchromatin and is present in only a single copy. Thus, D4Z4 has lost both the opportunity for repeat-induced silencing as well as its normal heterochromatic genomic environment, both changes that would be predicted to render the transgene more potent than 4q35-linked FSHD alleles (Fig. 7). This may explain why the DUX4 transgene has a much stronger dominant-lethal phenotype in this mouse model than in human patients with FSHD. It may also explain the presence of phenotypes in unexpected tissues, for example skin, however vector-specific sequences may also contribute to tissue-specific aspects of the leakiness, for example the minimal promoter might be leaky in skin cells, whereas the endogenous human D4Z4 might not show skin expression. For obvious reasons, outside of muscle biopsies, a careful analysis of tissues in which DUX4 expression can be detected in humans has never been reported. With regard to pathologies in testes, this was somewhat surprising as DUX4 has been reported in human testes (Snider et al., 2010). The closest mouse homologue, Dux, was not reported expressed in testes (Clapp et al., 2007), however a different double homeobox protein, Duxbl, which lacks the c-terminal sequence conserved between DUX4 and mouse Dux (Leidenroth and Hewitt, 2010) important for toxicity (Bosnakovski et al., 2008a), is expressed in testes (Wu et al., 2010). The presence of testis pathology in iDUX4(2.7) mice suggests that if DUX4 has a specific function in testes, then this is probably a human-specific activity.
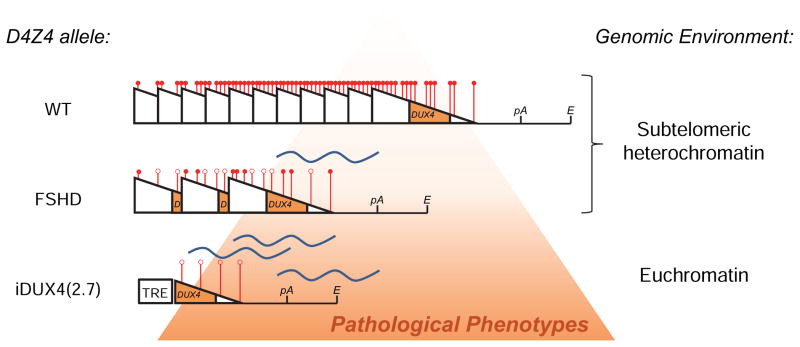
Figure 7. The iDUX4(2.7) allele vs. human D4Z4 alleles.
D4Z4 is indicated by a triangle. Methylation state is indicated at 6 representative sites for each array, with a red ball indicating methylation and an open circle indicating lack of methylation. In humans, when D4Z4 is present in a large tandem array within subtelomeric heterochromatin at 4q, a repeat-induced silencing mechanism leads to heterochromatinization of the array (indicated by compaction of the triangles and their respective methylation marks) and hypermethylation of DNA (indicated by red circles), which effectively silences the locus. When the array number is reduced in an FSHD allele, a loss of repeat-induced silencing leads to an opening up of chromatin (more space between the triangles), and a relative demethylation, leading to some transcription of DUX4. In the iDUX4(2.7) mouse, the single copy (which is not subject to repeat-induced silencing) and location within euchromatin, results in even greater opening, a complete absence of methylation, and greater transcription / transcription in additional tissues, leading to a more severe, lethal, phenotype.
It is notable that the retinae of the iDUX4(2.7) animals demonstrate pathological retinal vascular changes like those seen in FSHD patients. It is not yet understood how the D4Z4 contraction leads to these changes in humans, however it has been proposed that the muscle pathology and the retinal pathology might both be attributed to an endothelial defect (Osborne et al., 2007). The iDUX4(2.7) mice also resemble FSHD patients in terms of DUX4 expression at the RNA and protein level: a spliced mRNA encoding DUX4 can be detected at very low levels in many tissues, but the DUX4 protein itself cannot, or is expressed in so few cells as to be difficult to distinguish from background in immunostaining experiments. Within the short lifespan of the rare live-born affected males, a muscular dystrophy does not develop. Whether or not MD would develop if the animals were to survive beyond 4–6 weeks, the absence of dystrophy is consistent with the lack of muscle pathology in young FSHD patients. The literature is inconsistent on fiber type changes in FSHD (Lin and Nonaka, 1991; Olsen et al., 2005), but neither do the mice show any signs of fiber type switching. Unlike DMD, in which pathological cycles of damage and regeneration are present from birth, FSHD muscles appear histologically normal prior to onset of the disease. The female mice do survive, but neither did they show signs of muscular dystrophy. However in heterozygous carrier females, the DUX4 transgene appears to be predominantly on the inactive X chromosome, as it is heavily methylated. As X-inactivation is random, this is best explained by positive selection for cells that silence the DUX4-bearing X chromosome.
To evaluate more directly the effect of DUX4 on primary muscle progenitors, we studied myogenic and fibroadiopgenic progenitors from iDUX4(2.7) males. DUX4 mRNA was indeed expressed at low levels in both myoblasts and FAPs, and was associated with a reduction in proliferative potential, more significantly to myoblasts. Doxycycline induction at high doses was clearly inhibitory to both myoblasts and FAPs, while low levels inhibited differentiation of myoblasts into multinucleated myotubes but not of FAPs into adipocytes. The DUX4 effects on primary muscle progenitors raise the question of whether a defect in regeneration contributes to muscle pathology in FSHD. Accordingly, when iDUX4(2.7) satellite cells were transplanted and recipients treated with a relatively low dose of doxycycline, their ability to generate new muscle tissue was severely impaired. The data do not formally differentiate between effects on progenitors vs. newly-formed myofibers – certainly when very high levels of DUX4 were delivered directly to mouse muscles tissues by adeno-associated virus, there was extensive degeneration (Wallace et al., 2011). However the data are consistent with the notion that DUX4 could impair muscle regeneration in FSHD. Most importantly, they represent a quantifiable assay that could be used to study the activity of pharmacological inhibitors of DUX4 in vivo.
This is the first animal model in which FSHD allele-specific DNA has been integrated into the genome and has generated a pathological phenotype. It clearly demonstrates that even low level expression from the terminal D4Z4 repeat is highly deleterious, is consistent with a novel model for FSHD in which muscle wasting is a consequence, at least in part, of impaired regeneration, and presents opportunities for testing activity of anti-DUX4 therapeutics in vivo.
EXPERIMENTAL PROCEDURES
Cloning of targeting construct and generation of iDUX4(2.7) mice
The genomic DNA encoding DUX4 from the terminal repeat together with downstream sequence (2.7 kb total) was obtained from pClneo-DUX4 (Gabriels et al., 1999) and subcloned into XhoI/NotI cloning sites of p2Lox, generating p2lox-DUX4(2.7). iDUX4(2.7) inducible mESC lines were generated by inducible cassette exchange recombination using an improved version of the A2Lox.cre mESC lines (ZX1 cells, which contain the second generation tetracycline-response element) as previously described (Iacovino et al., 2011). iDUX4(2.7) mice were derived at the University of Texas UT Southwestern Transgenic Core Facility by blastocyst injection of iDUX4(2.7) ES cells. Mice were maintained under the guidance of the University of Texas UT Southwestern and University of Minnesota Institutional Animal Care and Use Committees.
RNA isolation and RT-PCR for DUX4
RNA was isolated from fresh tissue samples dissected from iDUX4(2.7) and control male mice. Tissues were homogenized in Trizol (Invitrogen) and RNA was precipitated according to the manufacturer’s directions. The high GC content of DUX4 required the use of a high temperature reverse transcriptase. 1.5 ug RNA was treated for 30 min with DNase (Promega) and the cDNA was produced using Thermoscript polymerase and oligo dT primer according to the manufacturer’s directions. DUX4 transcript was analyzed by semiquantitative PCR using Takara LA Taq with GC rich buffer II and the following conditions: primers PLH394F: GCTGGAAGCACCCCTCAGCGAGGAA and PLH395R: TCCAGGTTTGCCTAGACAGCGTC; denaturation at 94 °C for 15 s, annealing at 57 °C for 15 s and elongation at 72 °C for 30 s. RNA experiments were performed with at least 3 biological replicates and representative results are shown.
Statistical analyses
GraphPad Prism was used with all samples to compute means, standard deviations and p-values by T test.
Histological analysis
Skinsamples were harvested from iDUX4(2.7) and control litermates (3 of each), fixed in 10% neutral buffered formalin containing 0.1 M CaCl2 for 2 days and embedded in paraffin. 4 μm sections were evaluated by hematoxylin and eosin (H&E) staining. Testes (from 6 iDUX4(2.7) and 3 littermate control males) were prepared in Bouin’s fixative and processed for sectioning and H&E staining as described (Hao et al., 2008).
Skeletal muscle contractile and histological analyses
EDL and soleus muscles were carefully dissected and maximal isometric force generating capacities measured. For histology, muscles were embedded in OCT. 10 μm sections were evaluated by hematoxylin and eosin (H&E) staining, NADH reactivity, and myosin heavy chain expression as described (Greising et al., 2012).
Retinal vascular imaging
Retinae were isolated immediately following euthanasia, fixed in Zamboni’s fixative for 24 h, washed and stored in 20% Sucrose + 0.5% Sodium Azide at 4 °C. Whole retinae were incubated with 5% donkey serum overnight and immunostained with rat anti-CD31/PECAM-1 (Santa Cruz Biotechnology) at 1:200 dilution for 12 h at room temp, followed by 6 h of washing and incubation with Cy2 conjugated secondary antibody (Jackson Immunoresearch) at 1:500 dilution for 12 h at room temp. Retina flat mounts were imaged using 2 micron thick z-stacks on a Fluoview FV1000 BX2 Upright Laser scanning confocal microscope (Olympus Corporation) with a 20X or 40X oil objective lens. Images were analyzed using Adobe Photoshop and Reindeer Games software to quantify different measures of retinal vasculature, as described previously (Gupta et al., 2002).
Isolation and differentiation of myoblasts and FAPs from muscle
Preparation of muscle samples was performed as described (Arpke et al., 2013). 106 total mononuclear cells were then plated in a T25 flask in F-10/Ham’s (Hyclone) medium containing 20% FBS (HyClone), 50 ng/μL human basic fibroblast growth factor (Peprotech), 1% penicillin/streptomycin (Gibco), and 1% Glutamax (Gibco) and cultured at 37°C under reduced oxygen conditions (5% O2, 5% CO2, 90% N2). After 4 days, primary cells were disassociated with 0.05% trypsin (Invitrogen) and resuspended in FACS staining medium. To isolate myoblast and FAP fractions, cells were stained with PDGFRα conjugated with PE (e-Biosciences, clone: APA5) and α7 integrin APC. Cells were sorted into a PDGFRα single-postive (FAP fraction) and an α7 integrin single-positive (myoblast) fraction and recultured in the same medium. Myoblasts were differentiated in 20% knockout serum replacer (Invitrogen,) as described in (Block et al., 2013). FAPs were differentiated in dexamethasone (Stem Cell Technologies) as described in (Lemos et al., 2012). To detect DUX4 by western blot or by immunostaining in primary myoblasts and myotubes, E5-5 DUX4 antibody (Abcam) was used as described (Geng et al., 2011). Myosin heavy chain staining used the MF20 antibody (Developmental Studies Hybridoma Bank).
Methylation analysis
Bisulfite sequencing was generally performed as described in (Hartweck et al., 2013). Tissues from 6 male mice at 6 weeks of age, and three females aged 6, 9 and 36 weeks were analyzed. DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen). 500 ng DNA was converted with the EZ DNA Methylation direct kit (Zymo research) and eluted in 20ul. Converted DNA (2 ul) was amplified using primer sets: DR3F: GTAGAGGGGATTTTTTAATTTGTTT; and DR3R: CAAACACCCCTTAACCCTAC using Takara ExTaq (Qiagen, Valencia, CA) according to manufacturers’ instructions with cycling parameters: Denature: 94°C 60 sec, Cycle: 94°C 10 sec, 60°C 15 sec, 72°C 20 sec, for 30 cycles; Final extension at 72°C 5 minutes. PCR products were purified from 2% agarose gels with Wizard SVgel and PCR Purification system (Promega) cloned with the TOPO T/A cloning kit (Invitrogen), and clones with 223 bp inserts, sequenced. To determine if frequency of methylated X chromosomes with the iDux4(2.7) transgene was greater than the expected frequency (50%), we pooled the female data and used the exact binomial test. This specific PCR represents methylated and unmethylated chromosomes equally (Hartweck et al., 2013).
Transplantation of satellite cells and fiber engraftment
Pax7-ZsGreen male mice, in which satellite cells are marked with green fluorescence (Bosnakovski et al., 2008c), were crossed to iDUX4(2.7) females to generate iDUX4(2.7) ; Pax7-ZsGreen males from which satellite cells were isolated by flow cytometry as described in (Arpke et al., 2013). In an independent replicate that gave similar results, satellite cells were also isolated based on surface markers as described previously (Chan et al. 2013). First dead cells were gated out with propidium iodide and then blood and endothelial lineages were gated out using anti-CD31 (eBioscience, clone 390) and rat anti-CD45 (eBioscience, Clone RA3-6B2) antibodies conjugated to phycoerythrin-Cy7. Finally satellite cells were sorted by being double positive for CD106 (eBioscience, biotinylated primary VCAM-1 antibody, Clone 429; followed by streptavidin secondary antibody conjugated with phycoerythrin) and α-7 integrin conjugated with allophycocyanin (AbLab, Clone R2F2).
Transplantation was according to (Arpke et al., 2013). Briefly, two days prior transplantation of cells, 4-month-old NSG-mdx4Cv mice were anesthetized with ketamine and xylazine and both hind limbs were subjected to a 1200 cGy dose of irradiation using an X-RAD 320 Biological Irradiator (Precision X-Ray, Inc., Branford, CT). On the day of transplantation, 1,800 sorted satellite cells were resuspended in 1.2% BaCl2 (RICCA Chemical Company) and injected into TA muscles of NSG-mdx4Cv mice. Every day post-transplantation we injected mice with PBS or doxycycline at 1 mg/kg or 5 mg/kg. Four weeks after transplantation, six TAs of each group were harvested, sectioned, and immunostained with a mouse monoclonal antibody to laminin (Sigma-Aldrich) and rabbit polyclonal antibody to dystrophin (Abcam) antibodies followed by goat anti-mouse IgG conjugated with Alexa Fluor 488 and goat anti-rabbit IgG conjugated with Alexa Fluor 555 (Life Science Technologies). Images were acquired on a Zeiss Axio Imager M1 Upright microscope with AxioCam HRc camera and the number of donor-derived fibers (dystrophin+) was determined.
Supplementary Material
Acknowledgments
This project was primarily supported by grants from the NIH (R01 AR055685), the Dr. Bob and Jean Smith Foundation, and the Friends of FSH Research to MK, and the Muscular Dystrophy Centre Core Laboratory P30 AR0507220. DB was supported by a Muscular Dystrophy Association Development Grant (MDA 4361) and a Marjorie Bronfman Research Fellowship from the FSH Society (FSHS-MGBF-016). RCRP was supported by NIH grants R01 AR055299 and U01 HL100407. KB was supported by NIH grant T32-AR07612 and DL by K02-AG036827. KG and DK were supported by NIH grants R01 HL68802, R01 HL103773. FKH was supported by NIH grant R01 HD053889.
Footnotes
AUTHOR CONTRIBUTIONS
DB derived the iDUX4(2.7) mouse and performed initial characterization and assays for DUX4 expression with assistance from JB. Studies of DUX4 expression and effects on differentiation of myoblasts and FAPs were performed by AD. LAH performed methylation studies. Transplantation assays were performed by AD and RW with assistance from RD. KAB and DAW performed muscle force and fiber assays. DV and KG performed morphometric analyses of retina. FKH performed analysis of testes. MK, AD, DB, LAH, KG, FKH, DAW, and RCRP wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agha-Mohammadi S, O’Malley M, Etemad A, Wang Z, Xiao X, Lotze MT. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med. 2004;6:817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- Arpke RW, Darabi R, Mader TL, Zhang Y, Toyama A, Lonetree CL, Nash N, Lowe DA, Perlingeiro RC, Kyba M. A New Immuno- Dystrophin-Deficient Model, the NSG-Mdx Mouse, Provides Evidence for Functional Improvement Following Allogeneic Satellite Cell Transplantation. Stem Cells. 2013 doi: 10.1002/stem.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, Rasmussen K, Frants RR. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve. 1995;2:S39–44. [PubMed] [Google Scholar]
- Block GJ, Narayanan D, Amell AM, Petek LM, Davidson KC, Bird TD, Tawil R, Moon RT, Miller DG. Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum Mol Genet. 2013;22:4661–4672. doi: 10.1093/hmg/ddt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Lamb S, Simsek T, Xu Z, Belayew A, Perlingeiro R, Kyba M. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp Neurol. 2008a doi: 10.1016/j.expneurol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppee F, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. The EMBO journal. 2008b;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RC, Kyba M. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008c;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer OF, Padberg GW, Ruys CJ, Brand R, de Laat JA, Grote JJ. Hearing loss in facioscapulohumeral muscular dystrophy. Neurology. 1991;41:1878–1881. doi: 10.1212/wnl.41.12.1878. [DOI] [PubMed] [Google Scholar]
- Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, Cagnin S, Vigano A, Colantoni L, Begum S, Ricci E, et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- Chen TH, Lai YH, Lee PL, Hsu JH, Goto K, Hayashi YK, Nishino I, Lin CW, Shih HH, Huang CC, et al. Infantile facioscapulohumeral muscular dystrophy revisited: Expansion of clinical phenotypes in patients with a very short EcoRI fragment. Neuromuscul Disord. 2013;23:298–305. doi: 10.1016/j.nmd.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic B, Yang B, Williamson RA, Sigmund CD. Appropriate tissue- and cell-specific expression of a single copy human angiotensinogen transgene specifically targeted upstream of the HPRT locus by homologous recombination. J Biol Chem. 2000;275:1073–1078. doi: 10.1074/jbc.275.2.1073. [DOI] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- Deidda G, Cacurri S, Piazzo N, Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD) J Med Genet. 1996;33:361–365. doi: 10.1136/jmg.33.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons RB, Gurwin EB, Bird AC. Retinal vascular abnormalities in facioscapulohumeral muscular dystrophy. A general association with genetic and therapeutic implications. Brain : a journal of neurology. 1987;110(Pt 3):631–648. doi: 10.1093/brain/110.3.631. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- Geng LN, Tyler AE, Tapscott SJ. Immunodetection of human double homeobox 4. Hybridoma (Larchmt) 2011;30:125–130. doi: 10.1089/hyb.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieron MA, Korthals JK, Kousseff BG. Facioscapulohumeral dystrophy with cochlear hearing loss and tortuosity of retinal vessels. American journal of medical genetics. 1985;22:143–147. doi: 10.1002/ajmg.1320220116. [DOI] [PubMed] [Google Scholar]
- Greising SM, Call JA, Lund TC, Blazar BR, Tolar J, Lowe DA. Skeletal muscle contractile function and neuromuscular performance in Zmpste24 −/− mice, a murine model of human progeria. Age (Dordr) 2012;34:805–819. doi: 10.1007/s11357-011-9281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Kshirsagar S, Chang L, Schwartz R, Law PY, Yee D, Hebbel RP. Morphine stimulates angiogenesis by activating proangiogenic and survival-promoting signaling and promotes breast tumor growth. Cancer Res. 2002;62:4491–4498. [PubMed] [Google Scholar]
- Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, Zhao GQ, Hamra FK. Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells. 2008;26:1587–1597. doi: 10.1634/stemcells.2007-0502. [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Anderson LJ, Lemmers RJ, Dandapat A, Toso EA, Dalton JC, Tawil R, Day JW, van der Maarel SM, Kyba M. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology. 2013;80:392–399. doi: 10.1212/WNL.0b013e31827f075c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells. 2011;29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TI, Chen JC, Rahimov F, Homma S, Arashiro P, Beermann ML, King OD, Miller JB, Kunkel LM, Emerson CP, Jr, et al. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum Mol Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Krom YD, Thijssen PE, Young JM, den Hamer B, Balog J, Yao Z, Maves L, Snider L, Knopp P, Zammit PS, et al. Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD. PLoS genetics. 2013;9:e1003415. doi: 10.1371/journal.pgen.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidenroth A, Hewitt JE. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol Biol. 2010;10:364. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, Frants RR, Padberg GW, Morava E, van der Maarel SM. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CM, de Knijff P, Padberg GW, Frants RR, van der Maarel SM. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos DR, Paylor B, Chang C, Sampaio A, Underhill TM, Rossi FM. Functionally convergent white adipogenic progenitors of different lineages participate in a diffused system supporting tissue regeneration. Stem Cells. 2012;30:1152–1162. doi: 10.1002/stem.1082. [DOI] [PubMed] [Google Scholar]
- Lin MY, Nonaka I. Facioscapulohumeral muscular dystrophy: muscle fiber type analysis with particular reference to small angular fibers. Brain & development. 1991;13:331–338. doi: 10.1016/s0387-7604(12)80128-8. [DOI] [PubMed] [Google Scholar]
- Lutz KL, Holte L, Kliethermes SA, Stephan C, Mathews KD. Clinical and genetic features of hearing loss in facioscapulohumeral muscular dystrophy. Neurology. 2013;81:1374–1377. doi: 10.1212/WNL.0b013e3182a84140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DB, Orngreen MC, Vissing J. Aerobic training improves exercise performance in facioscapulohumeral muscular dystrophy. Neurology. 2005;64:1064–1066. doi: 10.1212/01.WNL.0000150584.45055.27. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- Padberg GW, Brouwer OF, de Keizer RJ, Dijkman G, Wijmenga C, Grote JJ, Frants RR. On the significance of retinal vascular disease and hearing loss in facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S73–80. [PubMed] [Google Scholar]
- Portales-Casamar E, Swanson DJ, Liu L, de Leeuw CN, Banks KG, Ho Sui SJ, Fulton DL, Ali J, Amirabbasi M, Arenillas DJ, et al. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci U S A. 2010;107:16589–16594. doi: 10.1073/pnas.1009158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small RG. Coats’ disease and muscular dystrophy. Transactions - American Academy of Ophthalmology and Otolaryngology American Academy of Ophthalmology and Otolaryngology. 1968;72:225–231. [PubMed] [Google Scholar]
- Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS genetics. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A, Laoudj-Chenivesse D, Vanderplanck C, Barro M, Charron S, Ansseau E, Chen YW, Mercier J, Coppee F, Belayew A. DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? Journal of cellular and molecular medicine. 2013;17:76–89. doi: 10.1111/j.1582-4934.2012.01647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touw K, Hoggatt AM, Simon G, Herring BP. Hprt-targeted transgenes provide new insights into smooth muscle-restricted promoter activity. Am J Physiol Cell Physiol. 2007;292:C1024–1032. doi: 10.1152/ajpcell.00445.2006. [DOI] [PubMed] [Google Scholar]
- Turki A, Hayot M, Carnac G, Pillard F, Passerieux E, Bommart S, Raynaud de Mauverger E, Hugon G, Pincemail J, Pietri S, et al. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free radical biology & medicine. 2012;53:1068–1079. doi: 10.1016/j.freeradbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, Harper SQ. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung SA, Masny PS, Figlewicz DA. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord. 2003a;13:322–333. doi: 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003b;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- Wu SL, Tsai MS, Wong SH, Hsieh-Li HM, Tsai TS, Chang WT, Huang SL, Chiu CC, Wang SH. Characterization of genomic structures and expression profiles of three tandem repeats of a mouse double homeobox gene: Duxbl. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:927–940. doi: 10.1002/dvdy.22210. [DOI] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS genetics. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.